Abstract
Enterovirus 118 (EV-118) within species HEV-C was detected in two 5-month-old boys with pneumonia in China. The EV-118 from both cases was genetically closer to ISR10 strain from Israel than to PER161 strain from Peru based on VP1 gene sequences. The complete genome of the detected EV-118 consists of 7,360 nucleotides, excluding the poly (A) tail. The 5′UTR contains 669 nucleotides, and 3′UTR consists of 73 nucleotides. A single open reading frame from base 670 to 7,287 that encodes a 2,206-amino-acid polyprotein was featured. The base composition of the full genome is 27.9 % A, 24.2 % C, 24.4 % G, and 23.6 % U. Phylogenetic analysis of the full genome sequences illustrated EV-118 was genetically closer to EV-109 and EV-105, and the Chinese strain differed from Peru strain. In summary, the presence of EV-118 was confirmed in pediatric pneumonia cases and complete genome sequences were identified for the first time in China.
Electronic supplementary material
The online version of this article (doi:10.1007/s11262-014-1044-1) contains supplementary material, which is available to authorized users.
Keywords: Human enterovirus, Pneumonia, Pediatric, China
Introduction
Enteroviruses cause a variety of diseases such as herpangina, hand-foot-and-mouth disease, diarrhea, neurologic diseases, and conjunctivitis as well as respiratory illnesses in infants or adults, resulting in bronchitis, bronchiolitis, and pneumonia. Over the past 3 years, human enterovirus 68 (EV-68) has been highlighted as an increasingly recognized cause of respiratory illnesses in outbreak events and case clusters, with the clinical presentation ranging from mild illness to complications requiring hospitalization and/or death [1, 2]. EV-104, EV-109, and EV-117 were also detected from the patients with acute respiratory tract illnesses [3–5]. EV-105, EV-116, and EV-118 were detected in the symptomatic children with fatal acute flaccid paralysis, gastroenteritis, and respiratory tract disease [6, 7]. Here we reported the detection and genomic characteristic of EV-118 in two pediatric cases with pneumonia in China.
Results and discussion
A 5-month-old boy (CQ5112) from Sichuan Province was admitted to the Children’s Hospital, Chongqing Medical University in May 2011, with diagnosis of interstitial pneumonia. He had a 1-week history of cough and wheezing, with clinical manifestation of fever (highest 38 °C), expectoration, diarrhea, rales, and coarse rhonchi in the bilateral pulmonary. Laboratory studies performed at admission to the hospital revealed a normal white blood cell (WBC) count (7.63 × 109/L) and C-reactive protein level (<8 mg/L), with a mild elevated platelet count (489 × 109/L). Sputum culture showed growth of Staphylococcus aureus and Haemophilus influenza.
Another nearly 5-month-old boy (CQ5185) was admitted to the hospital in June 2011, also from Sichuan Province. He was diagnosed with severe bronchiolitis with a 2-week cough and 1-week wheezing. He also had fever (highest temperature of 38.6 °C), expectoration, diarrhea, rales, and coarse rhonchi in the bilateral pulmonary as well as dyspnea. Laboratory studies performed on admission revealed an elevated WBC (13.03 × 109/L) and platelet count (490 × 109/L). All the other laboratory tests were normal, and the culture of the sputum was negative. Both patients were discharged from the hospital and remained healthy without respiratory symptoms after 2 months of regular follow-up.
After parents or guardians gave written informed consent for their children to participate in the study, the nasopharyngeal aspirates were collected, from which RNA/DNA was extracted. The study protocol was reviewed and approved by the Ethics Review Committee of Chongqing Medical University.
The detection of enterovirus was performed by real-time PCR to amplify the partial 5′UTR of enterovirus as previously described [8, 9], and both patients yielded positive results. The 330-bp amplicons were sequenced and BLAST analysis (www.ncbi.nlm.nih.gov) revealed their high similarities to EV-109 and EV-104.
The VP1 genes of CQ5112 and CQ5185 samples were further amplified and sequenced (GenBank accession numbers JX678287 and JX678288). BLAST analysis demonstrated their high similarities to EV-118 (the prototype strain ISR10, GenBank accession number: JQ768163). The VP1 nucleotide sequences of CQ5112 and CQ5185 demonstrated similarities of 97.6 and 97.3 % to ISR10 strain from Israel, and 91.1 and 91.5 % to PER161 strain from Peru (GenBank accession number: JX393301), respectively (Table 1). Phylogenetic tree was constructed based on the VP1 gene nucleotide sequences using the maximum-likelihood method, with all members of the HEV-C species available included for analysis. All EV-118, EV-109, EV-105, EV-117, and EV-104 strains formed a separate cluster (Cluster 2.1, Fig. 1). The two strains (CQ5112 and CQ5185) were genetically closer to ISR10 than to PER161. Based on the VP1 amino acid sequence, higher identities were presented between the two strains and ISR10 (98.9 and 99.6 %) than between the two strains and PER161 (95.9 and 97.2 %). Three amino acid differences occurred in the BC loop, at position 90, 92, and 94, while no differences found in the DE loop (Supplemental Fig. 2). BC loop in VP1 region has been shown to be important for the reactivity of type-specific antibodies, and these variations may affect the different antigenic properties in different areas.
Table 1.
Identities of nucleotide and amino acid sequences of CQ5185, PER161, and ISR10
| Gene | Strain | CQ5185 | PER161 | Positiona |
|---|---|---|---|---|
| 5′UTR | PER161 | 92.8 | – | 45–669 |
| VP4 | PER161 | 90.8 (100) | – | 670–876 |
| ISR10 | 95.6 (100) | 92.2 (100) | ||
| VP2 | PER161 | 91.6 (98.8) | – | 877–1,689 |
| ISR10 | 98.0 (99.2) | 92.2 (99.6) | ||
| VP3 | PER161 | 90.5 (99.1) | – | 1,690–2,412 |
| ISR10 | 96.8 (100) | 90.3 (99.1) | ||
| VP1 | PER161 | 91.8 (97.2) | – | 2,413–3,300 |
| ISR10 | 97.2 (99.6) | 91.6 (96.9) | ||
| 2A | PER161 | 91.7 (100) | – | 3,301–3,747 |
| 2B | PER161 | 89.6 (96.9) | – | 3,748–4,038 |
| 2C | PER161 | 91.2 (97.5) | – | 4,039–5,025 |
| 3A | PER161 | 92.0 (97.7) | – | 5,026–5,289 |
| 3B | PER161 | 89.3 (100) | – | 5,290–5,355 |
| 3C | PER161 | 90.4 (93.4) | – | 5,356–5,904 |
| 3D | PER161 | 90.4 (96.7) | – | 5,905–7,287 |
| 3′UTR | PER161 | 96.6 | – | 7,288–7,346 |
There were no sequences of 5′UTR, 2A–2D, 3A–3D, and 3′UTR for ISR10 strain
aThe position was according the CQ5185 strain (JX678288)
Fig. 1.
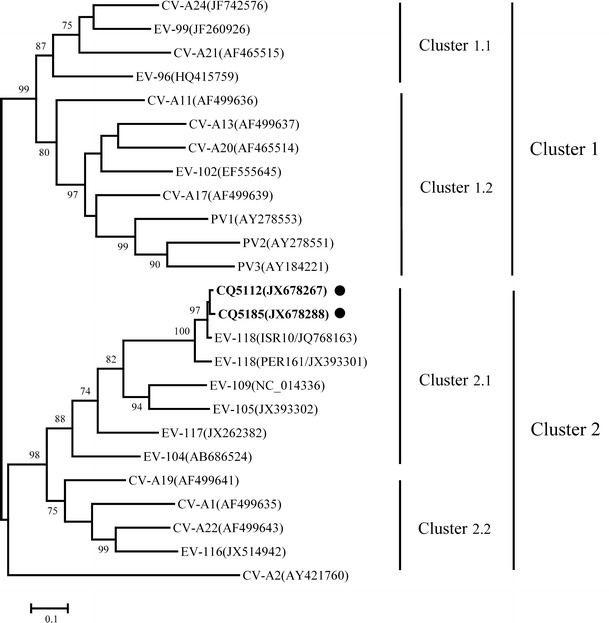
Phylogenetic trees were constructed based on the VP1 gene nucleotide sequences using the maximum-likelihood method with 1,000 bootstrap replicates by MEGA 5. The strains (CQ5112 and CQ5185) labeled by black solid circles were isolated from our study. CV-A2 was used as the outgroup strain. CV-A coxsackievirus A; PV poliovirus; EV enterovirus
The complete genome sequence of EV-118 was determined from CQ5185 as follows: cDNA was synthesized by using SuperScript® III First-Strand Synthesis System for Reverse Transcription Polymerase Chain Reaction (RT-PCR) (Invitrogen, America). End-specific nucleotide sequences were determined by using the 5′RACE (Rapid Amplification of cDNA Ends, Invitrogen) and 3′RACE (Core Set Ver 2.0, TaKaRa). The primers used were provided in the Supplemental Table 1. The complete genome of EV-118 consists of 7,360 nucleotides, excluding the poly (A) tail. The 5′UTR contains 669 nucleotides, and 3′UTR consists of 73 nucleotides. EV-118 features a single open reading frame from base 670 to 7,287 that encodes the 2,206-amino-acid polyprotein. The base composition of the full genome of EV-118 is 27.9 % A, 24.2 % C, 24.4 % G, and 23.6 % U. Based on the comparison of the full genome, EV-118 shares 64–79 % nucleotide sequence identities with other HEV-C coding regions, including EV-109 (79 %), EV-104 (73 %), coxsackievirus A19 (CV-A19) (69 %), CV-A22 (69 %), and CV-A1 (69 %). A phylogenetic tree was constructed based on the complete nucleotide sequences with maximum-likelihood method, which illustrated that CQ5185 and PER161 clustered with EV-109, EV-105, EV-104, and EV-117, and were genetically closer to EV-109 and EV-105 (Supplemental Fig. 2). The CQ5185 strain differed from the PER161 strain, being supported by a 100 bootstrap value.
The sequence identities of all genes between CQ5185 and PER161 were around 90 %, lower than those between CQ5185 and ISR10, in which comparison was made only based on the VP1-VP4 gene sequences (Table 1). There were 53 different amino acid sites (97.5 % identity) between CQ5185 and PER161, mainly focusing on 3D gene, VP1 gene, 2C gene, and 3C gene, which may be explained by the regional disparity.
The presence of other respiratory pathogens, including respiratory syncytial virus, adenovirus, influenza virus, human bocavirus, metapneumonia, coronavirus, parainfluenza virus (PIV), and rhinovirus were screened as previously described [8, 9]. Patient CQ5185 was positive for PIV-3, while Patient CQ5112 was detected as negative. The two cases had similar clinical syndrome. In addition to presenting with respiratory illnesses (fever, cough, expectoration, or dyspnea), gastrointestinal symptoms were also featured. Pneumonia and severe capillary bronchitis were displayed in both patients, however, due to the co-infection with other respiratory viruses and bacteria; the causal inference between EV-118 and respiratory disease could not be established at this moment.
In summary, we report the detection of EV-118 from the pediatric patients with pneumonia in China, in which finding highlights the necessity of surveillance of other enteroviruses, including EV-118, in causing respiratory illnesses in children. The EV-118 from both cases was genetically closer to the ISR10 strain from Israel than to the PER161 strain from Peru based on VP1 gene sequences. Based on the complete genome sequencing, EV-118 is genetically most close to EV-109 and EV-105, and the Chinese EV-118 strain differs from PER161 strain. The clinical characteristics of single EV-118 infection and the pathogenic role of this virus in respiratory infection need to be further investigated in the future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Alignment of the amino acid of VP1 gene at position 80–161 from CQ5185, PER161 and ISR10. The amino acid in the BC and DE loops are indicated in box. (TIFF 2671 kb)
Phylogenetic trees were constructed based on the partial complete gene nucleotide sequences (at position 45–7347, according the CQ5815 strain) using the maximum-likelihood method with 1,000 bootstrap replicates by MEGA 5. The strain CQ5185 were isolated from our study. CV-A2 was used as the outgroup strain. CV-A, coxsackievirus A; PV, poliovirus; EV, enterovirus. (TIFF 635 kb)
Acknowledgments
This study is supported by the China Mega-Project on Infectious Disease Prevention (No. 2013ZX10004202-002) and National Natural Science Fund for Young Scholars (81222037).
Conflict of interest
The authors do not have any commercial or other association that might pose a conflict of interest. All authors read and approved the final manuscript.
Footnotes
Qing-Bin Lu and Ying Wo have contributed equally to this work.
Contributor Information
Wei Liu, Phone: +86-10-63896082, FAX: +86-10-63896082, Email: lwbime@163.com.
Wu-Chun Cao, Phone: +86-10-63896082, FAX: +86-10-63896082, Email: caowc@bmi.ac.cn.
References
- 1.Imamura T, Fuji N, Suzuki A, Tamaki R, Saito M, Aniceto R, Galang H, Sombrero L, Lupisan S, Oshitani H. Emerg. Infect. Dis. 2011;17:1430–1435. doi: 10.3201/eid1708.101328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tokarz R, Firth C, Madhi SA, Howie SRC, Wu W, Sall AA, Haq S, Briese T, Lipkin WI. J. Gen. Virol. 2012;93:1952–1958. doi: 10.1099/vir.0.043935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tapparel C, Junier T, Gerlach D, Belle SV, Turin L, Cordey S, Mühlemann K, Regamey N, Aubert J-D, Soccal PM, Eigenmann P, Zdobnov E, Kaiser L. Emerg. Infect. Dis. 2009;15:719–726. doi: 10.3201/eid1505.081286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yozwiak NL, Skewes-Cox P, Gordon A, Saborio S, Kuan G, Balmaseda A, Ganem D, Harris E, Derisi JL. J. Virol. 2010;84:9047–9058. doi: 10.1128/JVI.00698-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daleno C, Piralla A, Scala A, Baldanti F, Usonis V, Principi N, Esposito S. J. Virol. 2012;86:10888–10889. doi: 10.1128/JVI.01721-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukashev AN, Drexler JF, Kotova VO, Amjaga EN, Reznik VI, Gmyl AP, Grard G, Taty Taty R, Trotsenko OE, Leroy EM, Drosten C. J. Gen. Virol. 2012;93:2357–2362. doi: 10.1099/vir.0.043216-0. [DOI] [PubMed] [Google Scholar]
- 7.Tokarz R, Hirschberg DL, Sameroff S, Haq S, Luna G, Bennett AJ, Silva M, Leguia M, Kasper M, Bausch DG, Lipkin WI. J. Gen. Virol. 2012;86:13756. doi: 10.1128/JVI.02341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunson RN, Collins TC, Carman WF. J. Clin. Virol. 2005;33:341–344. doi: 10.1016/j.jcv.2004.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiveljung-Lindell A, Rotzén-Östlund M, Gupta S, Ullstrand R, Grillner L, Zweygberg-Wirgart B, Allander T. J. Med. Virol. 2009;81:167–175. doi: 10.1002/jmv.21368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the amino acid of VP1 gene at position 80–161 from CQ5185, PER161 and ISR10. The amino acid in the BC and DE loops are indicated in box. (TIFF 2671 kb)
Phylogenetic trees were constructed based on the partial complete gene nucleotide sequences (at position 45–7347, according the CQ5815 strain) using the maximum-likelihood method with 1,000 bootstrap replicates by MEGA 5. The strain CQ5185 were isolated from our study. CV-A2 was used as the outgroup strain. CV-A, coxsackievirus A; PV, poliovirus; EV, enterovirus. (TIFF 635 kb)


