Abstract
In this study, a novel duplex nanoparticle-assisted polymerase chain reaction (nanoPCR) assay was developed to detect porcine epidemic diarrhea virus (PEDV) and porcine transmissible gastroenteritis virus (TGEV). Two pairs of primers were designed based on the conserved region within the N gene of PEDV and TGEV. In a screening of 114 clinical samples from four provinces in China for PEDV and TGEV, 48.2 and 3.5 % of the samples, respectively, tested positive. Under optimized conditions, the duplex nanoPCR assay had a detection limit of 7.6 × 101 and 8.5 × 101 copies μL−1 for PEDV and TGEV, respectively. The sensitivity of the duplex nanoPCR assay was ten times higher than that of a conventional PCR assay. Moreover, no fragments were amplified when the duplex nanoPCR assay was used to test samples containing other porcine viruses. Our results indicate that the duplex nanoPCR assay described here is useful for the rapid detection of PEDV and TGEV and can be applied in clinical diagnosis.
Keywords: Duplex nanoPCR, N gene, PEDV, TGEV, Clinical diagnosis
Introduction
Porcine epidemic diarrhea virus (PEDV) and porcine transmissible gastroenteritis virus (TGEV) are the etiological agents of porcine epidemic diarrhea and transmissible gastroenteritis, respectively. The clinical signs and pathological lesions of PEDV infection are similar to those of TGEV; the diseases are characterized by acute watery diarrhea, vomiting, dehydration, and weight loss [1]. PEDV and TGEV can cause viral enteritis and severe diarrhea with high morbidity and mortality in suckling piglets [2, 3]. PEDV and TGEV have been associated with significant economic losses in farms [4, 5]. PEDV was first observed among pigs in England in 1971 [6], whereas TGEV was isolated for the first time in 1946 [7]. Currently, PEDV and TGEV occur in parts of Europe, North America, and Asia [8–10].
PEDV and TGEV are enveloped viruses belonging to the Nidovirales order, Coronaviridae family, Coronavirinae subfamily, and Alphacoronavirus genus [11]. PEDV and TGEV are transmitted primarily by the fecal–oral route [12], although airborne transmission of PEDV was recently confirmed experimentally [13]. Since PEDV and TGEV belong to the same genus, they have similar genomic structure; both viruses are large, single-stranded, positive-sense RNA viruses that express structural proteins, including membrane (M) and nucleocapsid (N) proteins.
The similar epidemiological and clinical features of PEDV and TGEV complicate diagnosis, which requires differential laboratory tests. Differential diagnosis of PEDV and TGEV relies mainly on conventional reverse transcription (RT)-PCR and serological assays [14, 15]. Nanoparticle-assisted polymerase chain reaction (nanoPCR) is a type of sensitive PCR assay in which nanofluids are formed after solid nanoparticles (1–100 nm diameter) are suspended in the PCR system. The thermal conductivity of nanofluids is greater than that of ordinary fluids, allowing extremely rapid changes in temperature so that target temperatures are reached more quickly [16].
Here, we describe the development of a duplex nanoPCR-based assay for rapid clinical detection of PEDV and TGEV. The nanoPCR assay was evaluated by differential laboratory testing of PEDV and TGEV, which demonstrated that it was more sensitive and specific than conventional PCR.
Materials and methods
Virus preparation
PEDV strain ZJ08, TGEV strain HB08, porcine circovirus type 2 (PCV-2), porcine reproductive and respiratory syndrome virus (PRRSV), classic swine fever virus (CSFV), porcine parvovirus (PPV), swine influenza virus (H1N1), and porcine pseudorabies virus (PRV) were obtained from the Animal Medical Center DBN Technology Group. Vero and ST cells were used to culture PEDV and TGEV, after which the virus titer of the supernatant was determined.
Clinical samples
Fecal and small intestine samples were collected from 114 piglets suspected of being infected with PEDV or TGEV. The clinical signs of the animals from which samples were collected included acute watery diarrhea and enteritis. All samples were collected in accordance with the International Guiding Principles for Biomedical Research Involving Animals. Milled clinical samples were repeatedly frozen and thawed, after which they were centrifuged at 8000×g at 4 °C for 5 min. The supernatant of each sample was frozen at −80 °C.
Preparation of DNA/cDNA template
RNA was extracted from the reference viruses with an RNAsimple Total RNA kit (Beijing Tiangen Biotech Company, Beijing, China) in accordance with the manufacturer’s instructions. DNA was extracted from PPV, PRV, and PCV-2 with a TIANamp Virus genomic DNA/RNA kit (Beijing Tiangen Biotech Company, Beijing, China). cDNA synthesis was performed using the TranScript Firststrand cDNA Synthesis SuperMix (Beijing TransGen Biotech Company, Beijing, China). DNA and cDNA were stored at −20 °C.
Primer design and recombinant plasmid generation
The published sequences of the conserved regions of the N genes of PEDV and TGEV were obtained from GenBank (accession numbers KT323979 and ABG89328.1, respectively). Primers were designed using Primer Premier 5.0 software to amplify the entire N genes of PEDV and TGEV. Based on the conserved region of the N genes of PEDV and TGEV, primers were designed to amplify a 277-bp amplicon from the PEDV N gene and a 181-bp amplicon from the TGEV-N gene. All primers were synthesized by BGI (Beijing, China). The primer sequences are listed in Table 1.
Table 1.
Primer sequences used in this study
| Virus | Primer | Primer sequence (5′–3′) | Gene | Length (bp) |
|---|---|---|---|---|
| PEDV | F | ATGGCTTCTGTCAGTTTTCAG | N | 1336 |
| R | AGTCAAACATTGTTTAATTTCCT | |||
| TGEV | F | GTATAACTAAACTTCTAAATGGCC | N | 1161 |
| R | ATCTCGTTTAGTTCGTTACCTC | |||
| PEDV | P1 | TTGTTGAACCTAACACACCTCC | N (401–677 bp) | 277 |
| P2 | ACAGCAGCCACCAGATCATC | |||
| TGEV | T1 | CAATAACAAGAAGGATGACAGTGT | N (546–727 bp) | 181 |
| T2 | ACCTGCAGTTCTCTTCCAGG |
The complete coding sequences of the N genes of PEDV and TGEV were inserted into the pMD18-T vector (TaKaRa Biotechnology Company, Dalian, China) as standards (Fig. 1). After identification and sequencing, recombinant plasmids pMD18-T-PEDV-N and pMD18-T-TGEV-N were amplified in Escherichia Coli DH5α and purified using the AxyPrep™ Plasmid Miniprep Kit (AXYGEN Biotechnology Company, Hangzhou, China). The plasmids were kept at −20 °C.
Fig. 1.
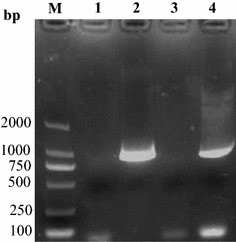
Positive recombinant plasmids pMD18-T-PEDV-N and pMD18-T-TGEV-N were identified by PCR. M DL 2000 marker, 1 PEDV negative control, 2 positive recombinant plasmid pMD18-T-PEDV-N, 3 TGEV negative control, 4 positive recombinant plasmid pMD18-T-TGEV-N
Optimization of the duplex nanoPCR assay
The duplex nanoPCR assay was performed to optimize the template concentration, amplification temperature, and primer volume. The 20-μL reaction volume included 1 μL of the plasmid template (pMD18-T-PEDV-N and pMD18-T-TGEV-N were mixed at a ratio of 1:1 by volume). Each pair of primers (P-N-1/P-N-2 and T-N-1/T-N-2 at 10 μM) was tested at volumes ranging from 0.2 to 1.4 μL in increments of 0.2 μL. The annealing temperature ranged from 50 to 59 °C. Amplified products were analyzed on 1.5 % agarose gels.
Sensitivity and specificity of the duplex nanoPCR assay
To evaluate the sensitivity of the duplex nanoPCR assay in comparison with that of conventional PCR, recombinant plasmids and total RNA extracted from infected cells were quantified and serially diluted by tenfold. Each sample dilution was tested as a template using the optimized duplex nanoPCR assay reaction parameters. The conventional PCR assay was performed using the same system and reaction parameters. Amplified products were analyzed on 1.5 % agarose gels.
Templates of the following viruses were used to evaluate the sensitivity of the duplex nanoPCR assay: PPV, PCV-2, PRRSV, PRV, CSFV, and H1N1. pMD18-T-PEDV-N plasmid DNA, pMD18-T-TGEV-N plasmid DNA, and infected cells served as positive controls. cDNA samples generated from RNA from Vero cells, ST cells, and samples from uninfected animals were used as negative controls.
Detection of PEDV and TGEV in clinical samples
A total of 114 samples were collected from piglets in Hebei, Zhejiang, Guangdong, and Beijing, China from October 2015 to March 2016. The presence of porcine viruses in the collected samples was determined using the duplex nanoPCR assay and conventional PCR.
Results
Optimization of duplex nanoPCR assay conditions
Optimized annealing temperatures and primer volumes for the PEDV and TGEV duplex nanoPCR assay were determined. Based on the results obtained with different annealing temperatures and primer volumes for the duplex nanoPCR assay, the optimal 20-μL reaction volume was established as containing 10 μL of 2× nanobuffer, 1.0 μL of primers P-N-1 and P-N-2 (10 μM), 0.8 μL of primers T-N-1 and T-N-2 (10 μM), 0.5 μL of Taq DNA polymerase (5 U μL−1), and 1 μL of template DNA and ddH2O. The reaction conditions were as follows: 5 min at 94 °C, followed by 30 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 15 s, with a final elongation step at 72 °C for 10 min. Amplified products were analyzed on 1.5 % agarose gels.
Sensitivity of the duplex nanoPCR assay
The sensitivity of the duplex nanoPCR assay was compared with that of conventional PCR using dilution series of recombinant plasmids and total RNA extracted from infected cells. In the duplex nanoPCR assay reactions, the plasmid detection limits for PEDV-N and TGEV-N were 7.6 × 101 and 8.5 × 101 copies/μL, respectively. The detection limits of the conventional PCR assay for PEDV-N and TGEV-N were 7.6 × 102 and 8.5 × 102 copies/μL, respectively. Therefore, the duplex nanoPCR assay was ten times more sensitive than the conventional PCR assay (Fig. 2). The virus RNA detection limits of the conventional PCR assay for PEDV and TGEV were 2.4 × 10−2 and 1.7 × 10−2 μg/mL, respectively, which were equivalent to virus titers of 101.75 and 101.25 TCID50/mL, respectively, of PEDV and TGEV, while the detection limits of the duplex nanoPCR assay for PEDV and TGEV were 100.5 and 100.5 TCID50/mL, respectively (Fig. 3). These results showed that the detection limits of the duplex nanoPCR assay for PEDV and TGEV were much higher than those of conventional PCR.
Fig. 2.
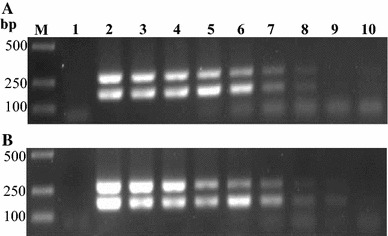
Comparison of the sensitivity of nanoPCR and conventional PCR assays. Serial tenfold dilutions of recombinant plasmids pMD18-T-PEDV-N and pMD18-T-TGEV-N were used as templates. The results of conventional PCR and nanoPCR assays are shown in (a) and (b), respectively. Lane M DL2000 marker, Lane 1 negative control, Lanes 2–10 pMD18-T-PEDV-N concentrations ranging from 7.6 × 108 to 7.6 × 100 copies/μL, pMD18-T-TGEV-N concentrations ranging from 8.5 × 108 to 8.5 × 100 copies/μL
Fig. 3.
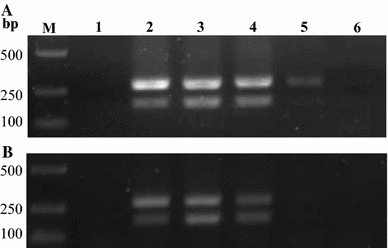
Comparison of the sensitivity of nanoPCR and conventional PCR assays. Tenfold serial dilutions of PEDV and TGEV RNA (24 and 17 μg/mL, respectively) were used as templates for nanoPCR and conventional PCR. The results of nanoPCR and conventional PCR assays are shown in (a) and (b), respectively. Lane M DL2000 marker, Lane 1 negative control, Lanes 2–6 serial dilutions of RNA templates
Specificity of the duplex nanoPCR assay
The specificity of the upstream and downstream primers was evaluated using other pig viruses, showing that there was no cross-reaction and only the expected bands were amplified (Fig. 4). These results demonstrate that the duplex nanoPCR assay is specific for PEDV and TGEV and can be applied to distinguish these viruses from other viruses.
Fig. 4.
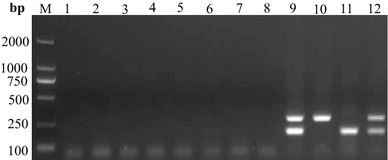
Specificity of the nanoPCR assay for the N genes of PEDV and TGEV. PEDV, TGEV, and other selected viruses were used as templates. Lane M DL2000 marker, Lane 1 cell negative control, Lane 2 uninfected tissue, Lane 3 PCV-2, Lane 4 PRRSV, Lane 5 PPV, Lane 6 PRV, Lane 7 H1N1, Lane 8 CSFV, Lane 9 recombinant plasmids, Lane 10 PEDV-infected tissue, Lane 11 TGEV-infected tissue, Lane 12 mixed PEDV and TGEV cell culture
Evaluation of clinical samples
Duplex nanoPCR and conventional PCR assays were used simultaneously to assess 114 clinical samples. The detection rates of the duplex nanoPCR and conventional PCR assays for PEDV were 48.2 % (55/114) and 45.6 % (52/114), respectively, while their detection rates for TGEV were 3.5 % (4/114) and 3.5 % (4/114), respectively; the co-infection rate of PEDV and TGEV was 2.6 % (3/114) (Table 2). These results indicate that the duplex nanoPCR and conventional PCR assays had similar sensitivity for TGEV in clinical samples, whereas the nanoplex PCR assay was slightly more sensitive than the conventional PCR assay for PEDV in clinical samples.
Table 2.
Detection of PEDV and TGEV in 114 clinical samples with duplex nanoPCR and conventional PCR assays
| Province or city | No. of clinical samples | Viruses detected | No. of positive samples detected by duplex nanoPCR | No. of positive samples detected by conventional | Positive (%) with duplex nanoPCR | Positive (%) with conventional PCR |
|---|---|---|---|---|---|---|
| Hebei | 91 | PEDV | 47 | 45 | 51.6 | 49.5 |
| TGEV | 4 | 4 | 4.4 | 4.4 | ||
| PEDV+TGEV | 3 | 3 | 3.3 | 3.3 | ||
| Beijing | 5 | PEDV | 2 | 2 | 40 | 40 |
| TGEV | 0 | 0 | 0 | 0 | ||
| PEDV+TGEV | 0 | 0 | 0 | 0 | ||
| Zhejiang | 3 | PEDV | 2 | 2 | 66.7 | 66.7 |
| TGEV | 0 | 0 | 0 | 0 | ||
| PEDV+TGEV | 0 | 0 | 0 | 0 | ||
| Guangdong | 9 | PEDV | 3 | 3 | 33.3 | 33.3 |
| TGEV | 0 | 0 | 0 | 0 | ||
| PEDV+TGEV | 0 | 0 | 0 | 0 | ||
| Others | 6 | PEDV | 1 | 0 | 16.7 | 0 |
| TGEV | 0 | 0 | 0 | 0 | ||
| PEDV+TGEV | 0 | 0 | 0 | 0 | ||
| Total | 114 | PEDV | 55 | 52 | 48.2 | 45.6 |
| TGEV | 4 | 4 | 3.5 | 3.5 | ||
| PEDV+TGEV | 3 | 3 | 2.6 | 2.6 |
Discussion
PEDV and TGEV cause significant economic losses in swine-raising countries. PEDV enteritis and TGEV enteritis result from destruction of enterocytes and villous atrophy of the intestinal mucosa, especially the jejunum and ileum [17]. PEDV and TGEV viruses are distinct species of the Alphacoronavirus genus that induce similar clinical signs and pathological lesions in newborn piglets; therefore, they are indistinguishable at clinical diagnosis [17, 18]. In recent years, nanoPCR has been developed significantly as a method of diagnosing swine diseases [19, 20]. Here, we used nanoPCR as a tool to distinguish PEDV and TGEV infections.
PEDV and TGEV can be specifically detected by the duplex nanoPCR assay described in this study by simultaneously amplifying more than one fragment during the same reaction. Multiplexing in this manner is useful because viruses such as PEDV, TGEV, PRV, and PRRSV may simultaneously infect pigs. Our duplex nanoPCR assay detected PEDV and TGEV with sensitivity comparable to that of conventional PCR. Most diagnostic procedures for coronaviruses, including RT-qPCR, are based on the nucleocapsid protein (N) gene, as it is the most abundantly expressed viral protein and can be detected soon after infection [21]. The duplex nanoPCR assay was successful only when PEDV and TGEV served as the template, indicating that the method is specific and applicable for differential diagnosis. To our knowledge, this is the first report regarding the establishment and optimization of a duplex nanoPCR assay for the N genes of PEDV and TGEV. The duplex nanoPCR assay reported here may be useful for the clinical diagnosis of PEDV and TGEV infection. In a screening of 114 samples collected from piglets in China, the duplex nanoPCR assay detected PEDV at a slightly higher rate in comparison with conventional PCR, while the assays had similar sensitivity for TGEV.
The duplex nanoPCR assay reported here was found to be a simple, sensitive, specific, and reliable method with the potential to be useful for routine molecular diagnosis and epidemiology.
Acknowledgments
This work was partly supported by the Agricultural Science and Technology Innovation Program (ASTIP-IAS15).
Compliance with ethical standards
Conflict of interest
The authors declare that they had no conflict of interest.
Ethical approval
This article does not contain any studies with human participants by any of the authors. The animal studies were approved by the Animal Medical Center DBN Technology Group.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
Yu Zhu, Lin Liang, Yakun Luo, and Guihua Wang contributed equally to this work.
Contributor Information
Xia Ai, Phone: + 86 22 23781297, Email: aixialucky@163.com.
Shangjin Cui, Phone: + 86 0 18518437100, Email: cuishangjin@caas.cn.
References
- 1.Meng F, Ren Y, Suo S, Sun X, Li X, Li P, Yang W, Li G, Li L, Schwegmann-Wessels C, Herrler G, Ren X. PLoS ONE. 2013;8:e57468. doi: 10.1371/journal.pone.0057468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enjuanes L, Smerdou C, Sanchez CM, Suñé C, Kelly S, Curtiss R, 3rd, Torres JM. Adv. Exp. Med. Biol. 1995;371B:1535–1541. [PubMed] [Google Scholar]
- 3.Jung K, Saif LJ. Vet. J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sestak K, Lanza I, Park SK, Weilnau PA, Saif LJ. Am. J. Vet. Res. 1996;57:664–671. [PubMed] [Google Scholar]
- 5.Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, Koster LG, Killian ML, Yoon KJ. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- 6.Wood EN. Vet. Rec. 1977;100:243–244. doi: 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- 7.Doyle LP, Hutchings LM. J. Am. Vet. Med. Assoc. 1946;108:257–259. [PubMed] [Google Scholar]
- 8.Hu X, Jr, Li N, Jr, Tian Z, Jr, Yin X, Jr, Qu L, Qu J. BMC Vet. Res. 2015;11:72. doi: 10.1186/s12917-015-0387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stadler J, Zoels S, Fux R, Hanke D, Pohlmann A, Blome S, Weissenböck H, Weissenbacher-Lang C, Ritzmann M, Ladinig A. BMC Vet. Res. 2015;11:142. doi: 10.1186/s12917-015-0454-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Shi BJ, Huang XG, Peng MY, Zhang XM, He DN, Pang R, Zhou B, Chen PY. J. Virol. Methods. 2013;194:107–112. doi: 10.1016/j.jviromet.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Carstens EB. Arch. Virol. 2010;155:133–146. doi: 10.1007/s00705-009-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowe J, Gauger P, Harmon K, Zhang J, Connor J, Yeske P, Loula T, Levis I, Dufresne L, Main R. Emerg. Infect. Dis. 2014;20:872–874. doi: 10.3201/eid2005.131628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alonso C, Goede DP, Morrison RB, Davies PR, Rovira A, Marthaler DG, Torremorell M. Vet. Res. 2014;45:73. doi: 10.1186/s13567-014-0073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SY, Song DS, Park BK. J. Vet. Diagn. Invest. 2001;13:516–520. doi: 10.1177/104063870101300611. [DOI] [PubMed] [Google Scholar]
- 15.Gerber PF, Gong Q, Huang YW, Wang C, Holtkamp D, Opriessnig T. Vet. J. 2014;202:33–36. doi: 10.1016/j.tvjl.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Rothberg L. Proc. Natl. Acad. Sci. USA. 2004;101:14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung K, Wang Q, Scheuer KA, Lu Z, Zhang Y, Saif LJ. Emerg. Infect. Dis. 2014;20:662–665. doi: 10.3201/eid2004.131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González JM, Gomez-Puertas P, Cavanagh D, Gorbalenya AE, Enjuanes L. Arch. Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan W, Li Y, Li P, Song Q, Li L, Sun J. J. Virol. Methods. 2015;220:18–20. doi: 10.1016/j.jviromet.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Y, Liang L, Zhou L, Zhao K, Cui S. J. Virol. Methods. 2015;219:46–50. doi: 10.1016/j.jviromet.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 21.Diel DG, Lawson S, Okda F, Singrey A, Clement T, Fernandes MH, Christopher-Hennings J, Nelson EA. Virus Res. 2016 doi: 10.1016/j.virusres.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]


