Abstract
Severe acute respiratory syndrome-associated coronavirus (SARS-CoV) structural proteins (S, E, M, and NC) localize in different subcellular positions when expressed individually. However, SARS-CoV M protein is co-localized almost entirely with S, E, or NC protein when co-expressed in the cells. On the other hand, only partial co-localization was observed when S and E, S and NC, or E and NC were co-expressed in the cells. Interactions between SARS-CoV M and other structural proteins but not interactions between S and E, S and NC, or E and NC were further demonstrated by co-immunoprecipitation assay. These results indicate that SARS-CoV M protein, similar to the M proteins of other coronaviruses, plays a pivotal role in virus assembly. The cytoplasmic C-terminus domain of SARS-CoV M protein was responsible for binding to NC protein. Multiple regions of M protein interacted with E and S proteins. A model for the interactions between SARS-CoV M protein and other structural proteins is proposed. This study helps us better understand protein-protein interactions during viral assembly of SARS-CoV.
Electronic supplementary material
The online version of this article (doi:10.1007/s11373-008-9278-3) contains supplementary material, which is available to authorized users.
Keywords: SARS-CoV, Membrane protein, Structural proteins, Co-localization, Co-immunoprecipitation
Introduction
Severe acute respiratory syndrome (SARS), a new infectious disease typically associated with fever, shortness of breath, cough, and pneumonia, first emerged in southern China in November 2002. Within months of the outbreak, SARS had spread globally, affecting over 8,000 patients in 29 countries with 774 fatalities [1]. The etiology of SARS is associated with a newly discovered coronavirus, SARS-associated coronavirus (SARS-CoV) [2–4]. SARS-CoV infects many organs, including lungs, liver, and immune cells [5, 6]. Subsequent studies have indicated that the SARS-CoV is of animal origin [7], and its precursor is still present in animal populations within the region. Although the global outbreak of SARS has been contained, there are serious concerns over its re-emergence. To date, no specific treatment exists for this disease. Thus, further basic and clinical research is required to control the disease.
SARS-CoV is phylogenetically distinct, and only distantly related to the other coronavirus clades [8, 9]. Coronaviruses are exceptionally large RNA viruses and employ complex regulatory mechanisms to express their genomes [10]. The genome structure, gene expression pattern and protein profiles of SARS-CoV are similar to those of other coronaviruses. Nine SARS-CoV specific mRNAs were synthesized in virus-infected cells [11]. These RNA were predicted to encode 4 structural proteins (spike, envelope, membrane, and nucleocapsid proteins), 16 non-structural proteins, and 8 accessory proteins. Previous studies on various coronaviruses indicated that the four structural proteins (S, E, M, and NC) play roles in virion morphogenesis [12, 13]. NC binds to viral RNA to form the nucleocapsid. Co-expression of M and E proteins together can form virus-like particles [14, 15]. Interactions between the M and E proteins and nucleocapsids result in virus budding through the cellular membrane [16]. Through the interaction with M protein, S protein is incorporated into the viral envelope [17, 18] and the mature virions are released from the cells. These studies suggest that coronavirus M protein [19, 20] plays a crucial role in the assembly of virus particles. Like other coronaviruses, SARS-CoV assembles at and buds into the lumen of the endoplasmic reticulum–Golgi intermediate compartment [21]. Accumulation of the viral envelope proteins at this compartment is a prerequisite for virus assembly [22]. Immuno-EM (electron microscopy) revealed that budding occurred at membranes of the ERGIC and the Golgi region as early as 3 h post infection, demonstrating that SARS-CoV replicated surprisingly fast. Previous data also suggests that SARS-CoV established replication complexes at ER-derived membranes [21]. Later on, viral nucleocapsids were transported to the budding sites in the Golgi region where the viral glycoproteins accumulate and particle formation occurs. Assembly of SARS-CoV RNA packaging signal into virus-like particles is nucleocapsid dependent [23]. In this report, the protein-protein interactions among SARS-CoV structural proteins were studied using confocal microscopy and immunoprecipitation followed by Western blotting analysis. Results from this study indicate that, similar to the M protein of other coronaviruses, SARS-CoV M protein plays a crucial role in the interactions between SARS-CoV structural proteins.
Materials and methods
Plasmid construction
The construction of plasmids expressing full-length spike and nucleocapsid proteins or encoding full-length and deletion mutants of membrane protein plus a V5 tag was described previously [24, 25]. Similar strategies were performed to prepare the plasmids expressing full-length spike, envelope, and nucleocapsid proteins with different tags using primers listed in Supplementary Table I. Vector pcDNA3.1/V5-His A (Invitrogen, CA, USA) was used to add a V5-His tag at the C-terminus of the expressed protein while vector pcDNA3-cMyc tag [26] was used to add a myc tag at the N-terminus of the expressed protein.
All the expression plasmids were verified by sequencing.
Protein expression in Vero E6 cells
Vero E6 cells were maintained in RPMI 1640 medium containing 10% fetal calf serum, 1% glutamine (200 mM, Biological Industries, Israel), and 100 μg/ml of penicillin/streptomycin (Gibco BRL, USA). The cells (2.5–2.7 × 105) were plated in the 35-mm dish. After an overnight incubation, cells were infected with a recombinant vaccinia virus carrying the T7 phage RNA polymerase gene [27]. Two hours after infection, the cells were transfected with 0.4 μg of plasmid DNA using Effectene transfection reagent (Qiagen, Germany). After 21 h of transfection, the recombinant proteins in the cells were analyzed.
Immunoprecipitation assay
The Vero E6 cells (1 × 106) were harvested 21 h after transfection and lysed in RIPA buffer (150 mM NaCl, 1% NP40, 0.5% deoxychloic acid, 0.1% SDS, 50 mM Tris, pH 7.5). After full-speed centrifugation for 5 min in a microcentrifuge, the supernatant was incubated with mouse anti-V5 monoclonal antibody (Invitrogen) or mouse anti-His monoclonal antibody (Santa Cruz Biotechnology, CA, USA) or mouse anti-myc monoclonal antibody (Oncogene, MA, USA) at 4°C overnight with shaking. The antigen-antibody complex was separated with pansorbin (Merck, Germany). The immunoprecipitated pellet was boiled for 10 min in sampling buffer and then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting. In each experiment, 10% of cell lysates were used for expression analysis (by Western blotting assay directly) while 90% of cell lysates were used for the co-immunoprecipitation assay [25].
Western blotting analysis
For Western blotting analysis, cells were dissolved in sample preparation buffer after washing with PBS twice. SARS-CoV M protein is not detected in SDS-PAGE after regular boiling treatment [24]. Therefore, treatments at room temperature were used for antigen preparation (sample buffer containing 50 mM of Tris–HCl (pH 6.8), 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, and 10% glycerol, without boiling) to detect the expression of SARS-CoV M protein. A 4.5% acrylamide stacking gel and 12% separating gel were used in this study. When proteins with smaller size were analyzed (e.g., membrane protein deletion mutants), a 15% separating gel was used. After SDS-PAGE, the gel was transferred to PVDF paper (Pall Corporation, NY, USA). All procedures were carried out at room temperature, according to previously published procedures [26, 28, 29], except that the first antibody used in this assay was mouse anti-V5 monoclonal antibody (Invitrogen) or mouse anti-His monoclonal antibody (Santa Cruz Biotechnology) or mouse anti-myc monoclonal antibody (Oncogene).
Confocal microscopy
About 2.5 × 105 cells were seeded into 35-mm culture dishes. After overnight incubation at 37°C, the cells were transfected with 0.4 μg of plasmid using the Effectene transfection kit (Qiagen, Germany). After transfection for 48 h, recombinant proteins in the cells were analyzed. Cells were fixed by acetone/methanol (1:1) at 0°C for 10 min. Fixed cells were washed with incubation buffer (0.05% NaN3, 0.02% saponin, 1% skim milk in PBS) twice for 5 min each time, then incubated with primary antibody (e.g., mouse anti-V5 monoclonal antibody, Invitrogen), which was diluted 200 fold, at 37°C for 30 min. Samples were washed with PBS three times (5 min each time at room temperature), then incubated with FITC-conjugated secondary antibody (e.g., goat anti-mouse IgG antibody, diluted 20×) at 37°C for 30 min. Samples were again washed with PBS three times (5–10 min each time at room temperature). DAPI (4′, 6-diamidino-2-phenylindole) (Merck, Germany) was used to stain DNA to localize the cell nuclei. Samples were then observed with confocal microscopy. To quantify the average percentage of co-localization, the Image J (NIH website) program was used [25].
Results
M protein co-localization in cultured cells
SARS-CoV structural proteins localized in different subcellular positions when they were expressed individually (Fig. 1). When SARS-CoV M protein was co-expressed with other structural proteins (S, E, or NC), they were almost entirely co-localized with the other proteins (Fig. 2a); R = 0.93, 0.90, 0.90 for co-localization of M and S proteins, M and NC proteins, and M and E proteins, respectively. This result suggests that M protein binds to other structural proteins in the cultured cells. When S and E, S and NC, or E and NC were co-expressed in the cells, only partial co-localization was observed (Fig. 2b); R = 0.54, 0.70, 0.85 for co-localization of E and S proteins, E and NC proteins, and NC and S proteins, respectively.
Fig. 1.
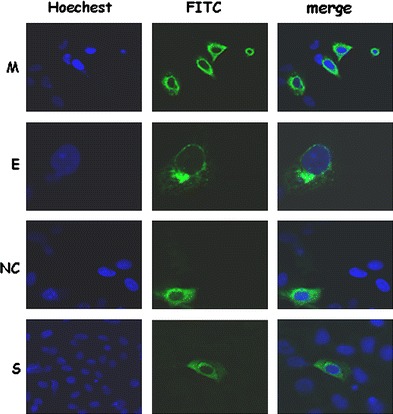
Differential subcellular localizations of SARS-CoV structural proteins S, E, M and NC) when they were expressed individually. Cells were transfected with the plasmid expressing M-V5, the plasmid expressing myc-E protein, the plasmid expressing NC protein, or the plasmid expressing S protein. After transfection, cells were fixed and stained with mouse anti-V5, mouse anti-myc, rabbit anti-NC, rabbit anti-S antibodies. Green color, M, NC, S, or E protein staining; blue color, DAPI staining
Fig. 2.
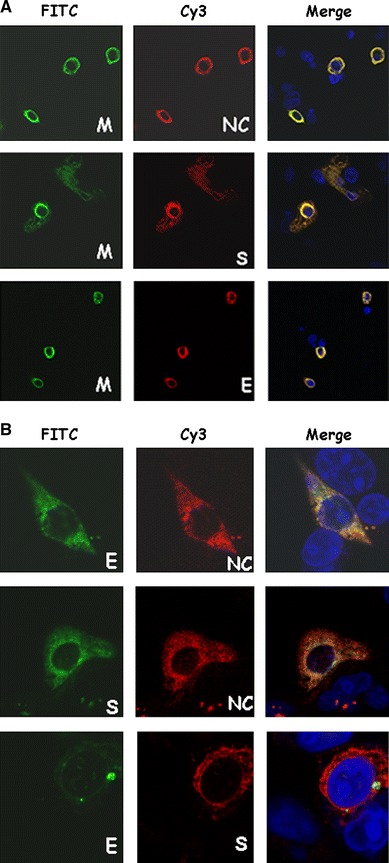
(a) Co-localization of SARS-CoV M protein with S, E, or NC protein when co-expressed in Vero E6 cells. Cells were co-transfected with plasmids expressing the M-V5 and NC (upper), S (middle), or myc-E (lower) proteins. After transfection, cells were fixed and stained with rabbit anti-NC and mouse anti-V5 antibodies (upper), or rabbit anti-S and mouse anti-V5 antibodies (middle), or mouse anti-myc and goat anti-mouse conjugated with Cy3 followed by anti-V5-FITC antibody (lower). Green color, M protein staining; red color, NC (or S, or E) protein staining; blue color, DAPI staining; yellow color, co-localization of M and other structural proteins. (b) Partial co-localization of E and NC, S and NC, or E and S proteins when they were co-expressed in Vero E6 cells. Cells were co-transfected with plasmids expressing myc-E and NC proteins (upper); S and myc-NC proteins (middle), and myc-E and S proteins (lower). After transfection, cells were fixed and stained with rabbit anti-NC (or anti-S) and mouse anti-myc antibodies
Binding between M and other structural proteins
To verify whether SARS-CoV M protein and other structural proteins could bind to each other within cells, we performed a co-immunoprecipitation experiment. The V5-tagged, full-length M protein and the myc-tagged E protein were co-expressed in Vero E6 cells by transient transfection. After transfection, cell lysates were immunoprecipitated with the anti-myc antibody, followed by Western blotting using the anti-V5 antibody. As shown in Fig. 3a, the V5-tagged M protein was immunoprecipitated by the anti-myc antibody in the presence (lane 8), but not in the absence (lane 6), of E protein. This result further confirmed that M and E proteins were bound to each other in the cultured cells. Similar co-immunoprecipitation experiments were performed to study the interactions between M and S proteins (Fig. 3b), and M and NC proteins (Fig. 3c). These results confirmed that M protein also bound S and NC proteins in the cultured cells.
Fig. 3.
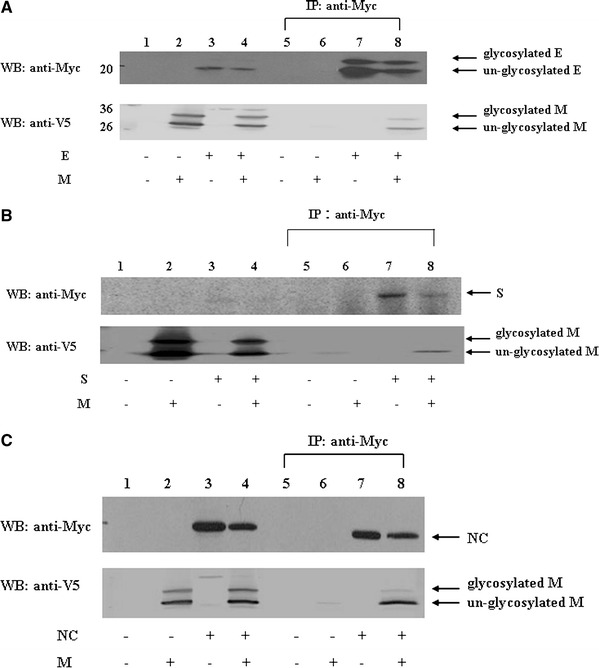
(a) SARS-CoV M interaction with E protein. Vero E6 cells were transfected with vector alone (lanes 1 and 5), plasmid encoding M-V5 (lanes 2 and 6), plasmid encoding myc-E (lanes 3 and 7), or co-transfected with both plasmids (lanes 4 and 8). Cell lysates were directly analyzed by Western blotting (lanes 1–4) or immunoprecipitated with the anti-myc antibody prior to Western blotting (lanes 5–8). (b) SARS-CoV M interaction with S proteins. Vero E6 cells were transfected with vector alone (lanes 1 and 5), plasmid encoding M-V5 (lanes 2 and 6), plasmid encoding myc-S (lanes 3 and 7), or co-transfected with both plasmids (lanes 4 and 8). Cell lysates were directly analyzed by Western blotting (lanes 1–4) or immunoprecipitated with the anti-myc antibody prior to Western blotting (lanes 5–8). (c) SARS-CoV M interaction with NC proteins. Vero E6 cells were transfected with vector alone (lanes 1 and 5), plasmid encoding M-V5 (lanes 2 and 6), plasmid encoding myc-NC (lanes 3 and 7), or co-transfected with both plasmids (lanes 4 and 8). Cell lysates were directly analyzed by Western blotting (lanes 1–4) or immunoprecipitated with the anti-myc antibody prior to Western blotting (lanes 5–8)
Under the same experimental conditions (Supplementary Fig. 1a), the interaction between E and M but not the interaction between E and S was detected. Similarly, the interaction between NC and M but not the interaction between NC and E was demonstrated (Supplementary Fig. 1b). Moreover, the interaction between NC and M but not the interaction between NC and S was detected (Supplementary Fig. 1c).
Deletion mapping and M protein interactions with other structural proteins
To identify which region(s) of SARS-CoV M protein were interacting with NC protein, a deletion mapping experiment was performed. As shown in Fig. 4a, the V5-tagged M protein without the first 100 amino acids was immunoprecipitated by the anti-myc antibody in the presence (lane 8), but not in the absence (lane 6) of NC protein, while the first 100 amino acids of M protein with V5 tag could not be immunoprecipitated by the anti-myc antibody in the presence of the NC protein (Fig. 4b). This result indicates that the cytoplasmic C-terminus domain of SARS-CoV M protein was responsible for the binding with NC protein. To determine whether different topologies of the C-terminus domain of M protein could still interact with NC protein, the first (amino acids 21–37) or the second (amino acids 46–68) transmembrane domain of M protein was deleted. M protein without the first transmembrane domain still left its C-terminus domain in the cytoplasm and it was not glycosylated, while M protein without the second transmembrane domain left its C-terminus domain in the endoplasmic reticulum lumen (Supplementary Fig. 2). As shown in Fig. 4c, M protein without the first transmembrane domain interacted with NC protein while M protein without the second transmembrane domain did not. To identify the regions within the cytoplasmic C-terminus domain of M protein responsible for binding to NC protein, different regions (amino acids 101–135, 136–170, and 171–221) within the cytoplasmic C-terminus domain of M protein were deleted separately. As shown in Fig. 4d, all three M deletion mutants still interacted with NC protein. Co-localization between the cytoplasmic C-terminus domain of M protein and NC protein was also demonstrated (upper panel, Fig. 7).
Fig. 4.
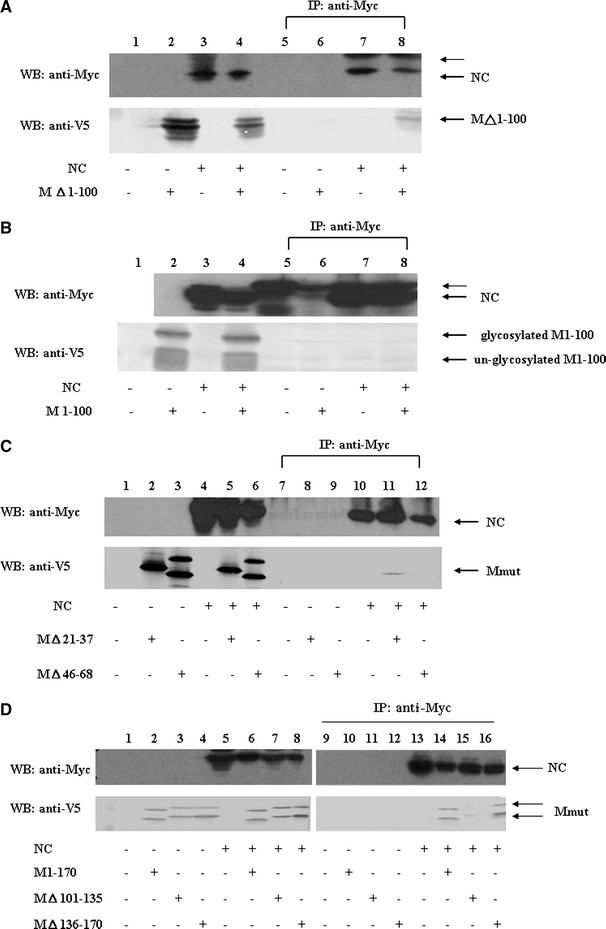
Immunoprecipitation and Western blotting analyses of SARS-CoV M protein cytoplasmic C-terminus domain interaction with NC protein. (a) Vero E6 cells were transfected with vector alone (lanes 1 and 5), plasmid encoding cytoplasmic C-terminus domain of M protein (i.e. M protein without its first 100 amino acids) with a V5 tag (lanes 2 and 6), plasmid encoding myc-NC (lanes 3 and 7), or co-transfected with both plasmids (lanes 4 and 8). Cell lysates were directly analyzed by Western blotting (lanes 1–4) or immunoprecipitated with the anti-myc antibody prior to Western blotting (lanes 5–8). The cytoplasmic C-terminus domain of M protein interacted with NC protein. The protein larger than NC protein marked by the thin arrow is the immunoglobulin heavy chain (lower panel). More than one band was detected when the plasmid encoding the cytoplasmic C-terminus domain of M protein was expressed, possibly due to sample preparation without boiling treatment. (b) Vero E6 cells were transfected with vector alone (lanes 1 and 5), plasmid encoding the first 100 amino acids of M protein with a V5 tag (lanes 2 and 6), plasmid encoding myc-NC (lanes 3 and 7), or co-transfected with both plasmids (lanes 4 and 8). Cell lysates were directly analyzed by Western blotting (lanes 1–4) or immunoprecipitated with the anti-myc antibody prior to Western blotting (lanes 5–8). M protein without its cytoplasmic domain did not interact with NC protein. The protein larger than NC protein marked by the thin arrow is the immunoglobulin heavy chain. (c) Vero E6 cells were transfected with vector alone, plasmid encoding M△21–37 plus a V5 tag (or the plasmid encoding M△46–68 plus a V5 tag), plasmid encoding myc-NC, or co-transfected with two plasmids (M△21–37 and myc-NC, M△46–68 and myc-NC). Cell lysates were directly analyzed by Western blotting (lanes 1–6) or immunoprecipitated with the anti-myc antibody prior to Western blotting (lanes 7–12). M protein without its first hydrophobic domain, but not M protein without its second hydrophobic domain interacted with NC protein. (d) Vero E6 cells were transfected with vector alone, plasmid encoding M1–170 with a V5 tag (or the plasmid encoding M△101–135 plus a V5 tag, or the plasmid encoding M△136–170 plus a V5 tag), plasmid encoding myc-NC, or co-transfected with two plasmids (M1–170 and myc-NC, M△101–135 and myc-NC, M△136–170 and myc-NC). Cell lysates were directly analyzed by Western blotting (lanes 1–8) or immunoprecipitated with the anti-myc antibody prior to Western blotting (lanes 9–18). All of the M cytoplasmic domain-deleted mutants interacted with NC protein
Fig. 7.
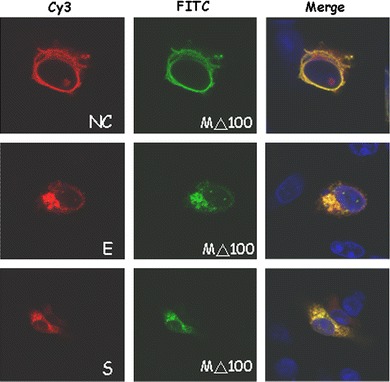
Co-localization of M△1–100 protein and NC (upper panel), E (middle panel), and S (lower panel) proteins. Similar to Fig. 2a, except the plasmid encoding M protein without the first 100 amino acids plus a V5 tag was used to replace the plasmid encoding M protein with a V5 tag
Deletion mapping was also performed to identify which domains of M protein interacted with E protein. The C-terminal fragment from amino acids 171–221 of M protein was truncated first. As shown in Fig. 5a, this truncated M protein still bound E protein. The first 50 amino acids of this truncated M protein were further deleted and still interacted with E protein. M protein without this region (amino acids 51–170) still interacted with E protein. These results suggest that multiple regions within M protein interacted with E protein. The second and third transmembrane regions (amino acids 46–68 and 78–100) of M protein by themselves, penetrated into the cell membrane, while the first transmembrane region (amino acids 14–36) was stabilized by interaction with the other transmembrane segments [25]. To determine whether the second or third transmembrane region alone interacted with E protein, these two transmembrane regions were fused with the first 115 amino acids of HCV core protein separately [25]. As shown in Fig. 5b, the second or the third transmembrane region of M protein alone was sufficient to bind E protein. Furthermore, the cytoplasmic C-terminus domain of M protein (i.e. M protein without its first 100 amino acids) also interacted with E protein (Fig. 5c). In this case, the unglycosylated, but not glycosylated, E protein was preferentially immunoprecipitated. Co-localization between the cytoplasmic C-terminus domain of M protein and E protein was also demonstrated (middle panel, Fig. 7).
Fig. 5.
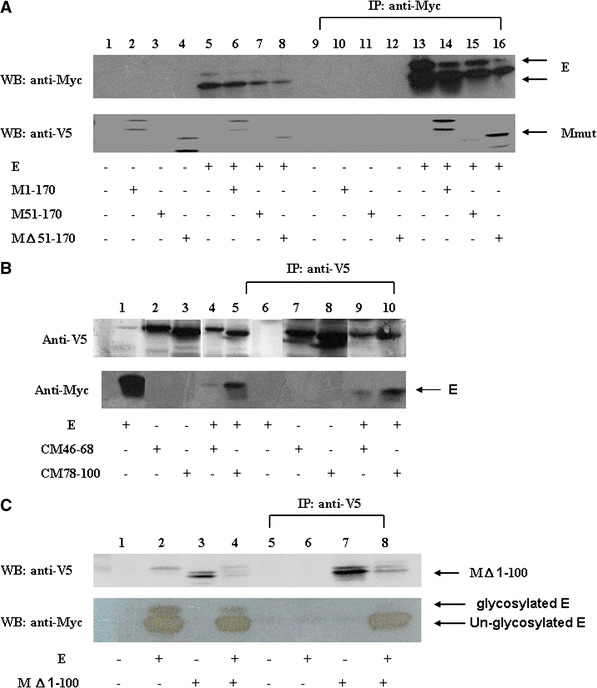
Immunoprecipitation and Western blotting analyses of SARS-CoV M protein fragment interactions with E protein. (a) Interactions between SARS-CoV E and different M fragments. Vero E6 cells were transfected with vector alone, with the plasmid encoding M1–170 with a V5 tag (or the plasmid encoding M51–170 plus a V5 tag, or the plasmid encoding M△51–170 plus a V5 tag), plasmid encoding myc-E, or co-transfected with two plasmids (M1–170 and myc-E, M51–170 and myc-E, M△51–170 and myc-E). Cell lysates were directly analyzed by Western blotting (lanes 1–8) or immunoprecipitated with the anti-myc antibody prior to Western blotting (lanes 9–18). (b) Vero E6 cells were transfected with the plasmid encoding myc-E protein, plasmid encoding the first 115 amino acids of HCV core protein and M protein amino acids 46–68 plus a V5 tag (the plasmid encoding the first 115 amino acids of HCV core protein and M protein amino acids 78–100 plus a V5 tag), or co-transfected with two plasmids. Cell lysates were directly analyzed by Western blotting (lanes 1–4) or immunoprecipitated with the anti-V5 antibody prior to Western blotting (lanes 5–8). Either the second or the third transmembrane domain of M protein was sufficient for interaction with E protein. (c) Interactions between SARS-CoV E and the cytoplasmic C-terminus domain of M protein. Vero E6 cells were transfected with vector alone, plasmid encoding myc-E protein, plasmid encoding M protein without the first 100 amino acids plus a V5 tag, or co-transfected with both plasmids. Cell lysates were directly analyzed by Western blotting (lanes 1–4) or immunoprecipitated with the anti-V5 antibody prior to Western blotting (lanes 5–8) (lower panel). More than one band was detected when the plasmid encoding the cytoplasmic C-terminus domain of M protein was expressed, possibly due to sample preparation without boiling treatment
The M mutant constructs were also used to map the binding region(s) of M protein with S protein. As shown in Fig. 6a, M protein containing only amino acids 51–170 interacted with S protein. M protein without this region also interacted with S protein (Fig. 6b). Again, the cytoplasmic C-terminus domain of M protein was also sufficient to interact with S protein (Fig. 6c). These results suggest that multiple regions within M protein interacted with S protein. Co-localization between the cytoplasmic C-terminus domain of M protein and S protein was also demonstrated (lower panel of Fig. 7).
Fig. 6.
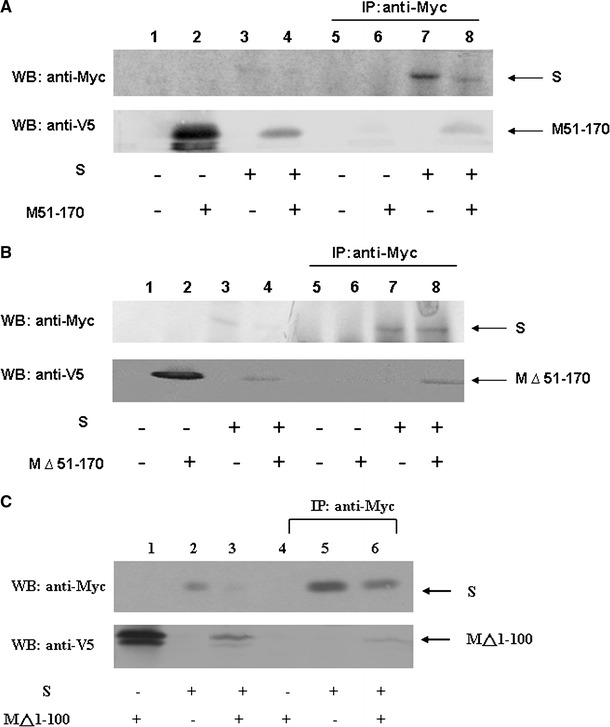
M protein interaction with S protein. (a) Vero E6 cells were transfected with vector alone (lanes 1 and 5), plasmid encoding M51–170 with a V5 tag (lanes 2 and 6), plasmid encoding myc-S (lanes 3 and 7), or co-transfected with both plasmids (lanes 4 and 8). Cell lysates were directly analyzed by Western blotting (lanes 1–4) or immunoprecipitated with the anti-myc antibody prior to Western blotting (lanes 5–8). (b) Similar to (a), except the plasmid encoding M△51–170 plus a V5 tag was used to replace the plasmid encoding M51–170 with a V5 tag. (c) Interactions between SARS-CoV S and cytoplasmic C-terminus domain of M protein. Similar to (a), except the plasmid encoding M protein without the first 100 amino acids plus a V5 tag was used to replace the plasmid encoding M51–170 with a V5 tag (lower panel). More than one band was detected when the plasmid encoding cytoplasmic domain of M protein was expressed possibly due to the sample preparation without boiling treatment. The results show that multiple regions of M protein interacted with S protein
Discussion
SARS-CoV structural proteins (S, E, M, and NC) localized to different subcellular positions when they were expressed individually (Fig. 1), similar to the results of a previous report [30]. SARS-CoV M protein co-localized almost entirely with S, E, or NC proteins when they were co-expressed within the cells (Fig. 2a). On the other hand, only partial co-localization was observed when S and E, S and NC, or E and NC were co-expressed in the cells (Fig. 2b). Furthermore, the interactions between M and the other structural proteins were demonstrated by co-immunoprecipitation (Fig. 3). The interactions of E and S, E and NC, NC and S proteins were not demonstrated by co-immunoprecipitation (Supplementary Fig. 1). These results suggest that SARS-CoV M protein plays a pivotal role in virus assembly. Interactions of S and E, S and NC, or E and NC could occur after binding with M protein.
Previous studies reported that virus-like particles (VLP) formed when either SARS-CoV M and E proteins [31] or M and NC proteins [32] were co-expressed in cells. We hypothesize that M protein plays a crucial role in virus assembly and our study results support this hypothesis.
SARS-CoV M protein appears to be a triple-spanning membrane protein [33], while NC protein is a cytoplasmic protein. Due to the topology of these two proteins, M protein is supposed to interact with NC through its cytoplasmic C-terminus domain. Indeed, as shown in Fig. 4, only the C-terminus domain of M protein residing in the cytoplasm interacted with NC protein. The result that three M deletion mutants (M protein without amino acids 101–135, 136–170, or 171–221) interacted with NC protein (Fig. 4d) suggests that almost the entire C-terminus domain of M protein is responsible for the interaction with NC protein. Our results agree with those of previous reports using in vitro GST pull-downed assays [34] or yeast two-hybrid and surface plasmon resonance techniques [35] to study the interactions between SARS-CoV M and NC proteins.
That at least two transmembrane domains and the cytoplasmic domain of M protein are sufficient for the interacting with E protein (Fig. 5) suggest extensive interactions between these two proteins. Compared with other constructs, the cytoplasmic C-terminus domain of M protein preferentially immunoprecipitated unglycosylated, but not glycosylated, E protein (Fig. 5c). This result supports a previously reported model for the topology of E protein [36]: unglycosylated E protein leaves both its N- and C-termini in the cytoplasm while glycosylated E protein leaves both its N- and C-termini in the endoplasmic reticulum lumen.
Similar to the interactions of M and S proteins in another coronaviruses [37], SARS-CoV M protein interacted with S protein through multiple regions (Fig. 6). A model for the interactions between SARS-CoV M protein and other structural proteins was proposed (Fig. 8). While SARS-CoV structural proteins (S, E, M, and NC) reside in different subcellular locales, M protein brings other structural proteins (NC, S, and E) together through interacting with them. After that, interactions of S and E, S and NC, or E and NC occur.
Fig. 8.
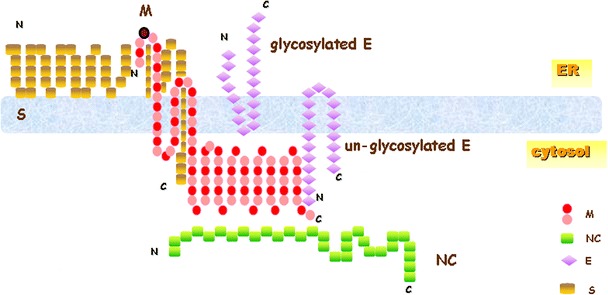
A proposed model for the interactions between the M protein and other structural proteins of SARS-CoV. M protein probably interacts with unglycosylated E protein through cytoplasmic C-terminus regions first. After glycosylation of E protein, interaction between M and E proteins are possibly through transmembrane regions, and the freed cytoplasmic region of M protein will then interact with NC protein. S protein could incorporate into virus-like particles formed by M and E proteins through interacting with multiple regions of M protein
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgement
This work has been supported by grants from Tzu Chi University (TCIRP95002-01 and TCIRP96004-05) and from National Science Council of Taiwan (NSC 96-3112-B-320-001) to Dr. Shih-Yen Lo.
References
- 1.Poon LL, Guan Y, Nicholls JM, Yuen KY, Peiris JS. The aetiology, origins, and diagnosis of severe acute respiratory syndrome. Lancet Infect Dis. 2004;4:663–671. doi: 10.1016/S1473-3099(04)01172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TC, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus AD, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 3.Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, Cheng VC, Chan KH, Tsang DN, Yung RW, Ng TK, Yuen KY. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, van Amerongen G, van Riel D, Laman JD, de Jong T, van Doornum G, Lim W, Ling AE, Chan PK, Tam JS, Zambon MC, Gopal R, Drosten C, van der Werf S, Escriou N, Manuguerra JC, Stohr K, Peiris JS, Osterhaus AD. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, Zou W, Zhan J, Wang S, Xie Z, Zhuang H, Wu B, Zhong H, Shao H, Fang W, Gao D, Pei F, Li X, He Z, Xu D, Shi X, Anderson VM, Leong AS. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Rao S, Jiang C. Molecular pathogenesis of severe acute respiratory syndrome. Microbes Infect. 2007;9:119–126. doi: 10.1016/j.micinf.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enserink M. Infectious diseases. Clues to the animal origins of SARS. Science. 2003;300:1351. doi: 10.1126/science.300.5624.1351a. [DOI] [PubMed] [Google Scholar]
- 8.Holmes KV, Enjuanes L. Virology. The SARS coronavirus: a postgenomic era. Science. 2003;300:1377–1378. doi: 10.1126/science.1086418. [DOI] [PubMed] [Google Scholar]
- 9.Lai M. SARS virus: the beginning of the unraveling of a new coronavirus. J Biomed Sci. 2003;10:664–675. doi: 10.1007/BF02256318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes K, Lai MM (1996) Coronaviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM (eds) Fields virology, vol 1. Lippincott-Raven Publishers, Philadelphia, pp 1075–1093
- 11.Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, Bayer S, Weissbrich B, Snijder EJ, Rabenau H, Doerr HW, Gorbalenya AE, Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen VP, Hogue BG. Protein interactions during coronavirus assembly. J Virol. 1997;71:9278–9284. doi: 10.1128/jvi.71.12.9278-9284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen VP, Hogue BG. Coronavirus envelope glycoprotein assembly complexes. Adv Exp Med Biol. 1998;440:361–365. doi: 10.1007/978-1-4615-5331-1_47. [DOI] [PubMed] [Google Scholar]
- 14.Vennema H, Godeke GJ, Rossen JW, Voorhout WF, Horzinek MC, Opstelten DJ, Rottier PJ. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Haan CA, Kuo L, Masters PS, Vennema H, Rottier PJ. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayanan K, Maeda A, Maeda J, Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J Virol. 2000;74:8127–8134. doi: 10.1128/JVI.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godeke GJ, de Haan CA, Rossen JW, Vennema H, Rottier PJ. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J Virol. 2000;74:1566–1571. doi: 10.1128/JVI.74.3.1566-1571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Opstelten DJ, Raamsman MJ, Wolfs K, Horzinek MC, Rottier PJ. Envelope glycoprotein interactions in coronavirus assembly. J Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rottier P, Brandenburg D, Armstrong J, van der Zeijst B, Warren G. In vitro assembly of the murine coronavirus membrane protein E1. Adv Exp Med Biol. 1984;173:53–64. doi: 10.1007/978-1-4615-9373-7_5. [DOI] [PubMed] [Google Scholar]
- 20.Rottier PJ, Welling GW, Welling-Wester S, Niesters HG, Lenstra JA, Van der Zeijst BA. Predicted membrane topology of the coronavirus protein E1. Biochemistry. 1986;25:1335–1339. doi: 10.1021/bi00354a022. [DOI] [PubMed] [Google Scholar]
- 21.Stertz S, Reichelt M, Spiegel M, Kuri T, Martinez-Sobrido L, Garcia-Sastre A, Weber F, Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride CE, Li J, Machamer CE. The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J Virol. 2007;81:2418–2428. doi: 10.1128/JVI.02146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh PK, Chang SC, Huang CC, Lee TT, Hsiao CW, Kou YH, Chen IY, Chang CK, Huang TH, Chang MF. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YN, Chen LK, Ma HC, Yang HH, Li HP, Lo SY. Thermal aggregation of SARS-CoV membrane protein. J Virol Methods. 2005;129:152–161. doi: 10.1016/j.jviromet.2005.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma HC, Fang CP, Hsieh YC, Chen SC, Li HC, Lo SY. Expression and membrane integration of SARS-CoV M protein. J Biomed Sci. 2008;15:301–310. doi: 10.1007/s11373-008-9235-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma HC, Ku YY, Hsieh YC, Lo SY. Characterization of the cleavage of signal peptide at the C-terminus of hepatitis C virus core protein by signal peptide peptidase. J Biomed Sci. 2007;14:31–41. doi: 10.1007/s11373-006-9127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma HC, Ke CH, Hsieh TY, Lo SY. The first hydrophobic domain of the hepatitis C virus E1 protein is important for interaction with the capsid protein. J Gen Virol. 2002;83:3085–3092. doi: 10.1099/0022-1317-83-12-3085. [DOI] [PubMed] [Google Scholar]
- 29.Ma HC, Lin TW, Li H, Iguchi-Ariga SM, Ariga H, Chuang YL, Ou JH, Lo SY. Hepatitis C virus ARFP/F protein interacts with cellular MM-1 protein and enhances the gene trans-activation activity of c-Myc. J Biomed Sci. 2008;15:417–425. doi: 10.1007/s11373-008-9248-9. [DOI] [PubMed] [Google Scholar]
- 30.Nal B, Chan C, Kien F, Siu L, Tse J, Chu K, Kam J, Staropoli I, Crescenzo-Chaigne B, Escriou N, van der Werf S, Yuen KY, Altmeyer R. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J Gen Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- 31.Ho Y, Lin PH, Liu CY, Lee SP, Chao YC. Assembly of human severe acute respiratory syndrome coronavirus-like particles. Biochem Biophys Res Commun. 2004;318:833–838. doi: 10.1016/j.bbrc.2004.04.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y, Yang ZY, Kong WP, Nabel GJ. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J Virol. 2004;78:12557–12565. doi: 10.1128/JVI.78.22.12557-12565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 34.Fang X, Ye L, Timani KA, Li S, Zen Y, Zhao M, Zheng H, Wu Z. Peptide domain involved in the interaction between membrane protein and nucleocapsid protein of SARS-associated coronavirus. J Biochem Mol Biol. 2005;38:381–385. doi: 10.5483/bmbrep.2005.38.4.381. [DOI] [PubMed] [Google Scholar]
- 35.Luo H, Wu D, Shen C, Chen K, Shen X, Jiang H. Severe acute respiratory syndrome coronavirus membrane protein interacts with nucleocapsid protein mostly through their carboxyl termini by electrostatic attraction. Int J Biochem Cell Biol. 2006;38:589–599. doi: 10.1016/j.biocel.2005.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Q, Liao Y, Torres J, Tam JP, Liu DX. Biochemical evidence for the presence of mixed membrane topologies of the severe acute respiratory syndrome coronavirus envelope protein expressed in mammalian cells. FEBS Lett. 2006;580:3192–3200. doi: 10.1016/j.febslet.2006.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Haan CA, Smeets M, Vernooij F, Vennema H, Rottier PJ. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. J Virol. 1999;73:7441–7452. doi: 10.1128/jvi.73.9.7441-7452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


