Abstract
Drosophila melanogaster silver gene is the ortholog of the coding gene of mammalian carboxypeptidase D (CPD). The silver gene gives rise to eight different splicing variants of differing length that can contain up to three homologous repeats. Among the protein variants encoded, the short form 1B alias DmCPD1Bs (D. melanogaster CPD variant 1B short) is necessary and sufficient for viability of the fruit fly. It has one single repeat, it is active against standard peptide substrates, and it is localized to the secretory pathway. In this work, the enzyme was found as a monomer in solution and as a homodimer in the crystal structure, which features a protomer with an N-terminal 311-residue catalytic domain of α/β-hydrolase fold and a C-terminal 84-residue all-β transthyretin-like domain. Overall, DmCPD1Bs conforms to the structure of N/E-type funnelins/M14B metallopeptidases, but it has two unique structural elements potentially involved in regulation of its activity: (i) two contiguous surface cysteines that may become palmitoylated and target the enzyme to membranes, thus providing control through localization, and (ii) a surface hot spot targetable by peptidases that would provide a regulatory mechanism through proteolytic inactivation. Given that the fruit fly possesses orthologs of only two out of the five proteolytically competent N/E-type funnelins found in higher vertebrates, DmCPD1Bs may represent a functional analog of at least one of the missing mammalian CPs.
Abbreviations: CD, catalytic domain; CP, carboxypeptidase; DmCPD1Bs, Drosophila melanogaster carboxypeptidase D variant 1B short; GEMSA, 2-guanidinoethyl-mercaptosuccinic acid; PDB, Protein Data Bank; TAFI(a), (activated) thrombin-activatable fibrinolysis inhibitor; TLD, transthyretin-like domain
Keywords: metallocarboxypeptidase, funnelin, splicing variant, activity regulation, development
Introduction
The silver (svr) gene was discovered in Drosophila melanogaster by Bridges in the early 1920s and it maps near the distal end of chromosome X.1, 2, 3, 4 Adult fruit flies with mutations in this gene display cuticles that are pale and silvery in color due to reduced melanization, a finding that gave rise to the name of the gene. Silver includes nine exons (1A, 1B, and 2–8), which code for eight different mRNA splicing variants. These result in two short protein variants of 433 (protein svr-PE alias “1A short”) and 435 [svr-PF alias “1B short” and DmCPD1Bs (D . melanogaster carboxypeptidase D variant 1B short)] residues and in six long forms spanning between 1259 and 1439 residues (svr-PB, svr-PC, svr-PD, svr-PG, svr-PH, and svr-PI; see UniProt sequence database entry P42787 and Fig. 1a).3, 4, 5, 6 The long variants possess the overall modular structure and sequence features of carboxypeptidase D (CPD), a glycosylated 180-kDa enzyme studied since the middle 1990s in fruit fly, mouse, rat, duck, bovine, chicken, and humans.7, 8, 9 CPD was localized to the trans-Golgi network, the secretory and reuptake pathways, and transiently on the cell surface.10 It was hypothesized to remove basic C-terminal residues from proteins and peptides supporting the endoproteolytic action of furin-like serine proprotein convertases of the subtilisin/kexin type. Thus, CPD would contribute to processing of hormones (e.g., adipokinetic hormone and bradykinin), growth factors, neurotransmitters, and other bioactive peptides (e.g., dynorphin B-14) that follow the secretory and endocytic pathways.10, 11, 12, 13 Moreover, it has been suggested that arginine released by CPD from peptide substrates may stimulate the production of nitric oxide, an important regulator of cellular processes such as neurotransmission, vasodilatation, and tumor progression and survival.14, 15
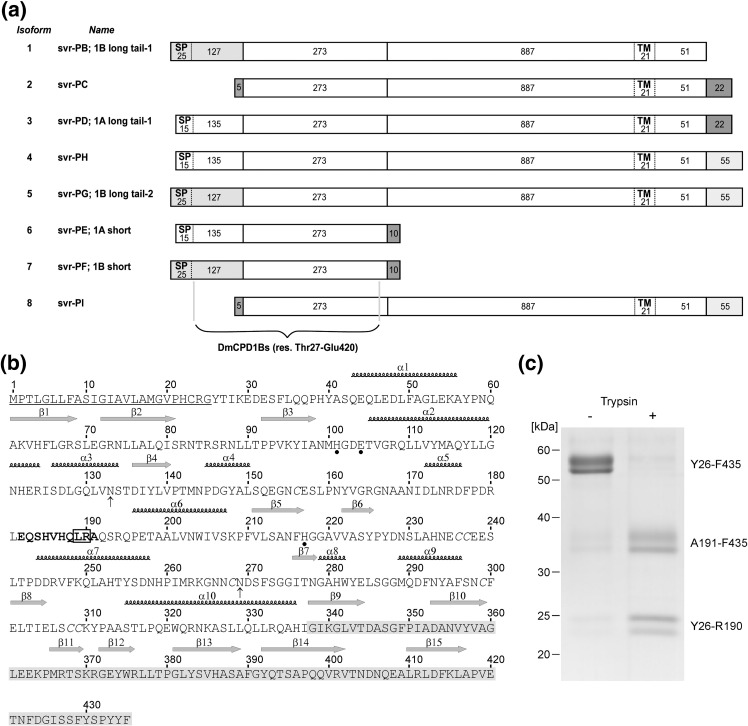
Fig. 1.
Sequence and hydrolytic cleavage of DmCPD1Bs. (a) Protein isoforms described for the silver gene as a result of differential splicing (UniProt database entry P42787). Each exon product is shown as a distinct bar with the residues it spans. The N-terminal fragments of 150 (encoded by exon 1A) and 152 (encoded by exon 1B) residues show ∼ 39% sequence identity; the extent of the signal peptide (SP) of the former was estimated with program SignalP at www.expasy.ch. The short fragments of 5 and 10 residues, as well as the C-terminal fragments of 22 and 55, are unrelated to any other sequence of the protein. The extent of the transmembrane region (TM) is according to the UniProt entry. Isoform 7 alias svr-PF, 1B short, herein DmCPD1Bs, spans 435 residues, of which 27–420 correspond to the first funnelin + TLD repeat studied herein. The flanking peptides protrude from the surface and are unstructured. The last residue encoded by exon 1B is Gln152. See also Ref. 5. (b) Full-length amino acid sequence of DmCPD1Bs in single-letter code. The 25-residue signal peptide is underlined, the residues involved in zinc coordination are labeled with full circles, and the two glycosylation sites are indicated by arrows. The cysteine pairing observed in the structure is Cys156-Cys309, Cys236-Cys237, and Cys268-Cys308. The sequential numbering employed throughout the text and the regular secondary-structure elements are shown above each sequence line. The TLD is hallmarked by a light gray background. The segment of the regulatory loop, which is disordered in the structure, is shown in boldface and the experimentally verified serine protease cleavage sites are framed. (c) Coomassie-stained SDS-PAGE analysis of the mature protein (residues Tyr26-Phe435) reveals two main bands at ∼ 52–58 kDa, which indicates heterogeneity in the glycosylation pattern. Incubation with trypsin produced a single cleavage within the regulatory loop at Arg190 and gave rise to two cleavage product pairs of ∼ 35 and ∼ 23 kDa. The cleavage products are already present in the non-treated enzyme due to incomplete inhibition of the P. pastoris serine protease during protein expression and purification.
CPD belongs to the funnelins, a tribe of zinc-dependent CPs also referred to as M14 metallopeptidases in MEROPS database.16, 17 Its hallmark is an ∼ 300-residue α/β-hydrolase domain first described for bovine CPA, an A/B-type funnelin. In contrast to the latter, N/E-type funnelins such as CPD possess an additional ∼ 80-residue C-terminal transthyretin-like domain (TLD).10, 12 In addition to CPD, seven further N/E-type funnelins are found in humans and mice, of which five show enzymatic activity (CPD, CPE, CPM, CPN, and CPZ) and three do not (AEBP1/ACLP, CPX1, and CPX2).9 In contrast, Drosophila comprises only CPD and CPM orthologs.5, 6 In vertebrates, CPM binds to cell membranes, but it also circulates in body fluids. It is assumed to participate in the extracellular processing of peptides and thus to have an activity comparable to CPD.18, 19, 20
CPD is unique in having three funnelin + TLD repeats followed by a transmembrane anchor and a small cytosolic domain, which may be required for intracellular routing and export to nascent secretory vesicles.9, 10, 16, 21, 22 Only the first two funnelin domains are catalytically competent in mammals and duck. This also happens in Drosophila variants encoded by exon 1B (see Refs. 5 and 6 and Fig. 1a). When isolated, the two funnelin repeats show different pH optima and specificity. The N-terminal repeat is more active at neutral pH and prefers arginine over lysine at the C-terminus of peptide substrates. In contrast, the second repeat prefers slightly acidic environments (pH 5–6) and excises lysines slightly more efficiently than arginines.6 Variants encompassing both repeats have broader pH optimum and substrate specificity than single-repeat variants. This is consistent with the conditions found throughout the secretory pathway, in which pH ranges from 5 to 7.5, 6 The third funnelin domain of CPD is catalytically incompetent as it lacks critical residues, but it may maintain the overall α/β-hydrolase scaffold.10 In the duck, it was reported to be recognized by the pre-S region of the large envelope protein of hepatitis B virus during infection.23, 24 This is reminiscent of ACE2, a CP belonging to another structural group, the cowrins,25 which is the receptor for human SARS coronavirus.26 In contrast to the long, three-repeat variants, the two Drosophila short forms have only the first repeat, and they are secreted and soluble (Fig. 1a and b). Among them, only DmCPD1Bs is catalytically active.5, 6 Although both short forms are present in the Golgi and may participate in physiological processes, the lack of membrane anchors makes them transient. Moreover, the endocytic vesicles contain only the long forms.5 In addition, while the membrane anchors can regulate the long forms through localization/compartmentalization, the soluble DmCPD1Bs may be regulated in a different way along the secretory pathway.
Drosophila silver alias CPD is essential for life as mutants that do not express functional CPD are not viable and die early in the larval stage. The enzyme plays a major role in melanization and sclerotization of the insect cuticle, wing morphogenesis, catecholamine metabolism, phagocytosis, engulfment, memory, and sensitivity to cold and ethanol.1, 2, 3, 4, 5, 6, 13, 27 Interestingly, mutants that contain a functional DmCPD1Bs but no three-repeat variant are viable, but they have a silvery body and minor defects in the wings. Therefore, this short form is necessary and sufficient for viability. In order to investigate the detailed molecular basis for its activity and regulation, we studied the structure and function of DmCPD1Bs and deduced a plausible hypothesis for its regulation in vivo.
Results and Discussion
Production and proteolytic susceptibility of DmCPD1Bs
Initial heterologous recombinant overexpression trials of DmCPD1Bs in the methylotrophic yeast, Pichia pastoris, rendered sufficient protein for structural studies. However, although we assayed several purification strategies, the presence of several bands in SDS-PAGE could not be avoided. N-terminal sequencing of these bands revealed that they were cleavage products of the target protein occurring after Leu189 that co-purified with the intact enzyme. Subsequent assays with different broad-spectrum inhibitors against the major classes of endopeptidases revealed that addition of phenylmethanesulfonyl fluoride (PMSF) during protein expression significantly limited degradation, giving rise to mainly two major bands (Fig. 1c, left lane). In contrast, inhibitors of metallo-, cysteine, and aspartyl proteinases had no appreciable effect. As a control, intact protein obtained in the presence of PMSF and purified was assayed for susceptibility to proteolysis by commercial trypsin and subtilisin. These experiments revealed similar degradation patterns to those observed during production in the absence of PMSF. In the case of subtilisin, cleavage occurred after Leu189, and for trypsin, cleavage occurred after Arg190 (Fig. 1c, right lane). According to the provisional genome sequence‡ (see Ref. 28), P. pastoris apparently encodes four soluble serine endopeptidases: a putative Kex2 subtilisin-like proprotein convertase, a sequence relative of the mammalian Omi/HtrA2 family of trypsin-like serine proteases, and two putative orthologs of Saccharomyces cerevisiae YscB alias cerevisin (UniProt P09232). The latter enzyme belongs to the subtilisin/kexin family of serine peptidases and preferentially cleaves after arginine, tyrosine, or leucine according to MEROPS database.29 Therefore, either of the cerevisin orthologs in P. pastoris may be responsible for the cleavage after Leu189 observed during recombinant overexpression of DmCPD1Bs.
The two variant bands obtained (Fig. 1c, left lane) corresponded to species of between 52 and 58 kDa and did thus not coincide with the theoretical molecular mass of the protein (47 kDa). Treatment with endoglycosidase F and N-terminal sequencing revealed that they were glycosylation variants of the intact target protein. There are two potential glycosylation sites in the enzyme at Asn133 and Asn269. We tried to reduce heterogeneity by constructing two mutants, N133Q and N269Q. However, the expression yield of both single mutants was dramatically lower than that of the wild type. In addition, both mutants still displayed heterogeneous glycosylation. The double mutant N133Q/N269Q could not be expressed. These results underpinned the relevance of sugars for this protein, in good agreement with the importance of protein glycosylation in the secretory pathway.30
Intact DmCPD1Bs has proteolytic activity
The catalytic efficiency of the purified intact protein against the standard chromogenic type B CP substrate, hippuryl-l-Arg (Table 1 ), was comparable to that of human CPE, human CPM, and human and bovine CPU (also known as activated thrombin-activatable fibrinolysis inhibitor, TAFIa). This is consistent with previous reports on DmCPD1Bs produced in baculovirus, which was also active.5, 6 Reported values for human CPN evince slightly lower activity for this substrate due to its preference for C-terminal lysine residues (Table 1). We further produced and purified human CPB to provide a standard for an A/B-type funnelin for comparison and it had about 10-fold higher efficiency. This is in accordance with previous studies, which showed that “digestive” funnelins are more efficient CPs than “regulatory” forms against small substrates.9, 34 Moreover, intact DmCPD1Bs previously incubated with commercial subtilisin or trypsin (see above) had an ∼ 70% decrease of activity. We conclude that cleavage compromises the proteolytic activity of DmCPD1Bs.
Table 1.
Kinetic constants for hydrolysis of hippuryl-l-Arg by different CPs
| CP | kcat (s− 1) | Km (μM) | kcat/Km (s− 1 mM− 1) |
|---|---|---|---|
| DmCPD1Bs | 38.8 ± 4.1 | 1112 ± 167 | 34.9 ± 8.9 |
| hCPB | 253.7 ± 11.9 | 485.3 ± 62.5 | 522.8 ± 89.8 |
| hTAFIa31 | 73.1 ± 5.7 | 3350 ± 1050 | 21.8 ± 8.6 |
| bTAFIa31 | 219.8 ± 26.7 | 7310 ± 2710 | 30.1 ± 14.8 |
| hCPE32 | 8.5 | 400 | 21.3 |
| hCPN14 | 4 | 650 | 6.2 |
| hCPM33 | 6.1 ± 0.1 | 290 ± 20 | 21 ± 1.0 |
Data are expressed as mean ± SD except for hCPE and hCPN, whose values were only reported as mean.
h, human; b, bovine.
Inhibition assays against DmCPD1B
DmCPD1Bs is inhibited by the small-molecule inhibitor 2-guanidinoethyl-mercaptosuccinic acid (GEMSA) with an apparent inhibition constant (K i) value of 0.67 ± 0.08 μM. This fits in the range reported for other funnelins of B-type specificity, which show K i values in the order of millimolar to nanomolar for this inhibitor.9 In contrast, DmCPD1Bs is not inhibited by protein inhibitors from potato, leech, tick, or mammal, even at micromolar concentrations. These inhibitors display high affinity for A/B-type funnelins, with low nanomolar inhibition constants, but they are inert against N/E-type funnelins.16, 35
Structure of DmCPD1Bs in complex with GEMSA
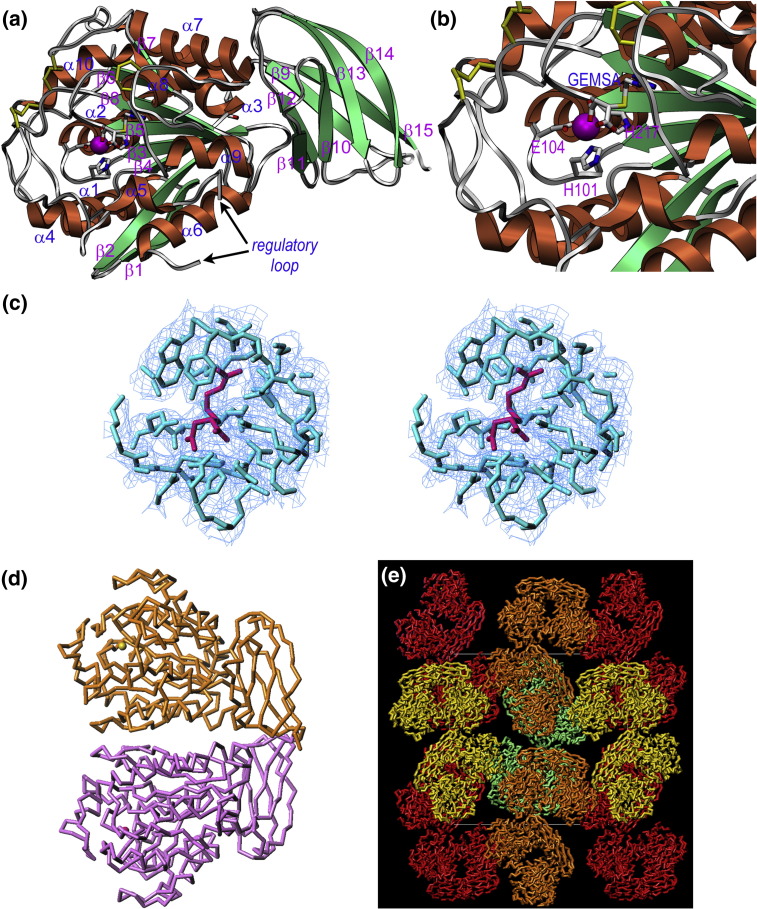
DmCPD1Bs was crystallized in the presence of GEMSA, which significantly improved the crystal diffraction possibly by providing rigidity, as found in the case of TAFI.36 Overall, the structure subdivides into an N-terminal catalytic funnelin domain (Ile28-His336) and a C-terminal TLD (Ile337-Glu420). The catalytic domain (CD) has a compact globular shape, which resembles the volume obtained when a cone is extracted from a sphere (Figs. 2a and b). There is a funnel-like opening to the left, within which the active-site cleft lies, at the base of the opening. The cleft is rather shallow, which is compatible with the capacity of funnelins to cleave a large variety of substrates, as only few contacts between the enzyme and the C-terminal stretch of a substrate are required to fix the latter to the active site. The DmCPD1Bs CD evinces an α8 + 2/β8 topology conforming to an α/β-hydrolase or PLEES fold37, 38 (Fig. 2a and b). It contains a central doubly wound eight-stranded β-sheet (β1–β8), which is strongly twisted and spans the molecule vertically bottom to top. The sheet has a mixed parallel/antiparallel topology and a strand connectivity + 1,+ 2, − 1x, − 2x, − 2, + 1x, − 2, and its core consists of four parallel coplanar central strands (β3–β5 and β8). The catalytic site is located at the C-terminal end of these strands, as observed in other funnelins.16 These four strands are flanked by two parallel strands at the top (β6–β7) and a β-ribbon (β1–β2) at the bottom (Fig. 2a and b). The sheet curvature leads to a concave front side, which shelters helices α5, α6, α8, and α9, as well as the active-site cleft. At the back of the molecule, the convex side of the sheet accommodates the surface N- and C-termini of the molecule, as well as helices α1–α4, α7, α8, and the C-terminal helix α10 of the CD, which runs left to right across the molecule along the convex face of the β-sheet and is bent by ∼ 40°. The funnel-like access to the active site is shaped by irregular segments of disparate length: the loop connecting strand β3 with helix α2 (Lβ3α2), Lβ5β6, Lβ6α7, Lα7β7, and, in particular, the 55-residue segment connecting strand β4 with helix α6 (segment β4α6 hereafter). This element contributes to the lower front of the molecule, shapes the funnel rim, and includes two short helices, α4 and α5. Between five and nine residues are missing in each of the four molecules present in the asymmetric unit within the surface-located Lα5α6 (hereafter referred to as “regulatory loop”) within segment β4α6. Two disulfide bonds also contribute to the funnel rim by cross-linking loop Lα4α5 with Lβ8α10 (Cys156-Cys309) and Lα7β7 with Lβ8α10 (Cys268-Cys308). In addition, a conspicuous disulfide bond between neighboring residues is found within Lβ6α7 (Cys236-Cys237; see Fig. 2a and b).
Fig. 2.
Structure of DmCPD1Bs. (a) Richardson plot of DmCPB1Bs in standard orientation16 showing the CD (left) and the TLD (right). The repetitive secondary-structure elements are shown as green arrows (strands β1–β8 in the CD and β9–β15 in the TLD) and coral ribbons (helices α1–α10) and labeled. The catalytic zinc ion is shown as a magenta sphere. The position of the regulatory loop is pinpointed by arrows. (b) Close-up view of (a) displaying the active-site cleft and the funnel-like border that gives access to it. The zinc-binding residues are presented as sticks and labeled, as is the bound GEMSA molecule. (c) Initial σA-weighted (2mFobs − DFcalc)-type electron density map centered on the GEMSA-binding site. This map was calculated without the inhibitor molecule (omit map) and is shown contoured at 0.75 σ. The final refined model is superimposed (GEMSA, magenta sticks; DmCPD1Bs, cyan sticks). (d) The quaternary structure of DmCPD1Bs in the crystal is a homodimer. (e) The crystal is made up of such dimers, shown in red, orange, yellow, and cyan. Two dimers are found in the crystallographic asymmetric unit. The unit-cell box is shown for reference.
The catalytic zinc ion resides at the bottom of the active-site cleft and is coordinated by the Nδ1 atoms of His101 and His217 and, bidentately, by Glu104 atoms Oɛ2 and Oɛ1 (Fig. 2b). These residues are provided by Lβ3α2 and by the end of strand β5. The protein was co-crystallized in the presence of the small-molecule inhibitor GEMSA (Fig. 2c), which mimics a product complex after substrate cleavage in which the C-terminal residue is still trapped in the S1′ pocket. The carboxymethylene group of the inhibitor binds the catalytic zinc ion asymmetrically through two carboxylate oxygens, thus giving rise to an overall 6-fold metal coordination. The other carboxylate group of GEMSA is anchored to Arg165 Nη2, Asn174 Nδ2, and, bidentately, to Arg175 Nη1 and Nη2. The guanidinoethylmercapto group, which imitates an arginine side chain, occupies the S1′ pocket and is surrounded by Ser224, Gly279, Trp282, Tyr283, Leu285, Gln290, and Thr303. The characteristic specificity for basic residues of N/E-type funnelins is provided in DmCPD1Bs by the side chain of Asp228, which binds one of the terminal guanidine nitrogen atoms of GEMSA. The role of the general acid/base, essential for catalysis in funnelins and other metal-dependent peptidases,16, 39 is fulfilled here by Glu305. The residues that give rise to the characteristic funnelin motif, HXXE + R + NR + H + Y + E,16 are found in DmCPD1Bs at positions 101, 104, 165, 174, 175, 217, 283, and 305, respectively.
The C-terminal TLD spans 84 residues in DmCPD1Bs and is rod-shaped (Fig. 2a). Its N- and C-termini are on opposite sides of the rod, which folds into an all-β seven-stranded β-barrel or β-sandwich, with two layers of three mixed (β1, β4, and β7) and four antiparallel strands (β2, β3, β5, and β6), respectively, which are glued by a hydrophobic core. These strands are arranged as two subsequent Greek-key-like elements related by a 2-fold axis perpendicular to the sandwich surface.21, 40, 41 This domain has topological similarity with transthyretin, a conserved plasma protein that tends to aggregate, thus giving rise to several forms of amyloidosis.42 However, transthyretin contains an eighth C-terminal β-strand that is absent in funnelins.21 The TLD is laterally attached to the CD and interacts with Lβ2β3, Lα6β5, Lα9β8, and Lβ6α7 of the latter through hydrophobic residues. It also interacts through a (bidentate) salt bridge formed by an aspartate (Asp244) at the beginning of helix α7 of the CD, which is conserved among N/E-type funnelins, and an arginine (Arg376) within the TLD. As in the case of related N/E-type funnelins, the function of TLD is uncertain. It may assist in folding, regulation of enzyme activity, or protein–protein interactions.16, 21, 22, 35
Hypothetical quaternary arrangement
Dynamic light-scattering and cross-linking experiments with glutaraldehyde showed that protein concentrations below 1 mg/mL rendered a monomer, whereas concentrations above 8 mg/mL gave rise to a monomer–dimer equilibrium. Size-exclusion chromatography indicated that the protein is monomeric throughout the concentration range assayed (0.5 to 18 mg/mL, as injected into the FPLC system). In the crystals, which resulted from highly concentrated protein solution (18 mg/mL), two pairs of dimers were found in the asymmetric unit (Fig. 2d and e). Inspection of the dimeric interface in the structure reveals that it measures ∼ 950 Å2, which is larger than the range described for experimentally validated “weak” transient homodimeric proteins (740 ± 140 Å2; Ref. 43). Such proteins form both monomers and dimers at physiological concentration. In DmCPD1Bs, the dimer is symmetric, so that the same structural segments of each protomer are involved in complex formation: helix α6, strands β1 and β2, and loops Lα5α6 and Lβ2β3 of each CD and loops Lβ11β12 and Lβ9β10 of each TLD. Two symmetric bidentate salt bridges (Asp353-Arg85) and a total of nine hydrogen bonds are observed at the interface. In addition, the two active-site clefts are located on opposite surfaces of the dimer; that is, there would be no steric hindrance for substrate binding (Fig. 2d). However, there is no concluding evidence that the enzyme is a functional dimer in vivo since the protein seems to be monomeric in solution and the dimer could be an artifact of the crystallization process. Moreover, we do not know whether the formation of a dimer could have functional implications. To date, no higher oligomerization states than a monomer have been reported for funnelins. Only CPN is believed to function in vivo as a tetramer comprising two N/E-type funnelin subunits linked to two non-catalytic regulatory domains,44 but it is not known how these subunits are arranged within the heterotetramer.
Implications for the other splicing variants of the silver gene
The splicing variants of silver give rise to two types of N-terminal repeat (Fig. 1a). These differ in the first 152 (encoded by exon 1B in DmCPD1Bs, svr-PB, and svr-PG) or 150 (encoded by exon 1A in svr-PD, svr-PE, and svr-PH) residues, which share ∼ 39% sequence identity.6 These stretches give rise to the lower half of the CD until the middle of segment β4α6. The second half of the first repeat until the end at Glu420 (numbering hereafter is according to DmCPD1Bs if not otherwise stated) is identical in all proteins. Svr-PC and svr-PI do not possess an ∼ 150-residue N-terminal segment but have a short five-residue tail. As they lack a signal peptide, it is questionable whether they are secreted. They may play an intracellular role instead.
A model of variant svr-PE, which had been shown to be catalytically incompetent by Fricker et al.,5, 6 based on the coordinates of DmCPD1Bs and a structure-based sequence alignment, reveals that the differences in sequence are compatible with maintenance of the overall structure upon minor readjustment of the main chain. Most changes are conservative or are compensated for by differences in size of the side chains in neighboring positions. However, there is a slight trend towards smaller side chains at positions contributing to the central hydrophobic core of the funnelin domain, which might lower thermal stability: Leu78Ala, Tyr96Leu, Ile97Val, Ile80Ala, Met100Ile, Leu111Val, Leu119Ala, Leu128Val, Met143Cys, and Ser151Ala. In addition, the presence of a histidine replacing an asparagine at position 121 would entail a minor rearrangement of the N-terminal stretch up to Phe34. In turn, the conservation of the N-glycoslyation site motif around Asn133 (Asn-Leu-Thr in exon 1A instead of Asn-Ser-Thr in exon 1B) suggests that the asparagine is also attached to a sugar moiety. However, the presence of a glutamine instead of a glycine at position 129 may entail a rotation of the sugar moiety around the linking bond, Nδ2–C1, which would provide space for the glutamine side chain. In addition, only a four-residue insertion is found within Lβ2β3, which should not disturb the interaction of the CD with the TLD (see above) due to the maintenance of leucine residues at positions 89 and 90.
The main difference between the exon-1A- and the exon-1B-encoded fragments is the substitution of glutamine (Gln99 in UniProt entry P42787 isoform 6) for the zinc-binding histidine at position 101 in DmCPD1Bs (Fig. 1b). Although glutamine is a rare zinc ligand in proteins,45, 46 in the case of funnelins, it may lead to metal-containing but catalytically impaired or even inert zinc sites, as reported for zinc-ligand mutants of human glyoxalase I.47 A glutamine at the position equivalent to residue 101 in DmCPD1Bs is also found in a series of (putative) proteins from related Drosophila species, namely, D. ananassae (UniProt B3N0D7), D. willistoni (B4NPC2), D. mojavensis (B4L2T3), D. sechellia (B4IEW7), D. grimshawi (B4JX73), D. erecta (B3NWB3), and D. virilis (B4M2C3). These entries span between 1302 and 1495 residues and could represent three-repeat orthologs of the silver gene product, also encoded by an exon that gives rise to a non-functional funnelin domain. In addition to fruit fly species, a glutamine was found in a potential three-repeat ortholog of the starlet sea anemone Nematostella vectensis (A7S4K6). In all these cases, it remains to be ascertained whether the glutamine-containing funnelin-like domains are functional, in particular taking into account that a recent report has shown that the third CPD domain greatly contributes to survival of transgenic fruit flies.13
The six long constructs all have identical sequences for the putative second and third repeats, the transmembrane helix, and the first 51 residues of the cytosolic domain (Fig. 1a). This means that they all have the second catalytically competent and the third silent repeats. They differ only in the very last segments of the cytosolic domains: svr-PB stops after the common stretch; svr-PC and svr-PD have additional 22 residues; and svr-PG, svr-PH, and svr-PI have a further 55 residues, which are unrelated to the others. These differences allow differential trafficking of the protein between the trans-Golgi network and the cell surface for the six membrane-anchored forms.5, 6
Comparison with duck CPD domain II
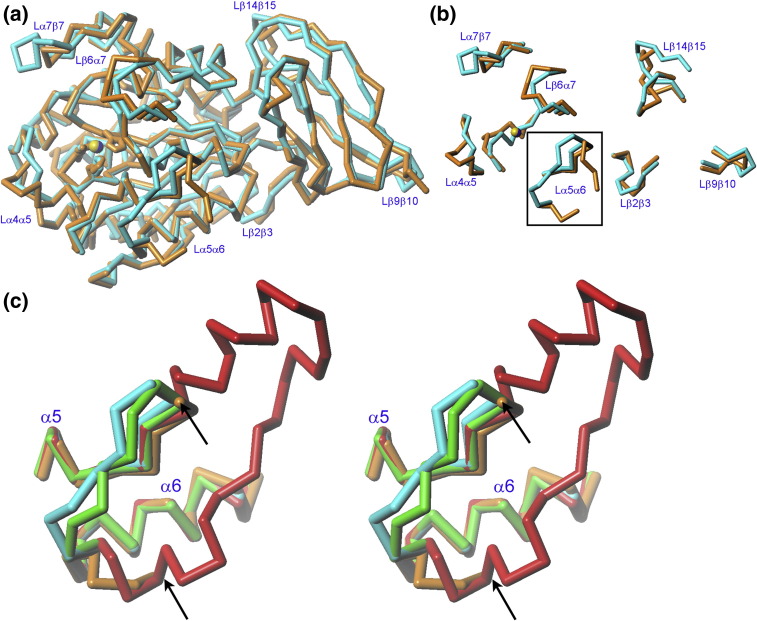
DmCPD1Bs shows 40% sequence identity with the second repeat of the duck ortholog, which is the only other CPD structure reported.6, 21, 48 This sequence similarity redounds to close structural similarity, as reflected by an rmsd of 0.97 Å for the 368 Cα atoms deviating less than 3 Å (see Fig. 3a). Significant deviations are found, however, within Lβ2β3, Lβ6α7, Lα7β7, and the regulatory loop, Lα5α6 (Fig. 3b). These loops contribute to shaping the funnel rim that allows access to the active site (see above). This rim is responsible for interaction with large protein substrates in funnelins.16 In addition, significant differences are found in two loops of the TLD: Lβ9β10 and, in particular, Lβ14β15, which is on the back surface of the molecule. This region might be important for function in N/E-type funnelin TLDs (see above) and structural differences might lead to disparate binding partners in duck CPD domain II and DmCPD1Bs. Overall, the most noteworthy difference is found in the regulatory loop, which is partially undefined in DmCPD1Bs (see above). As the protein found in the crystals is intact, this flexibility does not arise from cleavage but from intrinsic disorder, which, in turn, might have functional implications (see below).
Fig. 3.
Structural comparison of DmCPD1Bs. (a) Superimposition of the Cα traces of DmCPD1Bs (orange; yellow zinc ion) and duck CDP domain II (PDB 1QMU; cyan; blue zinc ion). (b) Detail of (a) showing only those loop segments that evince major conformational differences between the two structures. The regulatory loop (Lα5α6) is framed. (c) Superimposition of the Cα traces of DmCPD1Bs (orange), duck CDP domain II (PDB 1QMU; cyan21, 48), human CPM (PDB 1UWY; green40), and human CPN (PDB 2NSM; red41) corresponding to segments equivalent to α5, Lα5α6, and α6 of the fruit fly enzyme. Orientation as in (a). The arrows point to the residues that flank the flexible region of the regulatory loop in DmCPD1Bs: up to nine residues are missing in each of the protomers.
Proposed mechanisms of regulation
Funnelins of A/B type are secreted as zymogens with an N-terminal 90- to 95-residue pro-domain in charge of latency maintenance until the environmental and temporal conditions for activity are given. This ensures that no undesired proteolytic activity occurs in the transit between the locus of biosynthesis and the final destination.9, 16, 49 In contrast, N/E-type funnelins are not secreted as latent zymogens, and the regulation of the catalytically active forms is believed to rely on their localization: most of these enzymes are bound to membrane or to the extracellular matrix (e.g., long CPD forms, CPM, and CPZ). However, soluble variants such as DmCPD1Bs may be regulated by other mechanisms.
CPD is palmitoylated at cysteine residues of its transmembrane domain in the duck, with implications for its localization and biological function.50 Covalent attachment of fatty acids enhances the hydrophobicity of proteins and contributes to their membrane association, thus providing a means of regulation through localization. The structure of DmCPD1Bs has several exposed cysteine residues at the protein surface shaping the funnel rim, which could be potentially palmitoylated. In particular, adjacent residues Cys136 and Cys137, which were disulfide-linked in the crystal structure (see above), could be present as free thiols in physiological conditions and become palmitoylated. This could be a way to anchor DmCPD1Bs to membranes and thus regulate its intracellular trafficking.
Another, more likely mechanism of regulation could be limited proteolysis within the regulatory loop. Intrinsically flexible regions on the molecular surface of a protein directly correlate with instability, conformational changes and lability, and proteolytic susceptibility,51 in particular in a protease-rich medium such as that of the secretory pathway. This would explain why the regulatory loop is a hot spot for cleavage by host proteases during recombinant heterologous overexpression (see above). The equivalent regions of all other N/E-type metallocarboxypeptidases thus far structurally analyzed, that is, duck CPD domain II, human CPN, and human CPM, are well ordered and rigid, and they significantly deviate from the conformation found in DmCPD1Bs, thus pointing to potential differences in the function of this loop (Fig. 3c). Single cleavage at Leu189 or Arg190 was found to inactivate the protein, possibly due to disruption of the regulatory loop itself and the preceding structural elements α5 and Lα4α5. Therefore, a similar cleavage in vivo by peptidases along the secretory pathway would provide a means of selective regulation of DmCPD1Bs activity.
Conclusions
In contrast to mammals, the fruit fly has only two regulatory funnelins/M14B metallopeptidases, one CPD and one CPM ortholog, which is consistent with the fact that Drosophila contains fewer genes. Hence, the presence of eight splicing variants of CPD, which give rise to both single-repeat soluble and two- and three-repeat membrane-anchored forms, may be conceived as a strategy to compensate for this shortage.5 This is reminiscent of the finding that only two matrix metalloproteinase paralogs are found in the fruit fly, while 23 have been described in humans.52 Of the five active N/E-type funnelins in mammals, CPN is secreted into plasma and forms a dimer of heterodimers. CPM is membrane-anchored and functions extracellularly. CPZ is partially targeted to the extracellular matrix and has a frizzled-like cysteine-rich domain that may make it a Wnt-binding protein.22 DmCPD1Bs could have a homologous function to CPE, which is likewise a single-repeat funnelin and is found in secretory vesicles.22 The fruit fly enzyme is active and cleaves synthetic substrates with similar efficiency to human CPE, although the former has a different pH optimum.6 DmCPD1Bs could function as a CPE-like enzyme in mildly acidic regions of the secretory pathway such as the trans-Golgi network. However, further studies are required to validate this hypothesis. Finally, the structure of DmCPD1Bs conforms to the overall fold of N/E-type funnelins, as may be the case for CPE, but evinces two unique features that may correlate with its regulation: (i) two contiguous surface-located cysteines that may become palmitoylated, thus targeting the enzyme to membranes, and (ii) a surface hot spot for proteolytic inactivation. In this way, DmCPD1Bs could participate in Drosophila development through proteolytic processing of substrates previously targeted by furin-like proprotein convertases and would itself be inactivated at later stages by limited proteolysis.
Materials and Methods
Protein expression and purification
A clone encoding protein svr-PB of Drosophila was kindly provided by Dr. Lloyd D. Fricker (Albert Einstein College of Medicine, New York) and was used as template to generate a PCR-construct encoding DmCPD1Bs without the signal peptide, that is, encompassing amino acids Tyr26 to Phe435 (UniProt P42787, isoform 7). This construct was cloned into the pPICZαA vector with a C-terminal His6-tag, and transformed into P. pastoris strain KM71H following the manufacturer's instructions (EasySelect Pichia Expression Kit, Invitrogen). Selected transformant colonies were used to inoculate 1-L shake-flask cultures, which were grown at 28 °C for 36 h in buffered glycerol-complex medium until OD600 (optical density at 600 nm) reached 20–30. Cells were collected by centrifugation at 3000g, gently resuspended in 100 mL fresh BMMY medium (buffered glycerol-complex medium supplemented with 1% of methanol instead of 1% of glycerol), and cultured at 28 °C for another 24 h for protein expression. PMSF (0.3 mM) was added to the culture every 3–4 h to prevent cleavage of the recombinant protein. For protein purification, the culture supernatant was equilibrated with 30% ammonium sulfate, bound to a hydrophobic chromatography column (butyl-Toyopearl 650 M, Tosoh Bioscience) connected to an ÄKTA Purifier system (GE Healthcare), and eluted with a decreasing gradient of ammonium sulfate. The eluted sample was subsequently dialyzed against 20 mM Tris–HCl, pH 8.0, and purified by anion-exchange chromatography (TSK-DEAE 5PW, Tosoh Bioscience) by using a linear gradient of 0.4 M ammonium acetate (0 to 30%). DmCPD1Bs was then loaded onto a HiLoad Superdex 75 26/60 column (GE Healthcare) previously equilibrated with 50 mM Tris–HCl and 250 mM NaCl, pH 7.5. Eluted fractions were analyzed by SDS-PAGE, and the purest samples containing the active enzyme were pooled. The protein was buffer-exchanged to 10 mM Tris–HCl and 50 mM NaCl, pH 7.5, and concentrated by using an Amicon Centricon centrifugal device (10-kDa cutoff, Millipore). The typical final yield was ∼ 0.3 mg of protein per liter of growth medium. Western blot using an anti-His monoclonal antibody (GE Healthcare) showed that this protein lacked the C-terminal His6-tag, possibly due to autolysis during production/purification, as described for other funnelins.53, 54 Point mutants N133Q, N269Q, and N133Q/N269Q were obtained by site-directed mutagenesis by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and produced and purified in the same way as the wild-type protein. Human pancreatic procarboxypeptidase B was overexpressed and purified as described for DmCPD1Bs but without addition of PMSF. The enzyme was activated by controlled trypsin digestion as previously described.55
Treatment with endoglycosidase
DmCPD1Bs was denatured in a solution of 0.1% SDS and 50 mM β-mercaptoethanol for 5 min at 100 °C. The protein was then incubated with endoglycosidase F (Sigma) for 3 h at 37 °C in the presence of 0.75% Triton X-100 in 50 mM phosphate buffer, pH 7.5, and further analyzed by Coomassie-stained 10% SDS-PAGE.
Proteolytic activity assays
Activity was assayed by monitoring the rate of hydrolysis of hippuryl-l-Arg (Sigma) at λ = 254 nm on a Cary 400 UV–Vis spectrophotometer (Varian) with 50 mM Tris–HCl and 0.15 M NaCl, pH 7.5, as buffer at 37 °C. Initial turnover rates were determined from the first 5–10% of the time trace of each reaction for substrate concentrations close to the K m value whenever possible. The kinetic parameters, k cat and K m, were calculated from at least six experimental points by direct fit to a Michaelis–Menten curve applying a nonlinear least-squares regression analysis with program GRAFIT. Three independent experiments were performed.
Inhibitory assays
The apparent K i of GEMSA (Calbiochem) against DmCPD1Bs was determined by measuring the inhibition of the hydrolysis of the chromogenic substrate N-(4-methoxyphenylazoformyl)-Arg-OH (Bachem) at λ = 350 nm by considering linear competitive kinetics as previously described.56 Assays were performed with 100 μM of substrate in 50 mM Tris–HCl, pH 7.5. Similar inhibitory experiments were carried out by using the protein CP inhibitors from potato (PCI), leech (LCI), tick (TCI), and human (latexin). These protein inhibitors were produced by heterologous overexpression in Escherichia coli and purified as published elsewhere.57, 58
Proteolytic susceptibility assays
DmCPD1Bs (1.3 μg) was incubated with either 30 ng of bovine trypsin (Sigma) or 70 ng of subtilisin Carlsberg (Sigma) for 30 min at 37 °C in 50 mM Tris–HCl, pH 7.5. Equivalent samples without serine protease were used as controls. Cleavage products were analyzed by Coomassie-stained 10% SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore), and subjected to automated Edman degradation analysis in a Procise 492 protein sequencer (Applied Biosystems) at the in-house proteomics service at the Universitat Autònoma de Barcelona. Digested samples and controls were tested for CP activity as described for the inhibitory assays. Each experiment was performed six times.
Dynamic light-scattering, cross-linking, and size-exclusion chromatography experiments
Dynamic light-scattering measurements were performed at six DmCPD1Bs concentrations (ranging from 0.5 to 18 mg/mL) in 10 mM Tris–HCl and 50 mM NaCl, pH 7.5, in a Zetasizer Nanoseries apparatus (Malvern Instruments). Each sample was measured 30 times and the results were analyzed with the Dispersion Technology Software v5.1. For the cross-linking experiments, DmCPD1Bs was prepared at six protein concentrations as above and treated with 0.8 mM glutaraldehyde for 2 min at room temperature in a final volume of 20 μL. The reaction was thereafter quenched with 12 μL of 50 mM Tris–HCl, pH 7.5. Samples were analyzed by Coomassie-stained 10% SDS-PAGE. Size-exclusion chromatography was carried out for the aforementioned protein concentrations in 10 mM Tris–HCl and 50 mM NaCl, pH 7.5, using a Superdex 200 5/150 GL column (GE Healthcare).
Crystallization and X-ray diffraction data collection
Crystallization assays followed the sitting-drop vapor diffusion method. Reservoir solutions were prepared by a Tecan robot and 100-nL crystallization drops were dispensed on 96 × 2-well MRC plates (Innovadyne) by a Cartesian (Genomic Solutions) or a Phoenix (Art Robbins/Rigaku) nanodrop robot at the High-Throughput Crystallography Platform (PAC) at the Barcelona Science Park. The best crystals appeared in a Bruker steady-temperature crystal farm at 4 °C with protein solution (18 mg/mL in 10 mM Tris–HCl and 50 mM NaCl, pH 7.5) and 8% polyethylene glycol 4000, 0.2 M KSCN, and 0.1 M sodium cacodylate, pH 6.5, as reservoir solution. These conditions were efficiently scaled up to the microliter range with 24-well Cryschem crystallization dishes (Hampton Research). Complex crystals with GEMSA were obtained by soaking (10 mM inhibitor in reservoir solution) for 5 days. Crystals were cryo-protected with 16.5% polyethylene glycol 4000, 15% glycerol, 0.2 M KSCN, and 0.1 M sodium cacodylate, pH 6.5. A complete diffraction data set at 2.7 Å resolution was collected at 100 K (Oxford Cryosystems 700 series cryostream) from a single liquid-N2 flash-cryo-cooled GEMSA-complexed crystal on an ADSC Q315R CCD detector at beam line ID23-1 of the European Synchrotron Radiation Facility (ESRF, Grenoble, France) within the Block Allocation Group “BAG Barcelona”. The crystal was primitive orthorhombic, with four molecules per asymmetric unit. Diffraction data were integrated, scaled, merged, and reduced with programs XDS59 and SCALA60 within the CCP4 suite of programs (see Table 2 ). The presence of pseudomerohedral twinning following twin law “− h,l,k” was identified by using XTRIAGE within the PHENIX suite of programs.63
Table 2.
Crystallographic data
| Space group | P212121 |
| Cell constants: a, b, c (Å) | 97.0, 135.7, 141.7 |
| Wavelength (Å) | 0.9724 |
| No. of measurements/unique reflections | 419,491/52,035 |
| Resolution range (Å) (outermost shell) | 49.0–2.70 (2.85–2.70) |
| Completeness (%) | 99.9 (100.0) |
| Rmergea | 0.099 (0.590) |
| Rr.i.m. (= Rmeas)a/Rp.i.m.a | 0.106 (0.629)/0.037 (0.217) |
| Average intensity over SD (〈[〈I〉/σ(〈I〉)]〉) | 22.5 (4.6) |
| B-factor (Wilson) (Å2)/average multiplicity | 54.0/8.1 (8.2) |
| Resolution range used for refinement (Å) | 8–2.70 |
| No. of reflections in working set/in test set | 51,270/765 |
| Crystallographic R-factor (Rfree)a | 0.217 (0.283) |
| No. of protein atoms/solvent molecules/ions/ligands | 12,113/211/4 Zn2+/4 glycerol, 4 N-acetyl-glucosamine, 4 GEMSA |
| rmsd from target values | |
| Bonds (Å)/angles (°) | 0.006/1.028 |
| Bonded B-factors (Å2) | 8.49 |
| Average B-factors for protein atoms (Å2) | 55.2 |
| Main-chain conformational angle analysisb | |
| Residues in favored regions/outliers/all residues | 1460/4/1523 |
Structure solution and refinement
The structure of DmCPD1Bs was solved by Patterson-search methods with program Phaser64 by using the coordinates of duck CPD domain II [Protein Data Bank (PDB) accession code 1H8L 21, 48] as searching model. Subsequently, manual model building on a Silicon Graphics workstation with program Turbo-Frodo65 alternated with crystallographic refinement (including TLS refinement) with program REFMAC5 within the CCP4 suite at initial stages and with program suite PHENIX under consideration of twinning (refined to a fraction of α = 0.079) at the final stages until the model was completed (see Table 2). This model contained protein residues Lys29-Val419 (numbering according Fig. 1b) for chain A, Ile28-Glu420 for chain B, Glu30-Val419 for chain C, and Thr27-Glu420 for chain D plus a zinc ion, an N-acetyl-glucosamine sugar moiety attached to Asn133, and a GEMSA molecule for each protomer. Although there was evidence that Asn270 was also glycosylated, the electron density maps were too weak to enable reliable modeling. Segments 183–190, 185–191, 184–188, and 182–190 were disordered in each of the four chains, respectively. SDS-PAGE analysis of washed crystals revealed only intact protein.
Miscellaneous
Figures were prepared with programs SETOR66 and Turbo-Frodo. Interface analysis was performed with the PISA server§. Model validation was performed with MolProbity62 and the WHATCHECK routine of program WHAT IF.67 The model of variant svr-PE was constructed with Turbo-Frodo based on the structure of DmCPD1Bs and a sequence alignment calculated with CLUSTAL-W,68 which was modified to include structural restraints. The dimer interface was calculated as half of the total surface area buried by the complex.
Accession code
The final coordinates of DmCPD1Bs are available from the PDB∥ (accession code 3MN8).
Acknowledgements
This study was supported by the following grants from European, Spanish, and Catalan public agencies:FP7-Health-2009-241919-LIVIMODE, FP7-HEALTH-F3-2009-223101 “AntiPathoGN”, FP7-HEALTH-2010-261460 “Gums&Joints”, BIO2007-68046, BIO2008-04080-E, BIO2009-10334, CSD2006-00015, PSE-010000-2009-8, 2009SGR218, 2009SGR1036, and XeRBa biotechnology network. We acknowledge the help provided by European Molecular Biology Laboratory and ESRF synchrotron local contacts. Funding for data collection was provided by ESRF. We thank Robin Rycroft for helpful suggestions to the manuscript and Esther Vázquez for help with DLS measurements.
Edited by R. Huber
Footnotes
Contributor Information
Francesc X. Avilés, Email: FrancescXavier.Aviles@uab.es.
F. Xavier Gomis-Rüth, Email: fxgr@ibmb.csic.es.
References
- 1.Morgan T.H., Bridges C.B., Sturtevant A.H. The genetics of Drosophila. Bibliogr. Genet. 1925;2:1–262. [Google Scholar]
- 2.Lindsley D.L., Zimm G.G. Academic Press, Inc.; San Diego, CA: 1992. The Genome of Drosophila melanogaster. [Google Scholar]
- 3.Settle S.H., Jr, Green M.M., Burtis K.C. The silver gene of Drosophila melanogaster encodes multiple carboxypeptidases similar to mammalian prohormone-processing enzymes. Proc. Natl Acad. Sci. USA. 1995;92:9470–9474. doi: 10.1073/pnas.92.21.9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tweedie S., Ashburner M., Falls K., Leyland P., McQuilton P., Marygold S. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sidyelyeva G., Baker N.E., Fricker L.D. Characterization of the molecular basis of the Drosophila mutations in carboxypeptidase D. Effect on enzyme activity and expression. J. Biol. Chem. 2006;281:13844–13852. doi: 10.1074/jbc.M513499200. [DOI] [PubMed] [Google Scholar]
- 6.Sidyelyeva G., Fricker L.D. Characterization of Drosophila carboxypeptidase D. J. Biol. Chem. 2002;277:49613–49620. doi: 10.1074/jbc.M209652200. [DOI] [PubMed] [Google Scholar]
- 7.Song L., Fricker L.D. Purification and characterization of carboxypeptidase D, a novel carboxypeptidase E-like enzyme, from bovine pituitary. J. Biol. Chem. 1995;270:25007–25013. doi: 10.1074/jbc.270.42.25007. [DOI] [PubMed] [Google Scholar]
- 8.Song L., Fricker L.D. Tissue distribution and characterization of soluble and membrane-bound forms of metallocarboxypeptidase D. J. Biol. Chem. 1996;271:28884–28889. doi: 10.1074/jbc.271.46.28884. [DOI] [PubMed] [Google Scholar]
- 9.Arolas J.L., Vendrell J., Aviles F.X., Fricker L.D. Metallocarboxypeptidases: emerging drug targets in biomedicine. Curr. Pharm. Des. 2007;13:349–366. doi: 10.2174/138161207780162980. [DOI] [PubMed] [Google Scholar]
- 10.Fricker L.D. Carboxypeptidases D and E. In: Dalbey R.E., Sigman D.S., editors. vol. 23. Academic Press, Inc.; London, UK: 2002. pp. 421–452. (The Enzymes. Co- and Posttranslational Proteolysis of Proteins). [Google Scholar]
- 11.Skidgel R.A., Erdös E.G. Cellular carboxypeptidases. Immunol. Rev. 1998;161:129–141. doi: 10.1111/j.1600-065x.1998.tb01577.x. [DOI] [PubMed] [Google Scholar]
- 12.Fricker L.D. 251. Metallocarboxypeptidase D. In: Barrett A.J., Rawlings N.D., Woessner J.F., editors. 2nd edit. vol. 1. Elsevier; London, UK: 2004. pp. 848–851. (Handbook of Proteolytic Enzymes). 2 vols. [Google Scholar]
- 13.Sidyelyeva G., Wegener C., Schoenfeld B.P., Bell A.J., Baker N.E., McBride S.M., Fricker L.D. Individual carboxypeptidase D domains have both redundant and unique functions in Drosophila development and behavior. Cell. Mol. Life Sci. 2010 doi: 10.1007/s00018-010-0369-8. In press. doi:10.1007/s00018-010-0369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdelmagid S.A., Too C.K. Prolactin and estrogen up-regulate carboxypeptidase-D to promote nitric oxide production and survival of mcf-7 breast cancer cells. Endocrinology. 2008;149:4821–4828. doi: 10.1210/en.2008-0145. [DOI] [PubMed] [Google Scholar]
- 15.Hadkar V., Skidgel R.A. Carboxypeptidase D is up-regulated in raw 264.7 macrophages and stimulates nitric oxide synthesis by cells in arginine-free medium. Mol. Pharmacol. 2001;59:1324–1332. doi: 10.1124/mol.59.5.1324. [DOI] [PubMed] [Google Scholar]
- 16.Gomis-Rüth F.X. Structure and mechanism of metallocarboxypeptidases. Crit. Rev. Biochem. Mol. Biol. 2008;43:319–345. doi: 10.1080/10409230802376375. [DOI] [PubMed] [Google Scholar]
- 17.Rawlings N.D., Barrett A.J., Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skidgel R.A. Basic carboxypeptidases: regulators of peptide hormone activity. Trends Pharmacol. Sci. 1988;9:299–304. doi: 10.1016/0165-6147(88)90015-6. [DOI] [PubMed] [Google Scholar]
- 19.Skidgel R.A. Structure and function of mammalian zinc carboxypeptidases. In: Hooper N.M., editor. Zinc Metalloproteases in Health and Disease. Taylor and Francis Ltd.; London, UK: 1996. pp. 241–283. [Google Scholar]
- 20.Skidgel R.A. 252. Carboxypeptidase M. In: Barrett A.J., Rawlings N.D., Woessner J.F., editors. 2nd edit. vol. 1. Elsevier; London, UK: 2004. pp. 851–854. (Handbook of Proteolytic Enzymes). 2 vols. [Google Scholar]
- 21.Gomis-Rüth F.X., Companys V., Qian Y., Fricker L.D., Vendrell J., Avilés F.X., Coll M. Crystal structure of avian carboxypeptidase D domain II: a prototype for the regulatory metallocarboxypeptidase subfamily. EMBO J. 1999;18:5817–5826. doi: 10.1093/emboj/18.21.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reznik S.E., Fricker L.D. Carboxypeptidases from A to Z: implications in embryonic development and Wnt binding. Cell. Mol. Life Sci. 2001;58:1790–1804. doi: 10.1007/PL00000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroki K., Cheung R., Marion P.L., Ganem D. A cell surface protein that binds avian hepatitis B virus particles. J. Virol. 1994;68:2091–2096. doi: 10.1128/jvi.68.4.2091-2096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eng F.J., Novikova E.G., Kuroki K., Ganem D., Fricker L.D. gp180, a protein that binds duck hepatitis B virus particles, has metallocarboxypeptidase D-like enzymatic activity. J. Biol. Chem. 1998;273:8382–8388. doi: 10.1074/jbc.273.14.8382. [DOI] [PubMed] [Google Scholar]
- 25.Gomis-Rüth F.X. Catalytic domain architecture of metzincin metalloproteases. J. Biol. Chem. 2009:15353–15357. doi: 10.1074/jbc.R800069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 27.Wright T.R.F. The genetics of biogenic amine metabolism, sclerotization, and melanization in Drosophila melanogaster. Adv. Genet. 1987;24:127–222. [PubMed] [Google Scholar]
- 28.De Schutter K., Lin Y.C., Tiels P., Van Hecke A., Glinka S., Weber-Lehmann J. Genome sequence of the recombinant protein production host Pichia pastoris. Nat. Biotechnol. 2009;27:561–566. doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- 29.Rawlings N.D., Morton F.R., Kok C.Y., Kong J., Barrett A.J. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36:D320–D325. doi: 10.1093/nar/gkm954. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ungar D. Golgi linked protein glycosylation and associated diseases. Semin. Cell Dev. Biol. 2009;20:762–769. doi: 10.1016/j.semcdb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Valnickova Z., Thaysen-Andersen M., Hojrup P., Christensen T., Sanggaard K.W., Kristensen T., Enghild J.J. Biochemical characterization of bovine plasma thrombin-activatable fibrinolysis inhibitor (TAFI) BMC Biochem. 2009;10:13. doi: 10.1186/1471-2091-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fricker L.D., Devi L. Comparison of a spectrophotometric, a fluorometric, and a novel radiometric assay for carboxypeptidase E (EC 3.4.17.10) and other carboxypeptidase B-like enzymes. Anal. Biochem. 1990;184:21–27. doi: 10.1016/0003-2697(90)90005-t. [DOI] [PubMed] [Google Scholar]
- 33.Deiteren K., Surpateanu G., Gilany K., Willemse J.L., Hendriks D.F., Augustyns K. The role of the S1 binding site of carboxypeptidase M in substrate specificity and turn-over. Biochim. Biophys. Acta. 2007;1774:267–277. doi: 10.1016/j.bbapap.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 34.McKay T.J., Phelan A.W., Plummer T.H., Jr Comparative studies on human carboxypeptidases B and N. Arch. Biochem. Biophys. 1979;197:487–492. doi: 10.1016/0003-9861(79)90271-6. [DOI] [PubMed] [Google Scholar]
- 35.Arolas J.L., Lorenzo J., Rovira A., Castella J., Aviles F.X., Sommerhoff C.P. A carboxypeptidase inhibitor from the tick Rhipicephalus bursa: isolation, cDNA cloning, recombinant expression, and characterization. J. Biol. Chem. 2005;280:3441–3448. doi: 10.1074/jbc.M411086200. [DOI] [PubMed] [Google Scholar]
- 36.Marx P.F., Brondijk T.H., Plug T., Romijn R.A., Hemrika W., Meijers J.C., Huizinga E.G. Crystal structures of TAFI elucidate the inactivation mechanism of activated TAFI: a novel mechanism for enzyme autoregulation. Blood. 2008;112:2803–2809. doi: 10.1182/blood-2008-03-146001. [DOI] [PubMed] [Google Scholar]
- 37.Ollis D.L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S.M. The α/β hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 38.Puente X.S., López-Otín C. The PLEES proteins—a family of structurally related enzymes widely distributed from bacteria to humans. Biochem. J. 1997;332:947–949. doi: 10.1042/bj3220947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bayés A., Fernández D., Solà M., Marrero A., García-Piqué S., Avilés F.X. Caught after the act: a human A-type metallocarboxypeptidase in a product complex with a cleaved hexapeptide. Biochemistry. 2007;46:6921–6930. doi: 10.1021/bi700480b. [DOI] [PubMed] [Google Scholar]
- 40.Reverter D., Maskos K., Tan F., Skidgel R.A., Bode W. Crystal structure of human carboxypeptidase M, a membrane-bound enzyme that regulates peptide hormone activity. J. Mol. Biol. 2004;338:257–269. doi: 10.1016/j.jmb.2004.02.058. [DOI] [PubMed] [Google Scholar]
- 41.Keil C., Maskos K., Than M., Hoopes J.T., Huber R., Tan F. Crystal structure of the human carboxypeptidase N (kininase I) catalytic domain. J. Mol. Biol. 2007;366:504–516. doi: 10.1016/j.jmb.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 42.Prapunpoj P., Leelawatwattana L. Evolutionary changes to transthyretin: structure–function relationships. FEBS J. 2009;276:5330–5341. doi: 10.1111/j.1742-4658.2009.07243.x. [DOI] [PubMed] [Google Scholar]
- 43.Nooren I.M.A., Thornton J.M. Structural characterisation and functional significance of transient protein–protein interactions. J. Mol. Biol. 2003;325:991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 44.Skidgel R.A., Erdös E.G. Structure and function of human plasma carboxypeptidase N, the anaphylatoxin inactivator. Int. Immunopharmacol. 2007;7:1888–1899. doi: 10.1016/j.intimp.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCall K.A., Huang C., Fierke C.A. Function and mechanism of zinc metalloenzymes. J. Nutr. 2000;130:1437S–1446S. doi: 10.1093/jn/130.5.1437S. [DOI] [PubMed] [Google Scholar]
- 46.Auld D.S. Zinc coordination sphere in biochemical zinc sites. BioMetals. 2001;14:271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- 47.Ridderstrom M., Cameron A.D., Jones T.A., Mannervik B. Involvement of an active-site Zn2+ ligand in the catalytic mechanism of human glyoxalase I. J. Biol. Chem. 1998;273:21623–21628. doi: 10.1074/jbc.273.34.21623. [DOI] [PubMed] [Google Scholar]
- 48.Aloy P., Companys V., Vendrell J., Avilés F.X., Fricker L.D., Coll M., Gomis-Rüth F.X. The crystal structure of the inhibitor-complexed carboxypeptidase D domain II and the modeling of regulatory carboxypeptidases. J. Biol. Chem. 2001;276:16177–16184. doi: 10.1074/jbc.M011457200. [DOI] [PubMed] [Google Scholar]
- 49.Avilés F.X., Vendrell J., Guasch A., Coll M., Huber R. Advances in metallo-procarboxypeptidases. Emerging details on the inhibition mechanism and on the activation process. Eur. J. Biochem. 1993;211:381–389. doi: 10.1111/j.1432-1033.1993.tb17561.x. [DOI] [PubMed] [Google Scholar]
- 50.Kalinina E.V., Fricker L.D. Palmitoylation of carboxypeptidase D. Implications for intracellular trafficking. J. Biol. Chem. 2003;278:9244–9249. doi: 10.1074/jbc.m209379200. [DOI] [PubMed] [Google Scholar]
- 51.Zappacosta F., Pessi A., Bianchi E., Venturini S., Sollazzo M., Tramontano A. Probing the tertiary structure of proteins by limited proteolysis and mass spectrometry: the case of Minibody. Protein Sci. 1996;5:802–813. doi: 10.1002/pro.5560050502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Llano E., Adam G., Pendas A.M., Quesada V., Sánchez L.M., Santamaria I. Structural and enzymatic characterization of Drosophila Dm2-MMP, a membrane-bound matrix metalloproteinase with tissue-specific expression. J. Biol. Chem. 2002;277:23321–23329. doi: 10.1074/jbc.M200121200. [DOI] [PubMed] [Google Scholar]
- 53.Lyons P.J., Callaway M.B., Fricker L.D. Characterization of carboxypeptidase A6, an extracellular matrix peptidase. J. Biol. Chem. 2008;283:7054–7063. doi: 10.1074/jbc.M707680200. [DOI] [PubMed] [Google Scholar]
- 54.Lin Q., Yu N.J., Spremulli L.L. Expression and functional analysis of Euglena gracilis chloroplast initiation factor 3. Plant Mol. Biol. 1996;32:937–945. doi: 10.1007/BF00020490. [DOI] [PubMed] [Google Scholar]
- 55.Barbosa Pereira P.J., Segura-Martin S., Oliva B., Ferrer-Orta C., Avilés F.X., Coll M. Human procarboxypeptidase B: three-dimensional structure and implications for thrombin-activatable fibrinolysis inhibitor (TAFI) J. Mol. Biol. 2002;321:537–547. doi: 10.1016/s0022-2836(02)00648-4. [DOI] [PubMed] [Google Scholar]
- 56.Fricker L.D., Snyder S.H. Purification and characterization of enkephalin convertase, an enkephalin-synthesizing carboxypeptidase. J. Biol. Chem. 1983;258:10950–10955. [PubMed] [Google Scholar]
- 57.Pallarès I., Bonet R., García-Castellanos R., Ventura S., Avilés F.X., Vendrell J., Gomis-Rüth F.X. Structure of human carboxypeptidase A4 with its endogenous protein inhibitor, latexin. Proc. Natl Acad. Sci. USA. 2005;102:3978–3983. doi: 10.1073/pnas.0500678102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanglas L., Aviles F.X., Huber R., Gomis-Ruth F.X., Arolas J.L. Mammalian metallopeptidase inhibition at the defense barrier of Ascaris parasite. Proc. Natl Acad. Sci. USA. 2009;106:1743–1747. doi: 10.1073/pnas.0812623106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kabsch W. Chapter 25.2.9: XDS. In: Rossmann M.G., Arnold E., editors. International Tables for Crystallography. Volume F: Crystallography of Biological Macromolecules. 1st edit. Kluwer Academic Publishers (for The International Union of Crystallography); Dordrecht, The Netherlands: 2001. pp. 730–734. [Google Scholar]
- 60.Evans P. Scaling and assessment of data quality. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 61.Mallorquí-Fernández N., Manandhar S.P., Mallorquí-Fernández G., Usón I., Wawrzonek K., Kantyka T. A new autocatalytic activation mechanism for cysteine proteases revealed by Prevotella intermedia interpain A. J. Biol. Chem. 2008;283:2871–2882. doi: 10.1074/jbc.M708481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 64.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carranza C., Inisan A.G., Mouthuy-Knoops E., Cambillau C., Roussel A. AFMB Activity Report 1996–1999. CNRS-UPR 9039; Marseille, France: 1999. Turbo-Frodo; pp. 89–90. [Google Scholar]
- 66.Evans S.V. SETOR: hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 67.Vriend G. WHAT IF: a molecular modelling and drug design program. J. Mol. Graphics. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 68.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]