Graphical abstract
Keywords: Cryptosporidium parvum, Outbreak, Dairy cattle, Diarrhea, Subtype
Highlights
-
•
An outbreak of severe diarrhea was caused by Cryptosporidium parvum IIdA19G1 in dairy calves.
-
•
Concurrence of rotavirus was present, but not as a significant cause of diarrhea in the investigation.
-
•
Cryptosporidium parvum infection was associated with the occurrence of watery diarrhea in calves.
-
•
Cryptosporidium ryanae and Cryptosporidium bovis infections were associated with the occurrence of moderate diarrhea.
Abstract
Neonatal diarrhea is one of the most important syndromes in dairy cattle. Among enteropathogens, Cryptosporidium spp. are primary causes of diarrhea, but outbreaks due to cryptosporidiosis are rarely reported in cattle. From January to April in 2016, severe diarrhea was observed in over 400 neonatal dairy calves on a large dairy farm in Jiangsu Province of East China. Approximately 360 calves died due to watery diarrhea despite antibiotic therapy. In this study, 18 fecal specimens were collected from seriously ill calves on this farm during the diarrhea outbreak, and analysed for common enteropathogens by enzymatic immunoassay (EIA). In a post-outbreak investigation, 418 and 1372 specimens collected from animals of various age groups were further analysed for rotavirus and Cryptosporidium spp. by EIA and PCR, respectively, to assess their roles in the occurrence of diarrhea on the farm. Cryptosporidium spp. were genotyped using established techniques. Initial EIA tests showed that 15/18 seriously ill calves during the outbreak were positive for Cryptosporidium parvum, while 8/18 were positive for rotavirus. The overall infection rate of Cryptosporidium in pre-weaned calves on the farm was 22.7%, with odds of the Cryptosporidium infection during the outbreak 4.4–23.5 times higher than after the outbreak. Four Cryptosporidium spp. were identified after the outbreak including C. parvum (n = 79), Cryptosporidium ryanae (n = 48), Cryptosporidium bovis (n = 31), and Cryptosporidium andersoni (n = 3), with co-infections of multiple species being detected in 34 animals. Infection with C. parvum (73/79) was found in the majority of calves aged ≤3 weeks, consistent with the age of ill calves during the outbreak. All C. parvum isolates were identified as subtype IIdA19G1. In the post-outbreak investigation, C. parvum infection was associated with the occurrence of watery diarrhea in pre-weaned calves, C. ryanae infection was associated with moderate diarrhea in both pre- and post-weaned calves, while no association was identified between rotavirus infection and the occurrence of diarrhea. Results of logistic regression analysis further suggested that C. bovis infection might also be a risk factor for moderate diarrhea in calves. Thus, we believe this is the first report of a major outbreak of severe diarrhea caused by C. parvum IIdA19G1 in dairy calves. More attention should be directed toward preventing the dissemination of this virulent subtype in China.
1. Introduction
In dairy cattle, diarrhea is considered one of the most common syndromes in neonates, resulting in high morbidity and mortality during the first 4 weeks of life (Uetake, 2013). Neonatal diarrhea is primarily caused by the four most prevalent enteropathogens in calves including rotavirus, coronavirus, Escherichia coli K99, and Cryptosporidium spp. (Meganck et al., 2014). Mono-infections with one pathogen or co-infections with two or more pathogens are commonly seen (Brar et al., 2017, Cai et al., 2017). Among them, Cryptosporidium spp. and rotavirus are most frequently identified in fecal specimens from neonatal calves all over the world (Cho et al., 2013, Al Mawly et al., 2015, Mohamed et al., 2017).
Cryptosporidium spp. are important protozoan parasites that infect a wide variety of vertebrates including humans and farm animals (Feng et al., 2018). The association of Cryptosporidium infection with bovine diarrhea was first reported in 1978 (Pohlenz et al., 1978). This was followed by direct demonstration of the occurrence of diarrhea in experimentally infected calves (Tzipori et al., 1983). Since then, cryptosporidiosis has been considered one of the most important causes of diarrhea and enteritis in neonatal calves globally (Blanchard, 2012, Cho and Yoon, 2014, Wang et al., 2017). Recently, several cryptosporidiosis outbreaks have been reported in neonatal calves in Australia, Estonia, India and China, resulting in high mortality and substantial economic losses (Izzo et al., 2011, Randhawa et al., 2012, Cui et al., 2014, Brar et al., 2017, Niine et al., 2018).
Four Cryptosporidium spp. including Cryptosporidium parvum, Cryptosporidium bovis, Cryptosporidium ryanae and Cryptosporidium andersoni are responsible for the majority of Cryptosporidium infections in cattle, with C. parvum dominating in pre-weaned calves, C. bovis and C. ryanae in post-weaned calves, and C. andersoni in adult cattle in most areas (Xiao, 2010). Among them, C. parvum accounts for over 90% of Cryptosporidium infections in neonatal calves, and is the only major pathogenic and zoonotic species in dairy cattle (Thomson et al., 2017). The pathogenicity of C. bovis and C. ryanae in post-weaned calves has not been established (Thomson et al., 2017).
Within C. parvum, many subtypes exist based on sequence characterizations of the 60 kDa glycoprotein (gp60) gene. Among them, calves in most industrialised countries are commonly infected with IIa subtypes, especially subtype IIaA15G2R1 (Xiao, 2010). In addition, IId subtypes have been identified in dairy calves in some countries such as Sweden, Turkey, Romania and Egypt (Bjorkman et al., 2015, Vieira et al., 2015, Ibrahim et al., 2016, Taylan-Ozkan et al., 2016). In contrast, pre-weaned dairy calves in China are commonly infected with C. bovis, and when they are infected with C. parvum, almost exclusively with IId subtypes, especially IIdA15G1 and IIdA19G1 (Feng and Xiao, 2017)
In 2016, a diarrhea outbreak was identified in neonatal dairy calves on a large dairy farm in Jiangsu Province, China. Approximately 60% of neonatal calves on this farm died due to severe watery diarrhea. This study aims to explore causes of the diarrhea outbreak, as well as the pathogenicity of the three most common Cryptosporidium spp. in dairy calves.
2. Materials and methods
2.1. Ethics statement
Fecal specimens from dairy cattle in this study were collected with the permission of the farm manager, and the cattle were handled in accordance with the Animal Ethics Procedures and Guidelines of the People’s Republic of China. The research protocol was reviewed and approved by the Ethics Committee of the South China Agricultural University.
2.2. Animals and specimens
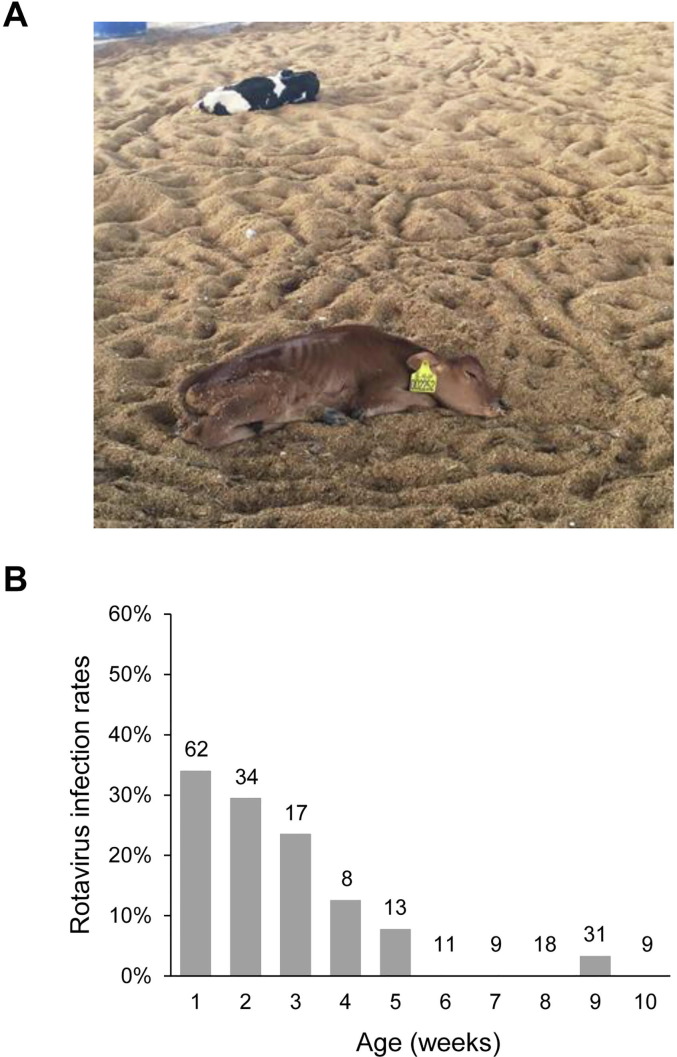
This study took place on a large dairy farm in Jiangsu Province in East China. There were approximately 5500 dairy cattle including ∼600 neonatal calves on this farm. The majority of animals on the farm were of the Holstein breed, with fewer than 50 of the Jersey breed. From January to April in 2016, a diarrhea outbreak in neonatal dairy calves was noticed on the study farm, with watery diarrhea occurring in over 400 pre-weaned calves (Fig. 1 A). Approximately 360 calves died at 10–20 days of age despite being treated with broad-spectrum antibiotics. To identify the cause of the outbreak, fecal specimens were collected from 18 seriously ill calves during the outbreak in April 2016 (Table 1 ).
Fig. 1.
A diarrhea outbreak in pre-weaned dairy calves on a farm in Jiangsu, China. (A) Two seriously ill calves during the outbreak. (B) Rotavirus infection rates by age in weeks after the outbreak.
Table 1.
Detection of common enteric pathogens in fecal specimens from 18 seriously ill calves during a diarrhea outbreak on a farm in Jiangsu, China using an enzymatic immunoassay (EIA) test and PCR.
| Specimen | Age (weeks) | EIA test |
Cryptosporidium PCR at the ssrRNA locus | Cryptosporidium sp. | C. parvum subtype | |||
|---|---|---|---|---|---|---|---|---|
| Rotavirus | Coronavirus | Escherichia coli K99 | Cryptosporidium | |||||
| 23151 | 2 | + | − | − | − | − | − | − |
| 23152 | 2 | + | − | − | − | − | − | − |
| 23153 | 2 | − | − | − | + | + | C. parvum | IIdA19G1 |
| 23154 | 2 | + | − | − | − | − | − | − |
| 23155 | 2 | + | − | − | + | + | C. parvum | IIdA19G1 |
| 23156 | 2 | + | − | − | + | + | C. parvum | IIdA19G1 |
| 23157 | 2 | + | − | − | + | + | C. parvum | IIdA19G1 |
| 23158 | 2 | − | − | − | + | + | C. parvum | IIdA19G1 |
| 23159 | 2 | + | − | − | + | + | C. parvum | IIdA19G1 |
| 23160 | 2 | + | − | − | + | + | C. parvum | IIdA19G1 |
| 23161 | 2 | − | − | − | + | + | C. parvum | IIdA19G1 |
| 23162 | 2 | − | − | − | + | + | C. parvum | IIdA19G1 |
| 23163 | 2 | − | − | − | + | + | C. parvum | IIdA19G1 |
| 23164 | 2 | − | − | − | + | + | C. parvum | IIdA19G1 |
| 23165 | 2 | − | − | − | − | + | C. parvum | IIdA19G1 |
| 23166 | 2 | − | − | − | + | + | C. parvum | IIdA19G1 |
| 23167 | 2 | − | − | − | + | + | C. parvum | IIdA19G1 |
| 23168 | 1 | − | − | − | − | + | C. parvum | − |
+, positive; −, negative.
To further investigate the transmission and pathogenicity of Cryptosporidium spp. after the outbreak, 1372 fecal specimens were collected from pre-weaned (<3 months), post-weaned (3–12 months), and adult (>24 months) dairy cattle on the farm in May 2016 (n = 356), August 2016 (n = 512), June 2017 (n = 304), and November 2017 (n = 200) (Table 2 ). Fecal specimens were divided into three groups: formed feces from animals with no diarrhea (n = 1126), loose feces from animals with moderate diarrhea (n = 149), and liquid feces from animals with watery diarrhea (n = 97). Feces were collected directly from the rectum of each cattle, using disposable gloves, into 50 mL centrifuge tubes. Approximately 1 g of each fecal specimen was transferred into a 1.5 mL tube and kept frozen at −80 °C, while the remaining feces were stored in 2.5% potassium dichromate at 4 °C.
Table 2.
Association between Cryptosporidium infection and occurrence of diarrhea in pre-weaned, post-weaned, and adult dairy cattle after a diarrhea outbreak on a farm in Jiangsu, China.
| Age | Animal group | Sample size | No. positive for Cryptosporidium (%) |
Cryptosporidium spp. |
|||||
|---|---|---|---|---|---|---|---|---|---|
| C. parvum | C. ryanae | C. bovis | C. andersoni | C. ryanae + C. bovis | C. parvum + C. bovis | ||||
| Pre-weaned | No diarrhea | 450 | 83 (18.4) | 46 | 14 | 15 | 0 | 6 | 2 |
| Moderate diarrhea | 101 | 35 (34.7) | 11 | 9 | 7 | 0 | 5 | 3 | |
| Watery diarrhea | 61 | 21 (34.4) | 16 | 4 | 0 | 0 | 1 | 0 | |
| Subtotal | 612 | 139 (22.7) | 73 | 27 | 22 | 0 | 12 | 5 | |
| Post-weaned | No diarrhea | 439 | 29 (6.6) | 2 | 10 | 5 | 2 | 7 | 3 |
| Moderate diarrhea | 46 | 9 (19.6) | 0 | 4 | 2 | 0 | 3 | 0 | |
| Watery diarrhea | 29 | 5 (17.2) | 0 | 3 | 2 | 0 | 0 | 0 | |
| Subtotal | 514 | 43 (8.4) | 2 | 17 | 9 | 2 | 10 | 3 | |
| Adults | No diarrhea | 194 | 2 (1.0) | 0 | 1 | 0 | 1 | 0 | 0 |
| Watery diarrhea | 1 | 0 (0) | 0 | 0 | 0 | 0 | 0 | 0 | |
| Subtotal | 195 | 2 (1.0) | 0 | 1 | 0 | 1 | 0 | 0 | |
| Unknown | No diarrhea | 43 | 10 (23.3) | 4 | 3 | 0 | 0 | 3 | 0 |
| Moderate diarrhea | 2 | 0 (0) | 0 | 0 | 0 | 0 | 0 | 0 | |
| Watery diarrhea | 6 | 1 (16.7) | 0 | 0 | 0 | 0 | 1 | 0 | |
| Subtotal | 51 | 11 (21.6) | 4 | 3 | 0 | 0 | 4 | 0 | |
| Total | 1372 | 195 (14.2) | 79 | 48 | 31 | 3 | 26 | 8 | |
2.3. Detection of common enteric pathogens by enzymatic immunoassay (EIA)
To perform a preliminary diagnosis of the cause of the calf diarrhea, fecal specimens of 18 seriously ill calves submitted by the farmer were tested by enzymatic immunoassay (EIA) for the four most common enteric pathogens in cattle. In this, 200 mg of frozen fecal specimen were tested for rotavirus, coronavirus, Escherichia coli K99, and Cryptosporidium spp. by using the Pathasure Enteritis EIA kit (Biovet, Saint-Hyacinthe, Canada). In addition, rotavirus in 418 fecal specimens collected after the diarrhea outbreak from both pre-weaned (n = 212) and post-weaned (n = 206) calves was tested using the ProSpecT Rotavirus Kit (Thermo-Oxoid, Basingstoke, UK).
2.4. DNA extraction
For all 1390 fecal specimens preserved in potassium dichromate, including the 18 specimens collected during the outbreak, 200 mg of feces were washed three times with distilled water by centrifugation at 2000g for 10 min. Genomic DNA was extracted from the washed specimens by using the FastDNA SPIN Kit for Soil (MP Biomedicals, CA, USA), and stored at −80 °C until being analysed by PCR.
2.5. Cryptosporidium detection, genotyping and subtyping
Cryptosporidium spp. in the specimens were detected by nested PCR amplification of a ∼830 bp fragment of the ssrRNA gene, and were genotyped by restriction fragment length polymorphism (RFLP) analysis of the secondary PCR products, using restriction enzymes SspI and MboII (New England BioLabs, MA, USA) (Feng et al., 2007). To identify the C. parvum subtype involved, a ∼850 bp fragment of the gp60 gene was amplified by nested PCR (Feng et al., 2009). Each specimen was analysed twice by PCR at each genetic locus. Reagent-grade water was used as the negative control, whereas Cryptosporidium canis DNA was used as the positive control for the ssrRNA PCR and Cryptosporidium hominis DNA as the positive control for the gp60 PCR.
2.6. Sequence analysis
Representative positive PCR products of the ssrRNA gene, including at least two of each Cryptosporidium spp. identified, were sequenced to confirm the RFLP results. For subtyping of C. parvum, all positive PCR products of the gp60 gene were sequenced. PCR products were sequenced in both directions on an ABI 3730 Genetic Analyzer (Applied Biosystems, CA, USA). The sequences obtained were assembled using ChromasPro (http://www.technelysium.com.au/ChromasPro.html), edited using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), and aligned with reference sequences in the GenBank database using ClustalX (http://clustal.org/) to determine Cryptosporidium spp. and subtypes.
2.7. Statistical analysis
Data in this study were statistically analysed using the software package R version 3.5.0 (https://www.r-project.org/). The χ 2 test was used to compare differences in Cryptosporidium or rotavirus infection rates between age groups or animals with or without diarrhea. To conduct a risk factor analysis, animals meeting the criteria of moderate or watery diarrhea were considered as cases of diarrhea. Logistic regression was used to assess the association between putative risk factors and the occurrence of diarrhea, using data from 886 calves of 0–6 months in age after the diarrhea outbreak. Initially, the strength of association was assessed using a univariate model at the liberal P ≤ 0.2. Variables significant at P ≤ 0.2 were thereafter used to build a multivariate model, with P < 0.05 being considered significant in the final model. Odds ratios (OR) with 95% confidence intervals (CI) were used to identify risk factors for diarrhea occurrence in calves.
3. Results
3.1. Occurrence of enteric pathogens in ill calves during the outbreak
In the initial EIA diagnosis of major enteric pathogens in 18 specimens from seriously ill calves (Fig. 1A), we found that 13 were positive for Cryptosporidium spp., eight for rotavirus, and all negative for coronavirus or E. coli K99 (Table 1). These 18 specimens were further analysed for Cryptosporidium spp. by the ssrRNA-based PCR, which led to the identification of C. parvum in 15 of them including all 13 EIA positive specimens. Fourteen of the C. parvum-positive specimens were successfully subtyped as IIdA19G1 by sequence analysis of the gp60 gene.
3.2. Occurrence of rotavirus in pre- and post-weaned dairy calves after the outbreak
Among the 1372 fecal specimens collected from calves after the diarrhea outbreak, 418 were tested for rotavirus by EIA. Altogether, 38/212 (17.9%) specimens from pre-weaned calves were positive, while 6/206 (2.9%) specimens from post-weaned calves were positive. The infection rate of rotavirus in pre-weaned calves was highest (33.9%) at 1 week of age, decreased afterwards in calves of 2–5 weeks of age, and mostly remained zero in calves older than 6 weeks (Fig. 1B). In pre-weaned calves, the highest infection rate for rotavirus occurred in calves with watery diarrhea (28.0%), followed by those without diarrhea (17.6%) and with moderate diarrhea (10.0%). In post-weaned calves, infection with rotavirus was similar among animals with watery diarrhea (2.6%), moderate diarrhea (3.1%) and no diarrhea (2.9%). Pairwise χ 2 analysis showed that none of the differences between any two of the symptom groups of pre- and post-weaned calves reached significance (P > 0.05; Table 3 ).
Table 3.
Statistical significance for associations between Cryptosporidium or rotavirus infections and occurrence of moderate or watery diarrhea in pre-weaned and post-weaned dairy calves on a farm in Jiangsu, China, as indicated by results of χ2 analysis.
| Age | Pathogens | Moderate diarrhea |
Watery diarrhea |
||||
|---|---|---|---|---|---|---|---|
| χ2 | OR (95% CI) | P | χ2 | OR (95% CI) | P | ||
| Pre-weaned | Cryptosporidium spp. | 12.877 | 2.3 (1.5, 3.8) | 0.000b | 8.464 | 2.3 (1.3, 4.1) | 0.003b |
| C. parvum | 0.040 | 1.1 (0.5, 2.2) | 0.842 | 12.911 | 3.1 (1.6, 6.0) | 0.000b | |
| C. ryanae | 6.937 | 3.0 (1.3, 7.3) | 0.008b | 1.000 | 2.2 (0.7, 6.9) | 0.171 | |
| C. bovis | 1.925 | 2.2 (0.9, 5.4) | 0.095 | NA | NA | NA | |
| C. ryanae + C. bovis | 3.822 | 3.9 (1.2, 12.9) | 0.019a | NA | NA | NA | |
| Rotavirus | 1.751 | 0.5 (0.2, 1.4) | 0.186 | 2.164 | 1.8 (0.8, 4.0) | 0.141 | |
| Post-weaned | Cryptosporidium spp. | 7.972 | 3.4 (1.5, 7.8) | 0.002b | 3.125 | 2.9 (1.0, 8.3) | 0.033a |
| C. ryanae | 4.042 | 4.1 (1.2, 13.6) | 0.013a | NA | NA | NA | |
| Rotavirus | 0.164 | 1.1 (0.2, 6.5) | 0.951 | 0.233 | 0.9 (0.1, 8.9) | 0.929 | |
NA, not available; OR, odds ratio; CI, confidence interval.
P < 0.05.
P < 0.01.
3.3. Occurrence of Cryptosporidium spp. in dairy cattle after the outbreak
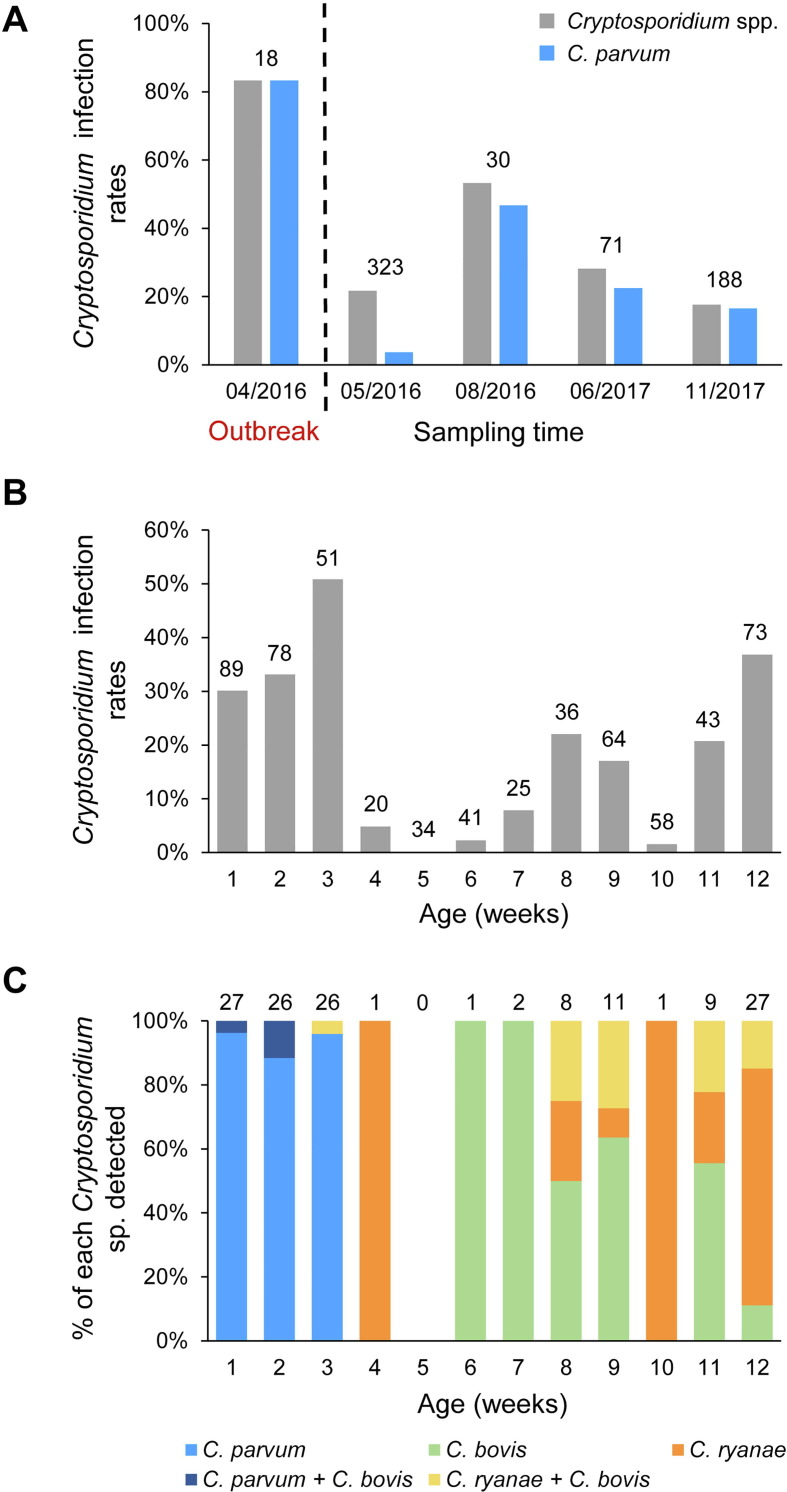
Of the 1372 fecal specimens collected after the diarrhea outbreak, 195 (14.2%) were positive for Cryptosporidium spp. by PCR analysis of the ssrRNA gene (Table 2). The highest infection rate was 22.7% (139/612) in pre-weaned calves, followed by post-weaned calves (8.4%; 43/514) and adult cattle (1.0%, 2/195) (Table 2). Pairwise χ 2 analysis among the three age groups all showed highly significant differences in infection rates (P < 0.001). In pre-weaned calves, the infection rate for Cryptosporidium spp. in April 2016 (83.3%) was significantly higher (P < 0.05) than those in May 2016 (21.7%), August 2016 (53.3%), June 2017 (28.2%), and November 2017 (17.6%) (Fig. 2 A). Odds of Cryptosporidium infection in calves during the diarrhea outbreak were 4.4–23.5 times higher than after the outbreak. The same trend was also observed in the occurrence of C. parvum; 83.3%, 3.7%, 46.7%, 22.5% and 16.5% of specimens were positive in April 2016, May 2016, August 2016, June 2017, and November 2017, respectively (Fig. 2A). The occurrence of C. parvum infection during the outbreak was also significantly higher (P < 0.05) compared with after the outbreak, with OR = 5.7–129.6.
Fig. 2.
Cryptosporidium infection in pre-weaned dairy calves on a farm in Jiangsu, China, with a diarrhea outbreak. (A) Temporal variation in infection rates of Cryptosporidium spp. and Cryptosporidium parvum. (B) Cryptosporidium infection rates by age in weeks after the outbreak. (C) Distribution of Cryptosporidium spp. by age in weeks after the outbreak.
3.4. Distribution of Cryptosporidium spp./subtypes after the outbreak
RFLP analysis was successful for all 195 ssrRNA PCR-positive specimens, allowing the identification of four Cryptosporidium spp. in dairy cattle. Overall, C. parvum was the most frequently identified species (n = 79), followed by C. ryanae (n = 48), C. bovis (n = 31) and C. andersoni (n = 3). Concurrent infections with mixed Cryptosporidium spp. were detected in 34 cattle and included C. ryanae/C. bovis (n = 26) and C. parvum/C. bovis (n = 8) (Table 2).
In DNA sequence analysis, the representative ssrRNA sequences generated from specimens of C. parvum (n = 6), C. ryanae (n = 1), C. bovis (n = 1) and C. andersoni (n = 1) were identical to the GenBank reference sequences KP793008, KP793012, KP793007 and KM110052, respectively. For the C. parvum-positive specimens, 77/87 specimens were successfully subtyped at the gp60 locus, and the sequences obtained were all identical to the reference sequence HQ009809 of the subtype IIdA19G1 in GenBank.
3.5. Distribution of Cryptosporidium spp. by age after the outbreak
In pre-weaned calves, infection rates for Cryptosporidium spp. varied greatly by age in weeks (Fig. 2B). The highest infection rate was 51.0% (26/51) at 3 weeks of age, whereas the lowest infection rate (0 or 0/34) was at 5 weeks of age. During the first 3 weeks, 73/79 Cryptosporidium-positive calves were infected with C. parvum (Fig. 2C). Afterwards, C. parvum disappeared in pre-weaned calves. In contrast, C. ryanae and C. bovis became the only two species identified in calves of 4–12 weeks of age, and co-infection with the two species was commonly seen in these animals.
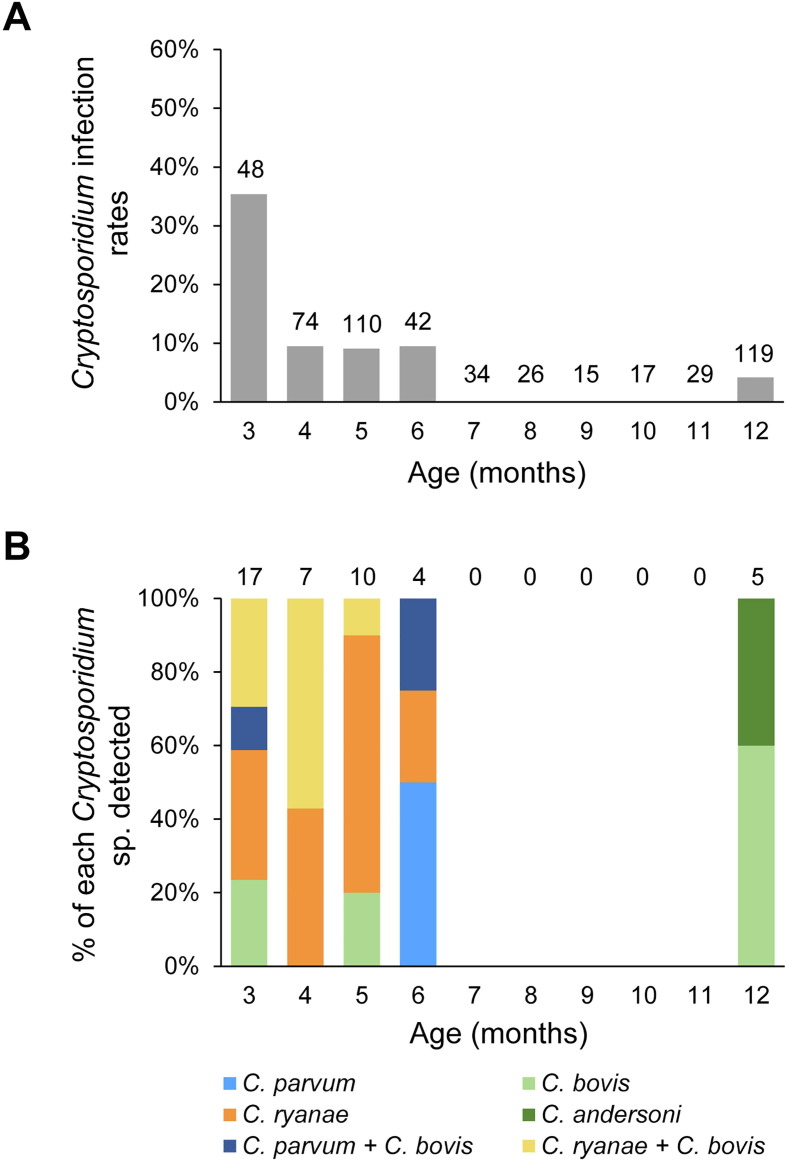
In post-weaned calves, Cryptosporidium infection rates varied from 0 to 35.4% at 3–12 months of age (Fig. 3 A). The infection rate was highest at 3 months of age, decreased to lower than 10% in calves of 4–6 months of age, and remained mostly near zero in calves older than 6 months. Cryptosporidium ryanae was the most common species identified in the 3–6 months age groups (Fig. 3B). Cryptosporidium bovis, the second most common species in post-weaned calves, was frequently seen in mixed infections with C. ryanae or C. parvum. In contrast to pre-weaned calves, C. parvum was rarely detected in post-weaned calves, being identified in only two calves. Thus, C. parvum was much less prevalent in post-weaned calves (0.4%) than in pre-weaned calves (11.9%; χ 2 = 59.831, P = 0.000). In addition, C. andersoni was detected in two calves of 12 months of age and one adult animal.
Fig. 3.
Cryptosporidium infection in post-weaned dairy calves on a farm in Jiangsu, China, after a diarrhea outbreak. (A) Cryptosporidium infection rates by age in months. (B) Distribution of Cryptosporidium spp. by age in months.
3.6. Correlation between Cryptosporidium spp. and diarrhea occurrence after the outbreak as indicated by χ2 analysis
In pre-weaned calves, Cryptosporidium infection rates in calves with moderate diarrhea (34.7%) or watery diarrhea (34.4%) were significantly higher than in calves without diarrhea (18.4%; P < 0.01) (Table 2, Table 3). For C. parvum, the infection rate in calves with watery diarrhea (26.2%) was higher than in calves with moderate diarrhea (10.9%) or no diarrhea (10.2%). The difference in infection rates for C. parvum was highly significant between calves with watery diarrhea and no diarrhea (χ 2 = 12.911, P = 0.000). Unlike C. parvum, the C. ryanae infection rate in calves with moderate diarrhea (8.9%) was significantly higher than that in calves without diarrhea (3.1%; χ 2 = 6.937, P = 0.008). In contrast, the presence of C. bovis was similar between calves with moderate diarrhea (6.9%) and no diarrhea (3.3%; P = 0.095).
In post-weaned calves, the highest Cryptosporidium infection rate was observed in calves with moderate diarrhea (19.6%), followed by those with watery diarrhea (17.2%) and no diarrhea (6.6%) (Table 2). The difference in Cryptosporidium infection rates was highly significant between calves with moderate diarrhea and no diarrhea (χ 2 = 7.972, P = 0.002) (Table 3). Among Cryptosporidium spp., only C. ryanae had sufficient positive specimens to perform the correlation analysis on occurrence of diarrhea, showing that the infection rate in animals with moderate diarrhea (8.7%) was significantly higher than those without diarrhea (2.3%; χ 2 = 4.042, P = 0.013).
3.7. Risk factors for occurrence of diarrhea after the diarrhea outbreak as indicated by univariate and multivariate analyses
In the univariate analysis of risk factors associated with the occurrence of diarrhea in calves of 0–6 months of age after the diarrhea outbreak, Cryptosporidium-positive animals had a significantly higher chance of having diarrhea (P = 0.000; Table 4 ), especially moderate diarrhea (P = 0.002). Among Cryptosporidium spp., C. parvum infection (P = 0.004) was a risk factor for the occurrence of watery diarrhea, while C. ryanae (P = 0.000) or C. bovis (P = 0.001) infection was a risk factor for the occurrence of moderate diarrhea. Surprisingly, we found no evidence showing age as a significant risk factor for the occurrence of diarrhea in pre- and post-weaned calves in this investigation (P = 0.779). Further analysis using the multivariate model showed that the association of C. ryanae or C. bovis infection and the occurrence of moderate diarrhea was just below statistical significance (P = 0.055 and 0.062, respectively; Table 4).
Table 4.
Association between Cryptosporidium spp. and occurrence of diarrhea in dairy calves after a diarrhea outbreak on a farm in Jiangsu, China, as indicated by results of univariate and multivariate analyses.
| Diarrhea status | Factor | Univariate model |
Multivariate model |
||||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | ||
| Watery diarrhea | Cryptosporidium infection | ||||||
| Yes | 1.8 | 1.1–2.9 | 0.023a | 1.2 | 0.6–2.4 | 0.583 | |
| C. parvum | 2.4 | 1.3–4.3 | 0.004b | 2.0 | 0.9–4.9 | 0.108 | |
| C. ryanae | 1.3 | 0.6–2.7 | 0.521 | NA | NA | NA | |
| C. bovis | 0.5 | 0.1–1.3 | 0.216 | NA | NA | NA | |
| No | 1 | Reference | 1 | Reference | |||
| Age | |||||||
| Pre-weaned (<3 months) | 1.1 | 0.8–1.5 | 0.779 | NA | NA | NA | |
| Post-weaned (3–6 months) | 1 | Reference | 1 | Reference | |||
| Moderate diarrhea | Cryptosporidium infection | ||||||
| Yes | 1.9 | 1.3–2.8 | 0.002b | 1.1 | 0.6–2.0 | 0.709 | |
| C. parvum | 0.9 | 0.5–1.7 | 0.850 | NA | NA | NA | |
| C. ryanae | 2.7 | 1.5–4.7 | 0.000b | 2.0 | 1.0–4.1 | 0.055 | |
| C. bovis | 2.7 | 1.5–4.8 | 0.001b | 2.0 | 1.0–4.1 | 0.062 | |
| No | 1 | Reference | 1 | Reference | |||
| Age | |||||||
| Pre-weaned (<3 months) | 1.0 | 0.8–1.3 | 0.916 | NA | NA | NA | |
| Post-weaned (3–6 months) | 1 | Reference | 1 | Reference | |||
| Any diarrhea | Cryptosporidium infection | ||||||
| Yes | 2.1 | 1.5–3.0 | 0.000b | 2.4 | 0.7–7.9 | 0.142 | |
| C. parvum | 1.6 | 1.0–2.5 | 0.066 | 0.7 | 0.2–2.5 | 0.629 | |
| C. ryanae | 2.5 | 1.5–4.1 | 0.001b | 1.2 | 0.4–3.5 | 0.760 | |
| C. bovis | 1.8 | 1.0–3.1 | 0.039a | 0.8 | 0.3–2.0 | 0.657 | |
| No | 1 | Reference | 1 | Reference | |||
| Age | |||||||
| Pre-weaned (<3 months) | 1.0 | 0.8–1.3 | 0.779 | NA | NA | NA | |
| Post-weaned (3–6 months) | 1 | Reference | 1 | Reference | |||
NA, not available; OR, odds ratio; CI, confidence interval.
P < 0.05.
P < 0.01.
4. Discussion
We identified cryptosporidiosis as the likely cause for the outbreak of watery diarrhea in pre-weaned calves described in this study. This was largely based on results of the initial EIA test, which identified the presence of C. parvum and rotavirus in the 18 seriously ill calves examined during the outbreak. Two other common enteropathogens for neonatal calf diarrhea, coronavirus and E. coli K99, were ruled out as the cause of the diarrhea. The role of rotavirus in the occurrence of the diarrhea outbreak appears to be small, as only fewer than half of the seriously ill calves had the infection and rotavirus infection was not significantly associated with the occurrence of moderate or watery diarrhea in calves during post-outbreak investigation. This was further supported by the peak occurrence of rotavirus infection in calves at 1 week of age, which is slightly earlier than the age range (10–20 days) of ill calves during the outbreak. The identification of cryptosporidiosis as the cause of the diarrhea outbreak was substantiated by genotyping of Cryptosporidium spp. in calves. All Cryptosporidium-positive ill calves had C. parvum instead of C. bovis, which is the dominant Cryptosporidium sp. in pre-weaned calves in China but mostly avirulent (Feng and Xiao, 2017). In addition, the occurrence of C. parvum in pre-weaned calves was significantly higher during the outbreak than after the outbreak (OR = 5.7–129.6), and C. parvum infection was associated with the occurrence of watery diarrhea in pre-weaned calves in a post-outbreak investigation. Nevertheless, rotavirus could have played a secondary role in the outbreak of watery diarrhea in pre-weaned dairy calves on the farm, as suggested by investigations of outbreaks of calf diarrhea in Australia and India (Izzo et al., 2011, Brar et al., 2017). In this study, pathological investigations were not conducted on dead calves due to the distant nature of the farm from research facilities. We became involved with the investigation when the outbreak was nearing the end, and the fecal specimens from the 18 seriously ill calves were received by express mail.
Compared with numerous reports of cryptosporidiosis outbreaks in humans, only a limited number of outbreaks have been documented in dairy calves (Reynolds et al., 1986, Aurich et al., 1990, Xiao et al., 1993). In these earlier studies, the detection of Cryptosporidium infections was based on microscopy or an EIA test. Although the species involved was unknown, it was reasonable to assume that these outbreaks were caused by C. parvum, which is well known to cause watery diarrhea (Meganck et al., 2014). In one recent study in Estonia, however, the C. parvum subtype IIaA18G1R1 was identified as the cause of a cryptosporidiosis outbreak in pre-weaned dairy calves (Niine et al., 2018). Similarly in China, the only reported bovine cryptosporidiosis outbreak was caused by the C. parvum subtype IIdA15G1 (Cui et al., 2014). That outbreak occurred on a dairy farm in Ningxia, northwestern China, leading to the death of 356 calves. Unlike the finding in Ningxia, the bovine cryptosporidiosis outbreak reported in the present study was caused by the C. parvum subtype IIdA19G1. Although C. parvum is still an emerging Cryptosporidium sp. in dairy cattle in China, IIdA15G1 and IIdA19G1 subtypes are dominant C. parvum subtypes in pre-weaned dairy calves (Feng and Xiao, 2017, Wang et al., 2017).
In this study, we also examined the transmission characteristics of Cryptosporidium spp. on this large dairy farm after the diarrhea outbreak. The overall infection rate of Cryptosporidium spp. was 14.2%, which is similar to the average prevalence rate (14.0%) in dairy cattle in China (Wang et al., 2017). As in most previous studies in China and elsewhere (Fayer et al., 2010, Venu et al., 2013, Wang et al., 2017), declining infection rates with increased age was also observed in the present study, with the infection rate in pre-weaned calves (22.7%) significantly higher than those in post-weaned calves (8.4%) and adult cattle (1.0%). Among the four Cryptosporidium spp. identified on the farm in the present study, C. parvum was the most frequently detected species in pre-weaned calves, C. ryanae and C. bovis were dominant in post-weaned calves, whereas C. andersoni was only identified in three older animals. This is largely in agreement with the age-related distribution of Cryptosporidium spp. in dairy cattle elsewhere in the world (Xiao, 2010). However, as in some locations of China (Cai et al., 2017, Wang et al., 2017), C. bovis and C. ryanae were also common in pre-weaned dairy cattle. In fact, C. ryanae was not only the most common species identified in post-weaned calves, but also more prevalent than C. bovis in pre-weaned calves in the present study. Due to the high infection rates of both C. bovis and C. ryanae, a high rate of co-infections with the two Cryptosporidium spp. was also observed in this study.
As expected, Cryptosporidium infection was significantly associated with the occurrence of calf diarrhea in the post-outbreak investigation (Cho et al., 2013, Meganck et al., 2014). In pre-weaned calves, the Cryptosporidium infection rate, especially the one for C. parvum, was significantly higher in animals with watery diarrhea than in those with moderate or no diarrhea in χ 2 analysis. Similar observations were also seen in dairy calves in numerous earlier studies (Aita et al., 2015, Cai et al., 2017). However, there was also an association between C. ryanae infection and the occurrence of moderate diarrhea in pre-weaned calves. This was also the case with co-infections of C. ryanae and C. bovis. The association between C. ryanae infection and the occurrence of moderate diarrhea was also seen in post-weaned calves. This is contrary to the findings elsewhere that C. ryanae has not yet been associated with any clinical signs in infected calves (Wang et al., 2017).
The pathogenicity of C. parvum and C. ryanae observed in this study was supported by results of the logistic regression analysis. Cryptosporidium parvum infection was significantly associated with the occurrence of watery diarrhea in a univariate analysis, whereas C. ryanae or C. bovis infection was associated with the occurrence of moderate diarrhea. Previously, the pathogenic potential of C. bovis was suggested in some earlier reports based on its detection in pre-weaned calves with diarrhea (Silverlas and Blanco-Penedo, 2013, Silverlas et al., 2013, Lee et al., 2016). The present study represents, to our knowledge, the first description of such an association through statistical analysis of epidemiological data.
In conclusion, we report a cryptosporidiosis outbreak caused by C. parvum subtype IIdA19G1 in neonatal calves with significant mortality on a large dairy farm in Jiangsu, China. This serves as new evidence of the virulence of this emerging subtype in the country. In addition, C. ryanae and C. bovis have been shown as potential causes for moderate diarrhea in dairy calves. The emerging nature of C. parvum in farm animals in China, the high mortality in this outbreak, and a lack of effective treatment for cryptosporidiosis highlight the need to develop control measures against the transmission of C. parvum on dairy farms.
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (31820103014 and 31425025).
References
- Aita J., Ichikawa-Seki M., Kinami A., Yaita S., Kumagai Y., Nishikawa Y., Itagaki T. Molecular characterization of Cryptosporidium parvum detected in Japanese black and Holstein calves in Iwate Prefecture and Tanegashima Island, Kagoshima Prefecture, Japan. J. Vet. Med. Sci. 2015;77:997–999. doi: 10.1292/jvms.15-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Mawly J., Grinberg A., Prattley D., Moffat J., Marshall J., French N. Risk factors for neonatal calf diarrhoea and enteropathogen shedding in New Zealand dairy farms. Vet. J. 2015;203:155–160. doi: 10.1016/j.tvjl.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurich J.E., Dobrinski I., Grunert E. Intestinal cryptosporidiosis in calves on a dairy farm. Vet. Rec. 1990;127:380–381. [PubMed] [Google Scholar]
- Bjorkman C., Lindstrom L., Oweson C., Ahola H., Troell K., Axen C. Cryptosporidium infections in suckler herd beef calves. Parasitology. 2015;142:1108–1114. doi: 10.1017/S0031182015000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard P.C. Diagnostics of dairy and beef cattle diarrhea. Vet. Clin. North Am. Food Anim. Pract. 2012;28:443–464. doi: 10.1016/j.cvfa.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar A.P.S., Sood N.K., Kaur P., Singla L.D., Sandhu B.S., Gupta K., Narang D., Singh C.K., Chandra M. Periurban outbreaks of bovine calf scours in Northern India caused by Cryptosporidium in association with other enteropathogens. Epidemiol. Infect. 2017;145:2717–2726. doi: 10.1017/S0950268817001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Guo Y., Pan B., Li N., Wang X., Tang C., Feng Y., Xiao L. Longitudinal monitoring of Cryptosporidium species in pre-weaned dairy calves on five farms in Shanghai, China. Vet. Parasitol. 2017;241:14–19. doi: 10.1016/j.vetpar.2017.05.005. [DOI] [PubMed] [Google Scholar]
- Cho Y.I., Han J.I., Wang C., Cooper V., Schwartz K., Engelken T., Yoon K.J. Case-control study of microbiological etiology associated with calf diarrhea. Vet. Microbiol. 2013;166:375–385. doi: 10.1016/j.vetmic.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.I., Yoon K.J. An overview of calf diarrhea - infectious etiology, diagnosis, and intervention. J. Vet. Sci. 2014;15:1–17. doi: 10.4142/jvs.2014.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Wang R., Huang J., Wang H., Zhao J., Luo N., Li J., Zhang Z., Zhang L. Cryptosporidiosis caused by Cryptosporidium parvum subtype IIdA15G1 at a dairy farm in Northwestern China. Parasite Vector. 2014;7:529. doi: 10.1186/s13071-014-0529-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayer R., Santin M., Dargatz D. Species of Cryptosporidium detected in weaned cattle on cow-calf operations in the United States. Vet. Parasitol. 2010;170:187–192. doi: 10.1016/j.vetpar.2010.02.040. [DOI] [PubMed] [Google Scholar]
- Feng Y., Li N., Duan L., Xiao L. Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: evidence for possible unique Cryptosporidium hominis transmission. J. Clin. Microbiol. 2009;47:153–157. doi: 10.1128/JCM.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Ortega Y., He G., Das P., Xu M., Zhang X., Fayer R., Gatei W., Cama V., Xiao L. Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet. Parasitol. 2007;144:1–9. doi: 10.1016/j.vetpar.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Feng Y., Ryan U.M., Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018;24:997–1011. doi: 10.1016/j.pt.2018.07.009. [DOI] [PubMed] [Google Scholar]
- Feng Y., Xiao L. Molecular epidemiology of cryptosporidiosis in China. Front. Microbiol. 2017;8:1701. doi: 10.3389/fmicb.2017.01701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim M.A., Abdel-Ghany A.E., Abdel-Latef G.K., Abdel-Aziz S.A., Aboelhadid S.M. Epidemiology and public health significance of Cryptosporidium isolated from cattle, buffaloes, and humans in Egypt. Parasitol. Res. 2016;115:2439–2448. doi: 10.1007/s00436-016-4996-3. [DOI] [PubMed] [Google Scholar]
- Izzo M.M., Kirkland P.D., Mohler V.L., Perkins N.R., Gunn A.A., House J.K. Prevalence of major enteric pathogens in Australian dairy calves with diarrhoea. Aust. Vet. J. 2011;89:167–173. doi: 10.1111/j.1751-0813.2011.00692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., VanBik D., Kim H.Y., Lee Y.R., Kim J.W., Chae M., Oh S.I., Goo Y.K., Kwon O.D., Kwak D. Multilocus typing of Cryptosporidium spp. in young calves with diarrhea in Korea. Vet. Parasitol. 2016;229:81–89. doi: 10.1016/j.vetpar.2016.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meganck V., Hoflack G., Opsomer G. Advances in prevention and therapy of neonatal dairy calf diarrhoea: a systematical review with emphasis on colostrum management and fluid therapy. Acta Vet. Scand. 2014;56:75. doi: 10.1186/s13028-014-0075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed F.F., Mansour S.M., El-Araby I.E., Mor S.K., Goyal S.M. Molecular detection of enteric viruses from diarrheic calves in Egypt. Arch. Virol. 2017;162:129–137. doi: 10.1007/s00705-016-3088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niine T., Dorbek-Kolin E., Lassen B., Orro T. Cryptosporidium outbreak in calves on a large dairy farm: Effect of treatment and the association with the inflammatory response and short-term weight gain. Res. Vet. Sci. 2018;117:200–208. doi: 10.1016/j.rvsc.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlenz J., Moon H.W., Cheville N.F., Bemrick W.J. Cryptosporidiosis as a probable factor in neonatal diarrhea of calves. J. Am. Vet. Med. Assoc. 1978;172:452–457. [PubMed] [Google Scholar]
- Randhawa S.S., Randhawa S.S., Zahid U.N., Singla L.D., Juyal P.D. Drug combination therapy in control of cryptosporidiosis in Ludhiana district of Punjab. J. Parasit. Dis. 2012;36:269–272. doi: 10.1007/s12639-012-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds D.J., Morgan J.H., Chanter N., Jones P.W., Bridger J.C., Debney T.G., Bunch K.J. Microbiology of calf diarrhoea in southern Britain. Vet. Rec. 1986;119:34–39. doi: 10.1136/vr.119.2.34. [DOI] [PubMed] [Google Scholar]
- Silverlas C., Blanco-Penedo I. Cryptosporidium spp. in calves and cows from organic and conventional dairy herds. Epidemiol. Infect. 2013;141:529–539. doi: 10.1017/S0950268812000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverlas C., Bosaeus-Reineck H., Naslund K., Bjorkman C. Is there a need for improved Cryptosporidium diagnostics in Swedish calves? Int. J. Parasitol. 2013;43:155–161. doi: 10.1016/j.ijpara.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylan-Ozkan A., Yasa-Duru S., Usluca S., Lysen C., Ye J., Roellig D.M., Feng Y., Xiao L. Cryptosporidium species and Cryptosporidium parvum subtypes in dairy calves and goat kids reared under traditional farming systems in Turkey. Exp. Parasitol. 2016;170:16–20. doi: 10.1016/j.exppara.2016.06.014. [DOI] [PubMed] [Google Scholar]
- Thomson S., Hamilton C.A., Hope J.C., Katzer F., Mabbott N.A., Morrison L.J., Innes E.A. Bovine cryptosporidiosis: impact, host-parasite interaction and control strategies. Vet. Res. 2017;48:42. doi: 10.1186/s13567-017-0447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzipori S., Smith M., Halpin C., Angus K.W., Sherwood D., Campbell I. Experimental cryptosporidiosis in calves: clinical manifestations and pathological findings. Vet. Rec. 1983;112:116–120. doi: 10.1136/vr.112.6.116. [DOI] [PubMed] [Google Scholar]
- Uetake K. Newborn calf welfare: a review focusing on mortality rates. Anim. Sci. J. 2013;84:101–105. doi: 10.1111/asj.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venu R., Latha B.R., Basith S.A., Sreekumar C., Raj G.D., Raman M. Factors influencing on prevalence of Cryptosporidium infection in south Indian dairy calves. J. Parasit. Dis. 2013;37:168–172. doi: 10.1007/s12639-012-0153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira P.M., Mederle N., Lobo M.L., Imre K., Mederle O., Xiao L., Darabus G., Matos O. Molecular characterisation of Cryptosporidium (Apicomplexa) in children and cattle in Romania. Folia Parasitol. (Praha) 2015:62. doi: 10.14411/fp.2015.002. [DOI] [PubMed] [Google Scholar]
- Wang R., Zhao G., Gong Y., Zhang L. Advances and perspectives on the epidemiology of bovine Cryptosporidium in China in the past 30 years. Front. Microbiol. 2017;8:1823. doi: 10.3389/fmicb.2017.01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 2010;124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- Xiao L., Herd R.P., Rings D.M. Concurrent infections of Giardia and Cryptosporidium on two Ohio farms with calf diarrhea. Vet. Parasitol. 1993;51:41–48. doi: 10.1016/0304-4017(93)90194-R. [DOI] [PMC free article] [PubMed] [Google Scholar]