Abstract
N and N bind by cross‐linking study (View Interaction: 1, 2, 3, 4)
Keywords: HCoV, human coronavirus, CoV, coronavirus, RNP, ribonucleoprotein, N protein, nucleocapsid protein, S, spike, M, membrane, E, envelope, Tm, melting temperature, SR-rich, serine–arginine-rich, IBV, avian infectious bronchitis virus, SARS, severe acute respiratory syndrome, MHV, murine hepatitis virus, SDS, sodium dodecyl sulfate, Human coronavirus, 229E strain, Nucleocapsid protein, Ribonucleocapsid, C-terminal domain, Oligomerization, Secondary structure, Stability
Short abstract
► The role of the C‐terminal tail of the HCoV‐229E N protein in oligomerization. ► A correlation between oligomerization and thermostability. ► The C‐terminal tail peptide interferes with the oligomerization. ► The development of drugs to disrupt the oligomerization of the viral N protein.
1. Introduction
Human coronavirus 229E (HCoV‐229E), belonging to the alphacoronaviruses, was first identified in the 1960s and has generally been associated with symptoms of the common cold [1, 2]. Although HCoV‐229E infections are generally mild, more severe upper and lower respiratory tract infections, such as bronchiolitis and pneumonia, have been well documented, particularly in infants, elderly individuals, and immunocompromised patients [1, 3, 4]. There have also been reports that clusters of HCoV‐229E infections cause pneumonia in otherwise healthy adults [2, 5]. Several emerging human coronaviruses have recently been discovered [6, 7, 8]. Between 2003 and 2004, the severe acute respiratory syndrome (SARS)–CoV caused a worldwide epidemic and had a significant economic impact in the countries where the disease outbreak occurred [8]. Phylogenetic analyses have shown that SARS–CoV is closely related to the sequences of the betacoronaviruses [9]. In 2004, another alphacoronavirus (HCoV‐NL63) was isolated from a 7‐month‐old child in the Netherlands suffering from bronchiolitis and conjunctivitis [7]. In 2005, Woo et al. described the discovery of a novel betacoronavirus, HKU1, which was found in patients with respiratory tract infections [10].
CoV particles have an irregular shape that consists of an outer envelope with distinctive, ‘club‐shaped’ peplomers, giving the virus a crown (corona) appearance [11]. The viral genome of coronaviruses consists of positive‐sense, single‐stranded RNA of approximately 30 kb, and it contains several genes encoding several structural and non‐structural proteins that are required for progeny virion production [1]. The virion envelope surrounding the nucleocapsid contains the following structural proteins: S (spike) protein, M (membrane), E (envelope), and N (nucleocapsid). Some variants have a third glycoprotein, HE (hemagglutinin‐esterase), which is present in most betacoronaviruses [12, 13]. A helical nucleocapsid exists in the center of the viral particle [14, 15, 16]. Nucleocapsid protein, the major structural protein of CoVs, binds the viral RNA genome to form the virion core, leading to the formation of a ribonucleoprotein (RNP) complex or to a long helical nucleocapsid structure [17, 18]. The formation of the RNP is important for maintaining the RNA in an ordered conformation suitable for replication and transcription of the viral genome [17, 19, 20, 21]. Previous studies have shown that the CoV N protein is involved in the regulation of cellular processes, such as gene transcription, actin reorganization, host cell cycle progression, and apoptosis [22, 23, 24, 25]. The CoV N protein has also been shown to act as an RNA chaperone [26]. Moreover, the N protein is an important diagnostic marker and immunodominant antigen in host immune responses [21, 27, 28].
The N protein of HCoV‐229E, which has a molecular weight of 50 kDa, is highly basic (pI, 10.0), and it shows strong hydrophilicity [29]. The HCoV‐229E N protein has 26–30% sequence homology with CoV N proteins from other strains or viruses, such as HCoV‐OC43 and SARS [30]. Despite their low sequence homology, CoV N proteins from different strains can show a high level of conservation in some motifs [30]. Chang et al. reported results from an order–disorder prediction and secondary structure prediction coupled with sequence alignment, which suggested that all CoV N proteins share the same modular organization [31]. Self‐association of the N protein is an important step in virus particle assembly for many CoVs [32]. Previous studies have shown that full‐length CoV N proteins can form high‐order oligomers, and the C‐terminal domains of the CoV N proteins are responsible for oligomerization [30, 33, 34, 35, 36, 37, 38]. Crystal structures of the C‐terminal domains of CoV N proteins have been published and suggest that the basic building block for coronavirus nucleocapsid formation is the dimeric assembly of the N protein [34, 39, 40, 41]. Luo et al. revealed that the CoV N protein might combine with viral genomic RNA to generate higher‐order oligomers, which could trigger the formation of the long nucleocapsid structure [32]. However, the oligomerization mechanism of the C‐terminal domain of the HCoV‐229E N protein remains unclear. The C‐terminal tail has been found to participate in the oligomerization of the SARS–CoV N protein since the removal of 40 aa from the C‐terminus apparently decreased the ability of the protein to oligomerize [32]. The extreme C‐terminal tail of the HCoV N protein was labeled as a separate functional domain [42]. We speculate that the C‐terminal tail might constitute an important molecular determinant of oligomerization for HCoV‐229E NP. However, the recombinant full‐length nucleocapsid N protein of the coronavirus is highly sensitive to proteolysis and aggregation that is difficult to analyze its oligomerization properties [21]. A comprehensive series of HCoV‐229E N protein mutants with truncated C‐terminal domains was generated based on the PrDOS prediction to clarify the role of the C‐terminal tail of the HCoV‐229E N protein in oligomerization (Fig. 1 ). According to the order–disorder profiles obtained from the protein disorder prediction system (PrDOS), the predicted structure of HCoV‐229 N protein contains one long ordered region (N245–350) located in the C‐terminal domain followed by three short regions predicted to be disordered (N351–370), ordered (N371–382), and disordered (N383–389) (Fig. 1) [30]. These truncations were systematically investigated by various biophysical and biochemical analyses. Understanding this mechanism would provide insight into the viral assembly process and could identify additional targets for drugs to combat CoVs through the disruption of N protein self‐association.
Figure 1.
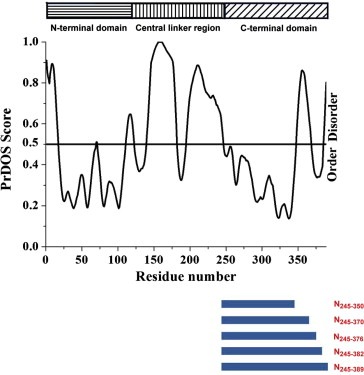
The order/disorder prediction for the full‐length HCoV‐229E N protein using the PrDOS program corresponding to the designations of the truncations, including N245–350, N245–370, N245–376, N245–382 and N245–389. PrDOS scores above a threshold value of 0.5 denote the disordered regions.
2. Materials and methods
The drugs and reagents were purchased from Sigma Chemical Co. All of oligoribonucleotides (or oligodeoxyribonucleotides) were synthesized using an automated DNA synthesizer and were purified by gel electrophoresis.
2.1. Expression and purification of the full‐length and truncated N proteins
The templates for the HCoV‐229E N protein were kindly provided by the Institute of Biological Chemistry, Academia Sinica (Taipei, Taiwan). To generate the truncated forms of the recombinant N proteins, the N protein gene was amplified by polymerase chain reaction (PCR) from the plasmid pGENT using various primers. The PCR products were digested with NdeI and XhoI, and the DNA fragments were cloned into pET21b (Novagen) using T4 ligase (NEB). The induction of protein expression was initiated by adding IPTG to 1 mM followed by incubation at 37 °C for 6 h. After harvesting the bacteria by centrifugation (6000 rpm, 30 min, 4 °C), the bacterial cells were lysed with lysis buffer (50 mM Tris–HCl, pH 7.3, 150 mM NaCl, and 15 mM imidazole). The lysate was clarified by centrifugation (15,000 rpm, 30 min, 4 °C) to obtain soluble proteins. The truncated C‐terminal domains of the N protein with an C‐terminal His6‐tag were purified using a Ni–NTA column (Novagen) with an elution gradient from 15 to 250 mM imidazole in the buffer solution. The fractions containing the target proteins were collected and dialyzed against a low‐salt buffer.
2.2. Circular dichroism (CD) spectroscopy
The CD spectra were obtained using a JASCO‐815 CD spectropolarimeter. The temperature was controlled by circulating water at the desired temperature in the cell jacket. Each protein was dissolved in 50 mM Tris–HCl, pH 7.3, and 150 mM NaCl. The CD spectra were collected between 250 and 190 nm with a 1 nm bandwidth at 1 nm intervals. All of the spectra were obtained from an average of five scans. The photomultiplier absorbance did not exceed 600 V during the analysis. The CD spectra were normalized by subtraction of a background scan with buffer alone. The mean residue ellipticity, [θ], was calculated based on the equation [θ] = MRW × θλ/10 × l × c, where MRW is the mean residue weight, θλ is the measured ellipticity in millidegrees at wavelength λ, l is the cuvette path length (0.1 cm), and c is the protein concentration in g/ml. The results were analyzed using the CDSSTR program to calculate the percentage of each type of secondary structure [43]. In addition, the T m was determined from the polynomial fitting of the observed curve and taken as the temperature corresponding to half denaturation of the N protein. The first derivative of the absorption with respect to temperature, dA/dT, of the melting curve was computer generated and used to determine the T m.
2.3. Chemical crosslinking assay
To investigate the oligomerization features of N proteins, a chemical crosslinking experiment was performed. A series of protein solutions containing N proteins were supplemented with various concentrations of glutaraldehyde, and the reaction mixture was incubated at room temperature for 5 min. The reaction was stopped by adding 1 M Tris–HCl at pH 7.3 (0.5%, v/v, final concentration) and placing it on ice. The sample was then analyzed by SDS–PAGE.
2.4. Fluorescence spectroscopy
In the peptide‐induced fluorescence quenching experiments, a final concentration of 5 μM N protein was added to a buffer that contained various concentrations of peptide, and the samples were incubated at 25 °C for various durations. The buffer consisted of 50 mM Tris–HCl (pH 7.5) and 150 mM NaCl. The tryptophan fluorescence was measured using a Hitachi F‐4500 fluorescence spectrophotometer that was equipped with a cuvette of a 1 cm light path. The excitation wavelength was 288 nm, and the emission data were collected between 300 and 400 nm. For the static measurements, all of the measurements were recorded in triplicate. To determine the binding constant between the peptide and the N proteins, the peptide‐induced fluorescence changes (ΔF) from three separate experiments at 1 h after the addition of the test peptide were averaged and fit with the Hill equation using the GraphPad Prism software program (San Diego, CA) as follows: ΔF/ΔF max = 1/[1 + (K d/X)n], where ΔF max is the saturating value of the fluorescence change, X is the drug concentration, K d is the dissociation constant, and n is the Hill coefficient [44].
2.5. Size‐distribution analysis by analytical ultracentrifugation
Sedimentation velocity experiments were performed using a Beckman Optima XL‐A analytical ultracentrifuge. The sample solutions (380 μl) and the buffer solutions (400 μl) were loaded into the double‐sector centerpiece separately and built up in a Beckman An‐50 Ti rotor. The experiments were performed at 20 °C with a rotor speed of 42,000 rpm. The protein samples were monitored by measuring the UV absorbance at 280 nm in continuous mode with a time interval of 420 s and a step size of 0.002 cm. Multiple scans at different time points were fit to a continuous size distribution model using the program SEDFIT [45] (Fig. S1). All of the size distributions were solved at a confidence level of P = 0.95, a best‐fit average anhydrous frictional ratio (f/f 0), and a resolution N of 250 sedimentation coefficients between 0.1 and 20.0 S. To precisely determine the dimer–tetramer dissociation constants (K d,24) of the C‐terminal domains of the HCoV‐229E N proteins in dimer–tetramer–oligomer equilibrium, sedimentation velocity experiments were performed for three different protein concentrations. The dimer–tetramer dissociation constant (K d,24) of the C‐terminal domains of the HCoV‐229E N proteins was analyzed using the SEDPHAT program with a monomer‐m‐mer‐n‐mer self‐association model [46]. The sedimentation velocity data collected for three different protein concentrations were globally fit with SEDPHAT. The solvent density and viscosity were calculated by the SEDNTERP program (Philo, J. website http://www.jphilo.mailway.com/default.htm).
3. Results
3.1. Oligomerization characterization of the HCoV‐229E nucleocapsid protein C‐terminus
The C‐terminal region of the HCoV N protein has been shown to mediate the self‐association of the protein. The self‐association mechanism of the full‐length HCoV‐229E N protein has been previously reported [21, 47]. To determine whether the C‐terminal tail region (N351–389) plays an important role in the oligomerization of the C‐terminal domain of the HCoV‐229E N protein, the oligomerization of several regions of the C‐terminal domain of the N protein were analyzed using analytical ultracentrifugation. The differences in the size distributions among these truncated N proteins were analyzed by sedimentation velocity experiments, and the dimer–tetramer dissociation constant (K d,24) for each was determined. The dimer–tetramer dissociation constant (Kd,24) reflects the affinity between two N protein dimers, with smaller numbers representing a higher tendency to oligomerize into tetramers. The truncated N245–350 protein predominantly displays a dimeric quaternary structure with a small amount of tetramers (Fig. 2 A), exhibiting a K d,24 value of 256 μM. With an extended C‐terminal tail that includes residue 370 or residue 376, the respective truncated N245–370 and N245–376 proteins also exist as dimers in solution with the major peaks exhibiting K d,24 values of 177 and 159 μM, respectively (Fig. 2B and C). With an extended C‐terminal tail that includes residue 382 or residue 389, the respective truncated N245–382 and N245–389 proteins demonstrated significant shifts in the equilibrium from dimers to tetramers and octamers, with significant decreases in the K d,24 values (3.83 and 3.50 μM, respectively) (Fig. 3 A and B). These results indicate that residues 377–389 at the end of the C terminus are necessary for the oligomerization of the HCoV‐229 N protein.
Figure 2.
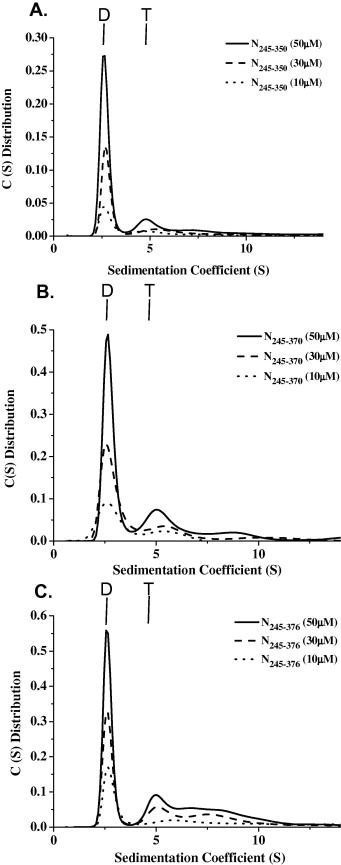
The continuous sedimentation coefficient distributions of the different HCoV‐229E N protein truncations (A) N245–350, (B) N245–370, and (C) N245–376. The protein concentrations were 50, 30 and 10 μM. The buffer consisted of 50 mM Tris–HCl (pH 7.5), 150 mM NaCl and 0.1% β‐mercaptoethanol. Dimer and tetramer are denoted as D and T.
Figure 3.
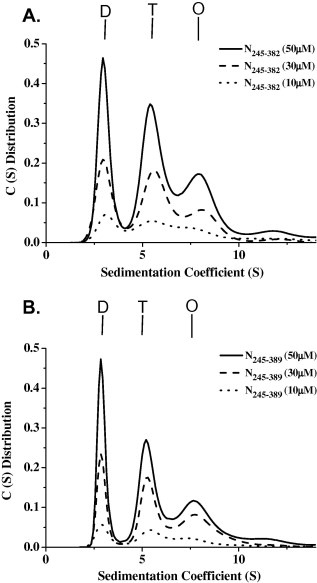
The continuous sedimentation coefficient distributions of the different HCoV‐229E N protein truncations (A) N245–382 and (B) N245–389. The protein concentrations were 50, 30 and 10 μM. The buffer consisted of 50 mM Tris–HCl (pH 7.5), 150 mM NaCl and 0.1% β‐mercaptoethanol. Dimer, tetramer, and octamer are denoted as D, T, and O.
3.2. Conformational and stability studies of the HCoV‐229E nucleocapsid protein C terminus
The conformation of the truncated C‐terminal domain of the HCoV‐229E N protein, including N245–350, N245–370, N245–376, N245–382, and N245–389, were monitored using CD spectroscopy. As shown in Fig. 4 A, the CD spectra of these truncated C‐terminal domains of the HCoV‐229E N protein were scanned from 190 to 250 nm at 25 °C. The CD spectrum of N245–350 showed well‐structured domains with α‐helical and β‐sheet secondary structures as well as two negative peaks at approximately 205 and 220 nm, respectively. The CD spectra of N245–370, N245–376, N245–382, and N245–389, which contain extended C‐terminal tails, showed increased intensities at approximately 205 and 220 nm, suggesting that they possessed a different secondary structure composition compared to N245–350. These CD spectra were further analyzed by the CDPro software to determine the quantitative percentages of the secondary structure (Table 1 ). N245–350 contains approximately 51% α‐helices, 22% β‐sheets, 12% turns, and 15% random coils at 25 °C. With an extended C‐terminal tail that includes residue 370, N245–370 showed a significant increase in undefined structural content with 14% turns and 17% random coils, consistent with the PrDOS prediction indicating that residues 351–370 are predicted to be disordered. Compared to N245–350, N245–370, and N245–376, N245–382 and N245–389 showed significant increases in the β‐sheet content because they may contain an ordered region in their C‐terminal tail as predicted by the PrDOS prediction.
Figure 4.
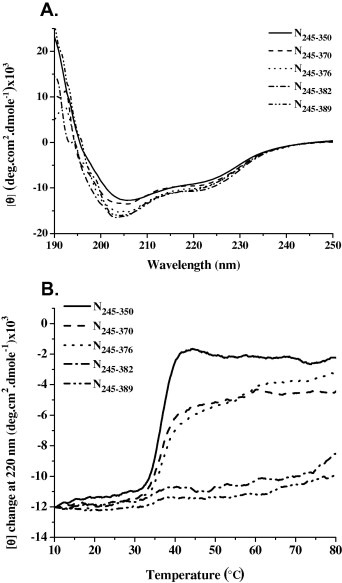
(A) The CD spectra of the HCoV‐229E N protein truncations N245–350, N245–370, N245–376, N245–382 and N245–389. The protein concentration was 5 μM, and the buffer consisted of 50 mM Tris–HCl (pH 7.5), 150 mM NaCl and 0.1% β‐mercaptoethanol. (B) The thermostability measurements of N245–350, N245–370, N245–376, N245–382 and N245–389 monitored by CD at 220 nm. The protein concentration was 7 μM, and the buffer consisted of 50 mM Tris–HCl (pH 7.5), 150 mM NaCl and 0.1% β‐mercaptoethanol.
Table 1.
The secondary structural content (%) of the truncated C‐terminal domain of the HCoV‐229E N protein as determined by CD analysis
| Construct | Helix | Sheet | Turn | Disordered | NRMSD |
|---|---|---|---|---|---|
| N245–350 | 51 a | 22 | 12 | 15 | 0.118 |
| N245–370 | 46 | 23 | 14 | 17 | 0.080 |
| N245–376 | 47 | 22 | 16 | 15 | 0.124 |
| N245–382 | 43 | 25 | 19 | 13 | 0.089 |
| N245–389 | 45 | 26 | 16 | 13 | 0.098 |
This value was determined by the CDSSTR program.
We also measured the melting temperatures (T ms) of the truncated C‐terminal domains of the HCoV‐229E N protein, including N245–350, N245–370, N245–376, N245–382, and N245–389, using CD in which the absorbance at 220 nm was analyzed as a function of temperature (Fig. 4B). The heat denaturation analysis showed that the T ms of N245–350, N245–370, and N245–376 were almost identical with values of 35.5, 36.2, and 36.7 °C, respectively. Interestingly, the T ms of N245–382 and N245–389, with values above 70 °C, were greater than N245–350, N245–370, and N245–376, indicating that oligomerization contributes significantly to the stability of the C‐terminal domain of the N protein. The melting temperatures (T ms) of the truncated C‐terminal domains of the HCoV‐229E N protein from CD studies were further confirmed by tryptophan (Trp) fluorescence analyses (Table S1).
3.3. Interference of the oligomerization of the HCoV‐229E nucleocapsid protein C terminus by a C‐terminal tail peptide
A previous observation indicated that a deletion mutant of the HCoV‐229E N protein C‐terminal domain lacking 13 amino acids from the C‐terminal tail appeared incapable of a high degree of oligomerization. To explore whether a C‐terminal tail peptide (residues 377–389) can compete with the oligomerization site of the C‐terminal domain of the HCoV‐229E N protein and interfere with its oligomerization, we synthesized a C‐terminal tail peptide (N377–389) and characterized the binding between N245–389 and this C‐terminal tail peptide. First, we utilized fluorescence to monitor the protein/peptide binding because N245–389 contains one tryptophan residue, which contributes to its intrinsic fluorescence. The fluorescence emission spectra for N245–389 showed the maximal emission wavelength at approximately 332 nm with a fluorescence intensity of 142.4 AU (Fig. 5 A). At the C‐terminal tail peptide concentrations of 5, 10, 20, 50, 75 and 100 μM, the C‐terminal tail peptide decreased the fluorescence intensity of N245–389 at approximately 332 nm by 8.7, 19, 23.5, 25.4, 26.9 and 28.9 AU, respectively, after 4 h of peptide addition. The N245–389 fluorescence decreased with increasing concentrations of the C‐terminal tail peptide, which suggests that this decrease reflected the interaction of N245–389 with the C‐terminal tail peptide. As shown in Fig. 5B, the fluorescence quenching of N245–389 by the C‐terminal tail peptide was analyzed with a Hill plot after the addition of the peptide. The dissociation constant of the C‐terminal tail peptide for N245–389 was calculated to be 7.43 μM.
Figure 5.
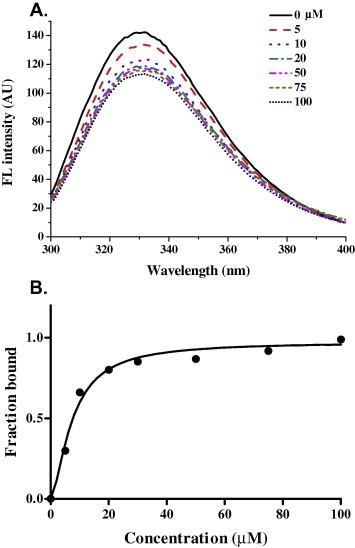
(A) The fluorescence spectra of N245–389 in Tris–HCl buffer with different concentrations of N377–389 peptide. The protein concentration was 5 μM, and the buffer consisted of 50 mM Tris–HCl (pH 7.3), 150 mM NaCl and 0.1% β‐mercaptoethanol. (B) Titration of the N245–389 protein with the N377–389 peptide in 50 mM Tris (pH 7.5), 150 mM NaCl and 0.1% β‐mercaptoethanol. The average of three experiments is shown. The data are expressed as a percentage of the maximal fluorescence change as determined by a fit to the Hill equation.
To quantify the effect of the C‐terminal tail peptide on the dimer–dimer association for N245–389, the dimer–tetramer dissociation constant (K d,24) for the N245–389 protein was determined in the absence and presence of the C‐terminal tail peptide. Sedimentation velocity (SV) experiments with increasing concentrations of the C‐terminal tail peptide were performed, and the data were globally fit to determine the dimer–tetramer dissociation constant of N245–389. Fig. 6 shows the distribution plots of the N245–389 protein in the absence and presence of the C‐terminal tail peptide. In the absence of the C‐terminal tail peptide, N245–389 formed a stable dimer, tetramer, and octamer with S‐values of approximately 2.85, 5.30 and 7.75, respectively, corresponding to the molecular masses of 31, 69, and 131 kDa, respectively. When the concentrations of the C‐terminal tail peptide were increased, the N245–389 tetramer and octamer peak decreased, whereas the N245–389 dimer peak increased (Fig. 6). When the molar ratio of N245–389/N377–389 was 0.25 and 0.1, the K d,24 value for N245–389 was approximately 6.1 and 9.7 μM, respectively, and it is significantly higher than that of N245–389 in the absence of the C‐terminal tail peptide. These results indicate that the N245–389 tetramer and octamer was significantly dissociated into dimers in the presence of the peptide (N377–389) and suggest that the C‐terminal tail peptide may complete the tetramer interface of N245–389 and interfere with the oligomerization of N245–389. We further analyze the effects of the peptide, N377–389, on the cell viability and viral titre of HCoV‐229E. The results showed that the cell viability was not affected by treatment with N377–389 alone (300 μM) for 24 h in A549 cell lines (Fig. S2A). In addition, viral titre of HCoV‐229E was inhibited by N377–389 at 300 μM, significantly (Fig. S2B).
Figure 6.
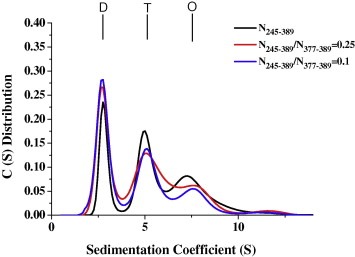
The continuous sedimentation coefficient distributions of the N245–382 protein in the presence of the N377–389 peptide. The protein concentration was 30 μM with two concentrations of the C‐terminal tail peptide, N377–389, at 120 and 300 μM. The buffer consisted of 50 mM Tris–HCl (pH 7.5), 150 mM NaCl and 0.1% β‐mercaptoethanol. Dimer, tetramer, and octamer are denoted as D, T, and O.
4. Discussion
The C‐terminal domains of the SARS–CoV and HCoV‐OC43 N proteins mediate the self‐association of the protein to form high‐order oligomers. These oligomers exist predominantly as dimers [47, 48]. The secondary structure alignment of the C‐terminal domains from the HCoV‐229E N protein with the corresponding proteins from SARS–CoV and IBV indicates that these proteins share very similar secondary structure profiles [49]. The crystal structures of the C‐terminal domains of SARS–CoV, IBV, and MHV N proteins show a similar general polypeptide fold, which strongly suggests that the dimerized N protein is the functional unit in vivo for the four groups of coronaviruses [35, 39]. The crystal structure of the C‐terminal domain shows a tightly intertwined twofold symmetric C‐terminal domain dimer, with a β‐hairpin (β1 and β2) from one subunit extending into the cavity of the opposite subunit, which forms an antiparallel β‐sheet with hydrogen bonds occurring across the dimer interface [39]. Chang et al. proposed that all coronaviruses employ the same interface mechanism for the dimerization of the N protein [48]. Based on the crystal structures of the N proteins from SARS–CoV, MHV, and IBV, the dimeric C‐terminal structural domain of the HCoV‐229E N protein has been mapped to N245–350. Here, analytical ultracentrifugation analysis consistently shows that the dimer appears to be the functional unit for the C‐terminal domain of the HCoV‐229E N protein. The dimeric N245–350 self‐associates into a small amount of tetramers. A crosslinking assay was also conducted to investigate oligomerization by the C‐terminal domain of the N protein. As shown in Fig. S3A, the crosslinking studies of N245–350 also detected dimers and tetramers.
The dimeric C‐terminal structural domain of the HCoV‐229E N protein (N245–350) is capped by the C‐terminal tail (N351–389). Compared to N245–350, the crosslinking results showed that N245–389 forms dimers and tetramers as well as high‐order oligomers (Fig. S3B). The analytical ultracentrifugation results were consistent with that of the chemical crosslinking analysis. Our results demonstrate that the C‐terminal tail plays a crucial role in N protein oligomerization. According to the PrDOS prediction, the C‐terminal tail (N351–389) is composed of disordered (N351–370), ordered (N371–382), and disordered (N383–389) regions. The truncations of the C‐terminal domain of the HCoV‐229E N protein, N245–370 and N245–376, which contain the first disordered region of the C‐terminal tail display a predominantly dimeric quaternary structure with a small amount of tetramers and high‐order oligomers and exhibit K d,24 values of 177 and 159 μM, respectively. Interestingly, when the dimeric C‐terminal structural domain of the HCoV‐229E N protein includes the C‐terminal tail to either residue 382 or 389, containing the short ordered region, the respective proteins, N245–382 and N245–389, demonstrated equilibrium shifts toward tetramers and octamers, indicating that the last 13 residues of the C‐terminal tail may play an important role in dimer–dimer association. A computer‐assisted prediction of the secondary structure based on the amino acid sequence predicted a short β‐strand at the end of the C‐terminal tail (Fig. S4), which is consistent with the CD results that showed that N245–382 and N245–389 contain higher β‐sheet content compared to the other C‐terminal domain truncations of the HCoV‐229E N protein. Therefore, we speculate that the hydrogen bonds across the tetramer interface formed by the main chain atoms of the short β‐strand may stabilize the oligomerization of the C‐terminal domain of the N protein through domain‐swapping (Fig. 7 ).
Figure 7.
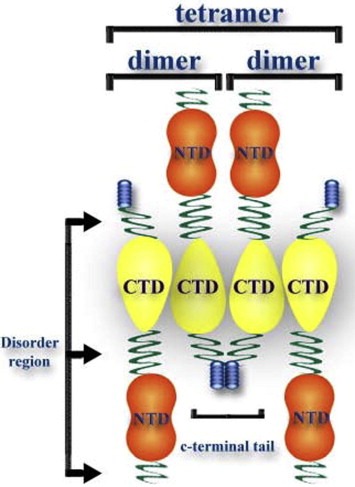
A schematic mechanism of tetramer formation by the N protein showing that the N proteins form a tetramer through the interactions between the C‐terminal tails of the dimer.
Oligomerization usually occurs through interfacial interactions in which subunits cooperatively interact with each other in several ways, including domain swapping and coiled–coil interaction [50]. Previous studies have shown that oligomerization makes a crucial contribution to the stability of proteins [51]. Many archaeal proteins have homo‐oligomeric structures, and some reports have shown a correlation between oligomerization and the hyperthermostability of archaeal proteins [52]. In this report, N245–382 and N245–389 show a very high degree of stability compared to the other C‐terminal domain truncations of the HCoV‐229E N protein due to a high degree of subunit interactions. The inhibition of viral N protein oligomerization by developing competing peptides and small organic compounds is an attractive therapeutic strategy against viral infection [53, 54, 55]. We showed that a peptide based on the C‐terminal tail interfered with the oligomerization of the C‐terminal domain of the HCoV‐229E N protein, N245–389 and performed the inhibitory effect on viral titre of HCoV‐229E. These results suggest that small molecules or peptides could be designed to target the oligomer interface as potential inhibitors of the CoV.
An amino acid sequence alignment of the C‐terminal domains from the HCoV‐229E N protein and corresponding proteins from SARS–CoV, IBV and HCoV‐OC43 using the MultAlin program reveals low sequence homology in the C‐terminal tail (Fig. S5) [49]. However, they share similar order–disorder profiles in the C‐terminal domain according to the PrDOS prediction (Fig. S6), suggesting that the oligomerization feature described above may be conserved across different groups of Coronaviridae. This study may assist the development of drugs to disrupt the oligomerization of the viral N protein and viral assembly.
Supporting information
Supplementary data 1
Supplementary Figures
Acknowledgments
This work was supported by the NSC Grant 100‐2113‐M‐005‐004‐MY3 to M.‐H.H. We thank Dr. Hui‐Chi Hung (Chung‐Hsing University) for her help on AUC experiments.
Appendix A 1.
1.1.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.febslet.2012.11.016.
Lo Yu-Sheng, Lin Shing-Yen, Wang Shiu-Mei, Wang Chin-Tien, Chiu Ya-Li, Huang Tai-Huang and Hou Ming-Hon (2013), Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein, FEBS Letters, 587, doi: 10.1016/j.febslet.2012.11.016
These authors equally contributed to this work.
References
- 1. St-Jean J.R., Jacomy H., Desforges M., Vabret A., Freymuth F., Talbot P.J., Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J. Virol., 78, (2004), 8824– 8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vabret A., Mourez T., Gouarin S., Petitjean J., Freymuth F., An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin. Infect. Dis., 36, (2003), 985– 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. El-Sahly H.M., Atmar R.L., Glezen W.P., Greenberg S.B., Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin. Infect. Dis., 31, (2000), 96– 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gagneur A., Sizun J., Vallet S., Legr M.C., Picard B., Talbot P.J., Coronavirus‐related nosocomial viral respiratory infections in a neonatal and paediatric intensive care unit: a prospective study. J. Hosp. Infect., 51, (2002), 59– 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wenzel R.P., Hendley J.O., Davies J.A., Gwaltney J.M. Jr., Coronavirus infections in military recruits. Three‐year study with coronavirus strains OC43 and 229E. Am. Rev. Respir. Dis., 109, (1974), 621– 624. [DOI] [PubMed] [Google Scholar]
- 6. Vabret A., Dina J., Gouarin S., Petitjean J., Corbet S., Freymuth F., Detection of the new human coronavirus HKU1: a report of 6 cases. Clin. Infect. Dis., 42, (2006), 634– 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vabret A., Mourez T., Dina J., van der Hoek L., Gouarin S., Petitjean J., Brouard J., Freymuth F., Human coronavirus NL63, France. Emerg. Infect. Dis., 11, (2005), 1225– 1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Skowronski D.M., Severe acute respiratory syndrome (SARS): a year in review. Annu. Rev. Med., 56, (2005), 357– 381. [DOI] [PubMed] [Google Scholar]
- 9. Drexler J.F., Genomic characterization of severe acute respiratory syndrome‐related coronavirus in European bats and classification of coronaviruses based on partial RNA‐dependent RNA polymerase gene sequences. J. Virol., 84, (2010), 11336– 11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Woo P.C., Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol., 79, (2005), 884– 895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leng Q., Bentwich Z., A novel coronavirus and SARS. N. Engl. J. Med., 349, (2003), 709– [DOI] [PubMed] [Google Scholar]
- 12. Hogue B.G., Brian D.A., Structural proteins of human respiratory coronavirus OC43. Virus Res., 5, (1986), 131– 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lai M.M., Cavanagh D., The molecular biology of coronaviruses. Adv. Virus Res., 48, (1997), 1– 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hiscox J.A., RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol., 5, (2007), 119– 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. You J.H., Reed M.L., Dove B.K., Hiscox J.A., Three‐dimensional reconstruction of the nucleolus using meta‐confocal microscopy in cells expressing the coronavirus nucleoprotein. Adv. Exp. Med. Biol., 581, (2006), 313– 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stohlman S.A., Fleming J.O., Patton C.D., Lai M.M., Synthesis and subcellular localization of the murine coronavirus nucleocapsid protein. Virology, 130, (1983), 527– 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Masters P.S., Parker M.M., Ricard C.S., Duchala C., Frana M.F., Holmes K.V., Sturman L.S., Structure and function studies of the nucleocapsid protein of mouse hepatitis virus. Adv. Exp. Med. Biol., 276, (1990), 239– 246. [DOI] [PubMed] [Google Scholar]
- 18. Masters P.S., Sturman L.S., Background paper. Functions of the coronavirus nucleocapsid protein. Adv. Exp. Med. Biol., 276, (1990), 235– 238. [DOI] [PubMed] [Google Scholar]
- 19. Pyrc K., Jebbink M.F., Berkhout B., van der Hoek L., Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J., 1, (2004), 7– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baric R.S., Nelson G.W., Fleming J.O., Deans R.J., Keck J.G., Casteel N., Stohlman S.A., Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol., 62, (1988), 4280– 4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang T.K., Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses. Proteomics, 5, (2005), 925– 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsieh P.K., Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus‐like particles is nucleocapsid dependent. J. Virol., 79, (2005), 13848– 13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P., Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol., 81, (2007), 548– 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Du L., Priming with rAAV encoding RBD of SARS–CoV S protein and boosting with RBD‐specific peptides for T cell epitopes elevated humoral and cellular immune responses against SARS–CoV infection. Vaccine, 26, (2008), 1644– 1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Surjit M., Liu B., Chow V.T., Lal S.K., The nucleocapsid protein of severe acute respiratory syndrome‐coronavirus inhibits the activity of cyclin–cyclin‐dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem., 281, (2006), 10669– 10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zuniga S., Sola I., Moreno J.L., Sabella P., Plana-Duran J., Enjuanes L., Coronavirus nucleocapsid protein is an RNA chaperone. Virology, 357, (2007), 215– 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chan K.H., Serological responses in patients with severe acute respiratory syndrome coronavirus infection and cross‐reactivity with human coronaviruses 229E, OC43, and NL63. Clin. Diagn. Lab. Immunol., 12, (2005), 1317– 1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mourez T., Vabret A., Han Y., Dina J., Legrand L., Corbet S., Freymuth F., Baculovirus expression of HCoV‐OC43 nucleocapsid protein and development of a Western blot assay for detection of human antibodies against HCoV‐OC43. J. Virol. Methods, 139, (2007), 175– 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kamahora T., Soe L.H., Lai M.M., Sequence analysis of nucleocapsid gene and leader RNA of human coronavirus OC43. Virus Res., 12, (1989), 1– 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chang C.K., Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci., 13, (2006), 59– 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chang C.K., Hsu Y.L., Chang Y.H., Chao F.A., Wu M.C., Huang Y.S., Hu C.K., Huang T.H., Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol., 83, (2009), 2255– 2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luo H., Chen J., Chen K., Shen X., Jiang H., Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: self‐association analysis and nucleic acid binding characterization. Biochemistry, 45, (2006), 11827– 11835. [DOI] [PubMed] [Google Scholar]
- 33. Chen C.Y., Chang C.K., Chang Y.W., Sue S.C., Bai H.I., Riang L., Hsiao C.D., Huang T.H., Structure of the SARS coronavirus nucleocapsid protein RNA‐binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol., 368, (2007), 1075– 1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fan H., Ooi A., Tan Y.W., Wang S., Fang S., Liu D.X., Lescar J., The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N‐terminal domain and multimerization properties. Structure, 13, (2005), 1859– 1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jayaram H., Fan H., Bowman B.R., Ooi A., Jayaram J., Collisson E.W., Lescar J., Prasad B.V., X‐ray structures of the N‐ and C‐terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol., 80, (2006), 6612– 6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nelson G.W., Stohlman S.A., Localization of the RNA‐binding domain of mouse hepatitis virus nucleocapsid protein. J. Gen. Virol., 74, Pt 9 (1993), 1975– 1979. [DOI] [PubMed] [Google Scholar]
- 37. Spencer K.A., Hiscox J.A., Characterisation of the RNA binding properties of the coronavirus infectious bronchitis virus nucleocapsid protein amino‐terminal region. FEBS Lett., 580, (2006), 5993– 5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang C.Y., Hsu Y.L., Chiang W.L., Hou M.H., Elucidation of the stability and functional regions of the human coronavirus OC43 nucleocapsid protein. Protein Sci., 18, (2009), 2209– 2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu I.M., Oldham M.L., Zhang J., Chen J., Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona‐ and arteriviridae. J. Biol. Chem., 281, (2006), 17134– 17139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saikatendu K.S., Joseph J.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P., Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N‐terminal domain of N protein. J. Virol., 81, (2007), 3913– 3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Takeda M., Chang C.K., Ikeya T., Guntert P., Chang Y.H., Hsu Y.L., Huang T.H., Kainosho M., Solution structure of the c‐terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL‐NMR method. J. Mol. Biol., 380, (2008), 608– 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Masters P.S., The molecular biology of coronaviruses. Adv. Virus Res., 66, (2006), 193– 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sreerama N., Woody R.W., A self‐consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem., 209, (1993), 32– 44. [DOI] [PubMed] [Google Scholar]
- 44. Hung H.C., Development of an anti‐influenza drug screening assay targeting nucleoproteins with tryptophan fluorescence quenching. Anal. Chem., 84, (2012), 6391– 6399. [DOI] [PubMed] [Google Scholar]
- 45. Schuck P., Size‐distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J., 78, (2000), 1606– 1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vistica J., Dam J., Balbo A., Yikilmaz E., Mariuzza R.A., Rouault T.A., Schuck P., Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem., 326, (2004), 234– 256. [DOI] [PubMed] [Google Scholar]
- 47. Yu I.M., Gustafson C.L., Diao J., Burgner J.W. 2nd, Li Z., Zhang J., Chen J., Recombinant severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein forms a dimer through its C‐terminal domain. J. Biol. Chem., 280, (2005), 23280– 23286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chang C.K., The dimer interface of the SARS coronavirus nucleocapsid protein adapts a porcine respiratory and reproductive syndrome virus‐like structure. FEBS Lett., 579, (2005), 5663– 5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Corpet F., Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res., 16, (1988), 10881– 10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Meier M., Stetefeld J., Burkhard P., The many types of interhelical ionic interactions in coiled coils – an overview. J. Struct. Biol., 170, (2010), 192– 201. [DOI] [PubMed] [Google Scholar]
- 51. Ogasahara K., Khechinashvili N.N., Nakamura M., Yoshimoto T., Yutani K., Thermal stability of pyrrolidone carboxyl peptidases from the hyperthermophilic Archaeon, Pyrococcus furiosus . Eur. J. Biochem., 268, (2001), 3233– 3242. [DOI] [PubMed] [Google Scholar]
- 52. Tanaka Y., Tsumoto K., Yasutake Y., Umetsu M., Yao M., Fukada H., Tanaka I., Kumagai I., How oligomerization contributes to the thermostability of an archaeon protein. Protein l‐isoaspartyl‐O‐methyltransferase from Sulfolobus tokodaii . J. Biol. Chem., 279, (2004), 32957– 32967. [DOI] [PubMed] [Google Scholar]
- 53. Hayouka Z., Inhibiting HIV‐1 integrase by shifting its oligomerization equilibrium. Proc. Natl. Acad. Sci. USA, 104, (2007), 8316– 8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pujol A., Deleu L., Nuesch J.P., Cziepluch C., Jauniaux J.C., Rommelaere J., Inhibition of parvovirus minute virus of mice replication by a peptide involved in the oligomerization of nonstructural protein NS1. J. Virol., 71, (1997), 7393– 7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shen Y.F., E339…R416 salt bridge of nucleoprotein as a feasible target for influenza virus inhibitors. Proc. Natl. Acad. Sci. USA, 108, (2011), 16515– 16520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data 1
Supplementary Figures


