Abstract
Fungi have been used since ancient times in food and beverage-making processes and, more recently, have been harnessed for the production of antibiotics and in processes of relevance to the bioeconomy. Moreover, they are starting to gain attention as a key component of the human microbiome. However, fungi are also responsible for human infections. The incidence of community-acquired and nosocomial fungal infections has increased considerably in recent decades. Antibiotic resistance development, the increasing number of immunodeficiency- and/or immunosuppression-related diseases and limited therapeutic options available are triggering the search for novel alternatives. These new antifungals should be less toxic for the host, with targeted or broader antimicrobial spectra (for diseases of known and unknown etiology, respectively) and modes of actions that limit the potential for the emergence of resistance among pathogenic fungi. Given these criteria, antimicrobial peptides with antifungal properties, i.e., antifungal peptides (AFPs), have emerged as powerful candidates due to their efficacy and high selectivity. In this review, we provide an overview of the bioactivity and classification of AFPs (natural and synthetic) as well as their mode of action and advantages over current antifungal drugs. Additionally, natural, heterologous and synthetic production of AFPs with a view to greater levels of exploitation is discussed. Finally, we evaluate the current and potential applications of these peptides, along with the future challenges relating to antifungal treatments.
Keywords: antifungal peptides, antimicrobial peptides, mycoses, antimicrobial resistance, production, new therapies
Introduction
Fungi are extraordinary, ubiquitous organisms that play critical roles in complex ecosystems. These eukaryotes range from giant forms to microscopic unicellular molds and yeasts. In recent years fungi have been recognized as an integral part of our commensal microbiota at different body sites (e.g., gut, oral cavity, skin, lung, vagina), although there is no consensus on what constitutes the standard mycobiome composition (Huffnagle and Noverr, 2013; Enaud et al., 2018; Kapitan et al., 2018) and some studies point out that gut fungi may come from oral and dietary sources (Auchtung et al., 2018). Indeed, fungi have been used as a source of food and for food processing for thousands of years (Campbell-Platt and Cook, 2008). Fungi are also routinely employed in many industrial processes including the production of peptides, enzymes, vitamins, organic acids, and antibiotics (Money, 2016; Mukherjee et al., 2018).
However, fungal infections, or mycoses, have become a serious threat to human health, causing a wide range of infections in humans. It is estimated that fungal diseases affect more than one billion people globally, of whom 150 million suffer from severe infections (Bongomin et al., 2017). These range from superficial and subcutaneous life quality-debilitating infections affecting the skin, keratinous tissues and mucous membranes (Kaushik et al., 2015), to systemic infections that can be life-threatening involving the brain, heart, lungs, liver, spleen, and kidneys (Rautemaa-Richardson and Richardson, 2017). The latter are especially worrying in the case of immunocompromised patients with HIV/AIDS or autoimmune diseases, and in those undergoing anticancer chemotherapy or organ transplantation.
The main human fungal pathogens are Candida albicans, Cryptococcus neoformans, and Aspergillus fumigatus, but, worryingly, non-albicans Candida spp. such as C. auris, in addition to other infectious agents such as Histoplasma capsulatum or Malassezia furfur are emerging. For a comprehensive review on main fungal pathogens affecting humans see (Roemer and Krysan, 2014).
Four major classes of antifungal agents dominate the market: azoles, which inhibit the synthesis of ergosterol; polyenes, which interact with fungal membrane sterols physicochemically; echinocandins that inhibit glucan synthesis; and fluorinated pyrimidines, which interfere with pyrimidine metabolism, leading to the inhibition of DNA and RNA biosynthesis (Roemer and Krysan, 2014). However, the high mortality of invasive fungal infections, the long course of treatments required, narrow spectrum activity and cross-resistance due to similar mechanisms of action across drugs has triggered the search for safer alternatives with reduced toxicity or other enhanced features. As eukaryotes, a particularly great challenge is to identify pathogen-specific targets not present in human cells.
Monoclonal antibodies, cytokine immunotherapy, vaccines and antimicrobial peptides (AMPs) have emerged as new biopharmaceuticals to prevent or treat fungal infections (Nicola et al., 2019). There is an increasing interest in peptides as promising novel antibiotic agents. Peptides can mimic natural ligands and therefore function as agonists or antagonists. Regarding their use as drugs, peptides are highly selective, effective and well-tolerated (Fosgerau and Hoffmann, 2015). Among the broader peptide category of antimicrobials, AMPs are gene-encoded conserved molecules produced by all organisms, from bacteria to humans. Compared with conventional antibiotics, which are generally targeted against bacteria or fungi, AMPs can exhibit broad antimicrobial activity including bacteria, fungi, parasites, viruses, protozoa and even some cancer cells (Hancock and Chapple, 1999). Being effective against this broad range of targets might imply different modes of action and prevent bacteria and fungi from developing resistance. AMPs produced by higher organisms are involved in the innate and secondary immune responses against microbes, while those produced by bacteria frequently kill other bacteria competing for the same ecological niche (Zhang and Gallo, 2016). AMPs also confer protection by contributing to gut homeostasis, and modulation of host inflammatory responses. Notably, AMPs with a narrow antimicrobial spectrum have particularly great therapeutic potential as they are less likely to cause disruption of the host microbiota.
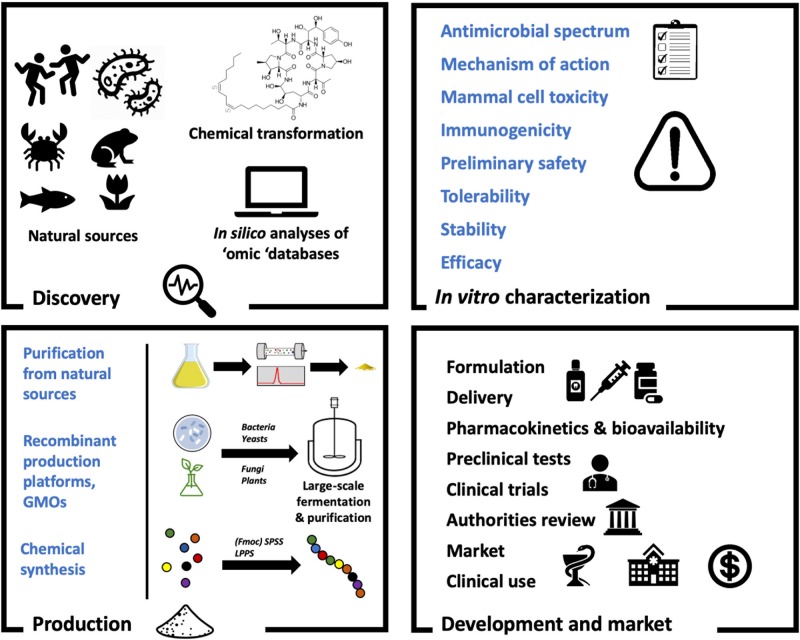
In this review we provide an overview of the bioactivity and classification of AMPs with antifungal activity, known as antifungal peptides (AFPs), as well as their mode of action and advantages over current antifungal drugs. Additionally, natural, heterologous and synthetic production of AFPs with a view to greater levels of exploitation is discussed. In this regard, Figure 1 shows a general overview on AFP development. Finally, we evaluate the current and potential applications of these peptides, together with future challenges in relation to antifungal treatments.
Figure 1.
The antifungal peptide development process. As with any drugs, AFPs must undergo several stages of development to reach clinical use. When the candidate molecule shows promise as a therapeutic (Discovery) it must be characterized (In vitro characterization). In order to facilitate this, sufficient amounts of the peptide must be available (Production). Finally, the molecule will be subjected to formulation processes and preclinical tests before going into clinical trials and receive approval (Development and market).
Types of Antifungal Peptides and Bioactivity
As of November 2019, there were 1,133 peptides with antifungal properties reported in the Antimicrobial Peptide Database (APD3) (Wang et al., 2016). AFPs have been classified following a number of different criteria, such as structure or mode of action. However, the most accepted classification is based on the peptide origin: natural, semisynthetic or synthetic (De Lucca, 2000). Here, we summarize some of the most important features of natural peptides and we describe how synthetic AFPs are designed.
Natural Peptides
Natural AFPs are produced by a number of different species of Bacteria, Archaea, and Eukarya isolated from natural sources (De Lucca and Walsh, 1999). Most natural AFPs have been discovered by testing their antagonistic activity in vitro against pathogenic fungi (Mania et al., 2010; Freitas and Franco, 2016; McNair et al., 2018). However, with the rise of sequencing technologies and the drop in associated costs, new strategies for prediction and discovery of new AFPs are emerging. New methods such as template-based, docking simulations, hidden Markov model and other sequence-based methods allow for novel in silico prediction of AFPs (Schneider and Fechner, 2005; Fjell et al., 2007, 2008; Robinson, 2011; Garrigues et al., 2017; Agrawal et al., 2018).
Natural AFPs are grouped in families according to their origin (Table 1). Those produced by bacteria and fungi are arguably of greatest interest with respect to medical applications (Essig et al., 2014). Although some other sources, such as plants, are a rich source of AFPs, they are mainly employed for other purposes, such as the control of phytopathogens, and thus, they are not further discussed in this review (De Lucca, 2000). Notably, the first archaeal AMP with antifungal and anti-biofilm capabilities against clinical fungal pathogens was recently reported (Roscetto et al., 2018).
Table 1.
Natural AFPs families with clinical applications.
| Origin | Family name | Chemical characteristics | Mode of action | Active against | Examples | References |
|---|---|---|---|---|---|---|
| Archaea | Cryptic CAMP-like | Cationic | Targets cell wall (still being investigated) | Candida spp. | VLL-28 | Roscetto et al., 2018 |
| Bacteria | Iturin | Small cyclic peptidolipids with a lipid-soluble β-amino acid linked to a peptide with D- and L- amino acids. | Lysis by pore formation in membranes |
niger
albicans F. oxysporum |
Iturin A Bacillomycin F Bacillomycin L |
Landy et al., 1948; De Lucca and Walsh, 1999; De Lucca, 2000 |
| Syringomycins | Small cyclic lipodepsipeptides | Forms voltage-sensitive ion channels Alters protein phosphorylation and H+-ATPase activity Pore formation |
Candida spp. Cryptococcus spp. Aspergillus spp. |
Syringomycin-E (SE) Syringostatin A Syringotoxin B |
Sinden et al., 1971; De Lucca and Walsh, 1999; De Lucca, 2000 | |
| Fungi | Nikkomycins | Peptide nucleosides | Inhibit chitin biosynthesis | Blastomyces dermatitidis, C. albicans | Nikkomycin X, Z | McCarthy et al., 1985; Hector et al., 1990; De Lucca and Walsh, 1999; De Lucca, 2000 |
| Polyoxins | Peptide nucleosides | Inhibit chitin biosynthesis | C. albicans | Polyoxin A, B, D | Suzuki et al., 1965; Isono et al., 1969; Hori et al., 1974; De Lucca and Walsh, 1999; De Lucca, 2000 | |
| Echinocandins | Cyclic hexapeptides with N-linked acyl lipid side chains | Inhibit glucan synthesis |
Candida spp. Aspergillus spp. |
Echinocandins, pneumocandins, aculeacins, mulundocandins, WF11899 | De Lucca and Walsh, 1999; De Lucca, 2000; Eschenauer et al., 2007 | |
| Aureobasidins | Cyclic depsipeptide | Lysis by altering actin assembly and delocalizing chitin in fungal walls / sphingolipid synthesis inhibition |
B. dermatitidis
Candida spp. |
Aureobasidin A | Endo et al., 1997; De Lucca, 2000 | |
| Leucinostatins | Contains five unusual amino acids, 4-methylproline (MePro), 2-amino-6-hydroxy-4-methyl- 8-oxodecanoic acid (AHMOD), threo-β-hydroxyleucine (HyLeu), three 2-aminoisobutyric acid (Aib), and β-alanine (b-Ala) |
Uncouplers mitochondria | Candida spp. | Leucinostatins A, B, D, H and K | De Lucca and Walsh, 1999; De Lucca, 2000; Abe et al., 2018 | |
| Amphibians | Magainins | Helical, amphiphilic | Lysis by dissipating ion gradient in cell membranes | C. albicans | Magainin 2 | De Lucca and Walsh, 1999; De Lucca, 2000 |
| Dermaseptins | Linear, cationic, lysine-rich | Lysis by interacting with lipid bilayers |
flavus
fumigatus F. oxysporum |
Dermaseptin | Mor et al., 1991; De Lucca and Walsh, 1999; De Lucca, 2000 | |
| Skin-PYY | Similar to neuropeptide NPY and gastrointestinal peptide PYY, C- terminal α-helix domain conserved | Membrane disruption |
C. neoformans
C. albicans A. fumigatus |
Skin-PYY | Vouldoukis et al., 1996; De Lucca, 2000 | |
| Plants | Defensins | Small, highly stable, cysteine-rich peptides | Membrane pore formation (carpet or toroidal pore), ion efflux, induction of reactive oxygen species and programmed cell death. | C. albicans, C. krusei, A. flavus and Fusarium solani | RsAFP1, RsAFP2, SPE10, NaD1 | Bondaryk et al., 2017; Sher Khan et al., 2019 |
| Insects | cecropins | Linear | Cell lysis | A. fumigatus | Cecropins A and B | De Lucca et al., 1998; Bondaryk et al., 2017 |
| Cysteine-rich peptides | Hairpin-like beta-sheet structure | Cell lysis | C. albicans | Defensins, Drosomycin, thanatin | Dimarcq et al., 1998; Bondaryk et al., 2017 | |
| Peptides from aquatic sources | Aciculitins | Cyclic peptides and lipid residues | Cell lysis | C. albicans | Aciculitins A-C | Bewley and Faulkner, 1994; Bondaryk et al., 2017 |
| Theonegramide | Glycopeptide with unusual amino acids | Unknown | C. albicans | Theonegramide | Bewley and Faulkner, 1994; Bondaryk et al., 2017 | |
| Laxaphycins | Cyclic peptides | Unknown | C. albicans | Laxaphycins A, B, D and E | Bondaryk et al., 2017 | |
| Defensins | β-sheet | Chitin binding | C. albicans | Tachycitin, “big defensin” | Bondaryk et al., 2017 | |
| Mammalian | α-defensins | β-sheet with cysteines forming intramolecular disulphide bonds | Cell lysis |
C. albicans
C. neoformans |
HNP-1, HNP-2, HNP-3, NP-1, NP-2, NP-3, NP-4 | De Lucca and Walsh, 1999; De Lucca, 2000 |
| β-defensins | β-sheet with cysteines with a disulphide motif different from α-defensins. Amino termini are blocked with a pyroglutamyl residue | Cell lysis | C. albicans | Tracheal antimicrobial protein (TAP) Gallinacins−1,−1α, 2 |
De Lucca and Walsh, 1999; De Lucca, 2000 | |
| Protegrins Cathelicidins |
Cationic, cysteine-rich β-defensins | Pore formations and lysis | C. albicans | Protegrins 1, 2 and 3 | De Lucca and Walsh, 1999; De Lucca, 2000; Bondaryk et al., 2017 | |
| Histatins | Basic and neutral helical peptides | Induction of cell death, osmosis stress | C. albicans | Histatins 1, 3, 5 | Koshlukova et al., 1999; De Lucca, 2000; Bondaryk et al., 2017 |
Typically, natural AFPs adopt an α-helix structure, β-hairpin or sheet (containing two cysteine residues) or mixed α-helix/β-sheet structures upon interaction with membranes. Some of these peptides are rich in specific amino acids and, as a result, are classified as glycine-rich, proline-rich, arginine-rich, histidine-rich, and tryptophan-rich (Bondaryk et al., 2017). However, the structure of most AFPs has not yet been determined.
Posttranslational modifications also play a major role in the final three-dimensional structure and bioactivity of AMPs but, unfortunately, cannot be predicted with current in silico tools (Agrawal et al., 2018). The most common posttranslational modifications in natural AFPs involve glycosylation or the addition of carbohydrates, normally observed in asparagine, or serine/threonine residues (Guo et al., 2012; Bednarska et al., 2017). The latter has been shown to improve antifungal activity in some cases. Other modifications include halogenation such as chlorination (Andreu and Rivas, 1998; Shinnar et al., 2003), phosphorylation, which can increase stability (McDonald et al., 2011) or hydroxylation, mainly observed at lysine, arginine, tryptophan, and phenylalanine residues with differing effects on antifungal activity (Houwaart et al., 2014; Akkam, 2016). Finally, cyclization of AMPs, which is not considered a posttranslational modification, improves antimicrobial activity in general, reduces toxicity and improves stability against proteases (Akkam, 2016).
Semisynthetic and Synthetic Peptides and Structure Activity Relationships (SAR) Criteria for Their Design
Antimicrobial semisynthetic and synthetic peptides are generally made with a view to improving pharmacological properties, reducing side effects and/or lowering the immunogenicity of natural peptides. Pharmaceutical formulation will also help to enhance their stability and bioavailability. As an example of synthetic transformation, it was found that the hemolytic activity of the natural echinocandin B AFP was significantly reduced by the replacement of the linoleoyl side-chain with either octyloxybenzoyl (cilofungin) or pentyloxyterphenyl (anidulafungin) side chains (Emri et al., 2013).
Structure-activity relationships (SAR) are a key element used for the design and development of synthetic peptides (Lum et al., 2015). There are several biophysical properties that can determine antifungal activity, such as net charge, stereospecificity, hydrophobicity, amphipathicity, secondary structure and peptide length, with some of these characteristics being interdependent. These properties have been extensively reviewed previously (Akkam, 2016). Most AFPs are cationic, but neutral and anionic AFPs have also been described. When present, cationic charges play a role in the electrostatic binding of AMPs to negatively charged membranes. Therefore, an increase in the positive net charge beyond a threshold might lead to a stronger activity on the membrane. However, positive charges are not a prerequisite for antimicrobial activity, and anionic peptides interact with the membrane peptide through specific amino acid distributions (Yeaman and Yount, 2003; Lakshminarayanan et al., 2014).
Most AFPs are non-stereospecific (Akkam, 2016). In the same way, there is no dominant conformation among AFPs. Thus, the main differences between peptides come from sequence and secondary structure variation (Emri et al., 2013). Hydrophobicity and amphipathicity are essential factors for peptide membrane interactions and membrane permeabilization (Lum et al., 2015), and important variables in the design of synthetic peptides. Hydrophobicity is defined as the percentage of hydrophobic residues within a peptide, ranging between 30 and 60% for most AMPs. An increase in hydrophobicity and amphipathicity usually correlates with an increase in antifungal activity, but also with higher hemolytic activity. The presence of tryptophan is also linked to hemolytic activity since it interferes with lipid polymorphism in the membranes (Schibli et al., 2002).
The peptide length of AFPs is important for the secondary structure and mode of action. Most AFPs have 11–40 residues. It has been described that 7–8 amino acids are needed to form amphipathic structures in AMPs (Bahar and Ren, 2013), while <20 amino acids limit the ability of a peptide to form transmembrane structures in the fungal membrane (Rothstein et al., 2001; Akkam, 2016). Nevertheless, longer lengths may also affect cytotoxicity, stability and manufacturing costs. In order to overcome these hurdles, short antimicrobial peptides (SAMPs), containing 2–10 amino acids, are attracting attention as less toxic and more stable alternatives. Overall, SAMPs have simpler amino acid composition and they are easier to synthesize and modify chemically with a view to improving toxicity, stability, half-life or specificity. Moreover, they are also less immunogenic (Duncan and O'Neil, 2013; Fox, 2013).
Some of the semisynthetic and synthetic peptides studied with a view to clinical applications are summarized in Table 2. Considering all of the factors mentioned above, different strategies have been employed for synthetic peptide design. Combinatorial libraries generate model peptides suitable for SAR studies (Blondelle and Lohner, 2000). The template-based strategy, known as de novo design, uses peptides with known antifungal activity as scaffolds (Lum et al., 2015). As an example, Agrawal et al., performed in silico studies by machine learning techniques based on AFP and non-AFP sequences revealing a higher frequency of certain residues (C, G, H, K, R, and Y) and the prevalence of R, V, K at N-terminus and C and H at the C-terminus of peptides (Agrawal et al., 2018).
Table 2.
Examples of semisynthetic and synthetic AFPs with clinical applications.
| Origin | Name | Structure | Mode of action | Active against | References |
|---|---|---|---|---|---|
| Semisynthetic | Cilofungin (LY 121019) | Lipopeptide | Glucan synthesis |
C. albicans
A. fumigatus |
Pfaller et al., 1989; De Lucca, 2000 |
| LY 303366 | Lipopeptide | Glucan synthesis | Candida spp. | Karlowsky et al., 1997; De Lucca, 2000 | |
| FK 463 | Lipopeptide | Glucan synthesis |
Candida spp. Aspergillus spp. |
De Lucca, 2000; Mikamo et al., 2000 | |
| L-693,989 | Lipopeptide | Glucan synthesis |
C. albicans
P. carinii |
Balkovec et al., 1992; De Lucca, 2000 | |
| PMAP-23 | α-helix | Membrane | C. albicans | Lee et al., 2002; Bondaryk et al., 2017 | |
| KU2 | α-helix | Membrane | C. albicans | Lum et al., 2015; Bondaryk et al., 2017 | |
| KU3 | α-helix | Membrane | C. albicans | Lum et al., 2015; Bondaryk et al., 2017 | |
| Synthetic | dF21-10K | Linear-kaxins | Membrane |
C. albicans
C. tropicalis |
Burrows et al., 2006; Bondaryk et al., 2017 |
| KSL-W | α-helix decapeptide | Membrane | C. albicans | Semlali et al., 2011; Bondaryk et al., 2017 | |
| B4010 | Tetravalent dendron—Polylysine dendrons (PLL) | Membrane | C. albicans | Lakshminarayanan et al., 2014; Freitas and Franco, 2016; Bondaryk et al., 2017 | |
| L1 | Polyamidoamine (PAMAM) dendrimers | Intercalation with DNA | Candida spp. | Ottaviani et al., 2016; Bondaryk et al., 2017 | |
| Killer peptide (KP) | Dimeric; β-sheet | Unknown |
Candida spp. Cryptococcus neoformans P. carinii Paracoccidioides brasiliensis A. fumigatus |
Magliani et al., 2011 |
Another strategy is to target virulence traits (Bondaryk et al., 2017). However, this approach is not efficient enough due to the expression of different virulence factors in fungi cells. For this reason, the generation of molecules with multiple ligands like dendrimers, or tree-like molecules, that have radially distributed layers of branches named generations, have been explored (Esfand and Tomalia, 2001).
Models of Mechanisms of Action
Whereas some peptides have primarily antifungal activity, such as lipopeptides (e.g., echinocandins) or histidine-rich (e.g., the linear histatins or branched HK), membrane-disrupting peptides (e.g., magainins, protegrins) have a broader antimicrobial spectrum, including bacteria, fungi and viruses. Here, we summarize the hypothetical mechanisms of action of AFPs with activity against fungal pathogens.
Peptides With Primarily Antifungal Properties
Inhibition of 1,3-β-Glucan Synthesis
β-glucan synthase is involved in cell wall integrity. Cyclic lipoproteins can non-competitively inhibit it, resulting in destabilization of the cell wall, leading to susceptibility to osmotic stresses and cell lysis. 1,3-β-glucans are involved in the division septum and assembly of the acropore wall as well; consequently, β-glucan synthase inhibitors affect these structures. β-glucan synthase is ubiquitous among fungi including Candida, Aspergillus, Cryptococcus, and Pneumocystis species. However, in mycelious fungi, it is found in tips of the growing hyphae, which makes them less sensitive. Inhibition of β-glucan synthase results in negative feedback, causing cell cycle arrest. Echinocandins and pneumocandins, aculeacins (A-D, F), mulundocandins and WF11899 (A, B, and C) as well as the killer toxin from Williopsis mrakii, WmKT are representative β-glucans synthase inhibitors (Guyard et al., 2002; Matejuk et al., 2010; van der Weerden et al., 2013).
Inhibition of Chitin Biosynthesis in the Cell Wall
Chitin, found in the fungal cell wall, is essential to maintain cell integrity and is absent in vertebrates. Aureobasidins are cyclic lipophilic 8-mer depsipeptides with an α-hydroxyacid that display two mechanisms of action: the disruption of cell wall/membranes by altering the assembly of actin and chitin (Endo et al., 1997) and the interruption of sphingolipids synthesis (Nagiec et al., 1997). Several members of the aureobasidin family exert anti-Candida activity. Nikkomycins are structural analogs of uridine diphosphate N-acetylglucosamine, a major constituent of chitin. These AFPs have been shown to inhibit the synthesis of chitin in C. albicans in both in vitro and in vivo studies (McCarthy et al., 1985) while human cells were not affected. They also show significant activity against C. immitis, B. dermatitidis, and moderate activity against H. capsulatum (Hector et al., 1990; Clemons and Stevens, 1997), but these agents are not active against filamentous fungi.
Selective Activity on Membranes
Rs-ARF2 is a 50 amino-acid residue plant defensin that has three-stranded β-sheets and an α-helix, structure that is stabilized by four disulfide bonds. Rs-ARF2 targets the fungus-specific membrane glucosylceramide inducing membrane permeability, which causes Ca2+ uptake, efflux of K+ and medium alkalinization. This defensin also induces the production of toxic reactive oxygen species intracellularly. Rs-AFP2 and analogs (with arginines replacing neutral amino acids) were found to inhibit A. flavus and Fusarium solani, C. albicans and C. krusei. C. glabrata, which does not contain this fungus-specific ceramide, is not inhibited. This peptide and its analogs (e.g., NaD1, Rs-ARF1, SPE10) have little cytotoxicity against mammalian cells at dosages that are inhibitory to fungal pathogens (Matejuk et al., 2010; Sher Khan et al., 2019).
Iturins, produced by Bacillus subtilis, are cyclic peptides with a lipophylic β-amino acid linked to a D and L amino acids, causing pore formation in membranes and leakage of key ions (Besson and Michel, 1984). Their antimicrobial activity is limited primarily to fungi, with little effect on bacteria. Unfortunately, iturins are toxic to mammalian cell membranes. In contrast to most antimicrobial peptides which are cationic, iturins can be anionic (bacillomycin L) or neutral (iturin A). One member of the family, bacillomycin F effectively inhibits A. niger, C. albicans, and C. tropicalis. Although this group of peptides is effective against dermatomycoses in humans and animals, they induce high levels of hemolysis (Latoud et al., 1986).
The mammalian peptide Histatin 5 contains seven histidines, four arginines, and three lysines which allow the peptide adopting an α-helical conformation in non-aquous environments (Raj et al., 1998). Histatin 5 binds to the Ssa2p, a 70-kDa cell wall protein required for the internalization of histatin 5 into cells (Li et al., 2006). The polyamine transporters Dur3 and Dur31 are necessary for the uptake of the peptide as well (Kumar et al., 2011), which needs to be translocated, not endocyted to exert an effect on C. albicans cells (Jang et al., 2010). If the cell is under respiratory metabolism, histatin 5 disrupts the integrity of mitochondrial membrane (Helmerhorst et al., 1999). Propidium iodide (PI) uptake and ATP release also occurs despite cell lysis does not appear to be induced by the peptide (Helmerhorst et al., 1999; Koshlukova et al., 1999). Subsequent ATP binding to surface P2X receptors induce signaling cascades leading to cell death.
Broad Spectrum Antimicrobial Peptides
Most AMPs affect a number of organisms including bacteria, fungi, and envelope-containing viruses. The most representative non-specific AMPs can be grouped as linear peptides (e.g., cecropins, magainins, cathelicidins, bombinins, lactoferrin-derived peptides) or cyclic peptides (e.g., mammalian defensins, poultry gallinacins, macrocyclic peptides, syringomycins, tachystatins) (Matejuk et al., 2010). Membrane disruption by the formation of toroidal pore is common to most peptides, resulting in leakage of essential molecules and ions with general loss of membrane functionality. Additional mechanisms include membrane thinning, formation of non-bilayer intermediates by interchelation of peptide-membrane, formation of anionic lipid-peptide domains, lipid flip-flop, demixing and clustering, and alteration of membrane potential (Shai, 2002; Bechinger and Lohner, 2006; Huang, 2006; Melo et al., 2009; Nguyen et al., 2011; Rautenbach et al., 2016). In addition, some of the peptides may have other effects such as DNA damage, apoptosis induction, inhibition of DNA replication and RNA or protein synthesis.
For supplementary reviews on the mechanisms of action and classification of antimicrobial peptides see reviews by Matejuk (Matejuk et al., 2010), van der Weerden (van der Weerden et al., 2013), and Rautenbach (Rautenbach et al., 2016).
Advantages of AFPs
Existing antifungal agents exhibit a diversity of drawbacks that reduce their efficiency as therapeutic tools against fungal infections. The existence of only a few approved classes of antifungal drugs and the increasing antifungal resistance further complicates the selection of an appropriate antifungal therapy (Sanguinetti et al., 2015; Pappas et al., 2018). Ergosterol synthesis, ergosterol in membranes and cell wall synthesis are the targets of azoles, polyenes and echinocandis, respectively. The low diversity of mechanisms of action of current treatments represents a problematic situation when fungal pathogens are multi-resistant to antifungals (Rautenbach et al., 2016). Furthermore, existing antifungal drugs are associated with adverse drug reactions such as hypokalemia, infusion reaction, nephrotoxicity, hepatotoxicity and gastrointestinal affections, among others (Chen et al., 2011; de Souza et al., 2016; Kyriakidis et al., 2017; Pappas et al., 2018). Some antifungal drugs affect common eukaryotic targets present both in pathogenic fungi and human cells (Rautenbach et al., 2016). This makes the development of novel efficient and non-toxic antifungal therapies more difficult than that of antibacterial ones.
AFPs have diverse advantages over the current antifungal drugs, which are directly related to their mechanisms of action and molecular targets. AFPs are particularly promising because they can recognize multiple microbial targets, thus reducing the possibility of resistance development (Rautenbach et al., 2016), a topic that is discussed in more detail in the following section. These microbial targets include fungal membranes, different cell wall components and molecules related to physiological processes, such as RNA, DNA and protein synthesis and cell cycle (van der Weerden et al., 2013; Bondaryk et al., 2017).
Regarding the absence of side effects, several AFPs target specific conserved fungal molecules, such as glucosylceramide, mannosyldiinositol phosphorylceramide, enzymes related to ergosterol or β-glucan synthesis, among others. This translates into high pathogen selectivity and reduces the probability of cytotoxicity against mammalian cells (Rautenbach et al., 2016). However, it does not ensure the absence of cytotoxicity (e.g., aculeacins, a type of echinocandin that targets the fungal 1,3-β-glucan synthase, displays hemolytic activity) (Matejuk et al., 2010). Over the last 13 years, three echinocandins, anidulafungin, caspofungin, and micafungin, have been approved in Europe and USA (Pound et al., 2011; Beyda et al., 2012). These peptides are poor substrates for cytochrome P450, which translates into both lower hepatotoxicity and pooled risk for discontinuation of treatment (3.7–4.8%) compared to other antifungals (Kyriakidis et al., 2017). The synthetic peptide killer peptide (KP) is another example that has demonstrated very promising antifungal activity without cytotoxicity against peripheral mononuclear blood cells in vitro or side effects in murine trials. This peptide seems to have a specific interaction with 1,3 β glucans and only the dimeric form of the peptide is active (Magliani et al., 2011).
In addition, AFPs show reduced cytotoxicity (Matejuk et al., 2010). Two reasons may explain this phenomenon. Firstly, there is a stronger interaction between the negatively charged fungal membrane (due to the higher content of phosphatidylinositol and phosphatidic acid) and the cationic charges of the peptides, in contrast to mammalian cell membranes, which are predominantly neutral to host mammals (due to the high content of phosphatidylcholine). Secondly, some AFPs target membrane lipids unique to fungi and absent from mammalian cells, which also reduces toxicity (Nguyen et al., 2011; Rautenbach et al., 2016).
Accumulating evidence demonstrates that the therapeutic activity of AMPs is multifactorial and not mediated only by their direct antimicrobial effect. Host defense peptides (HPDs), like defensins and cathelicidins families, often exert angiogeninc, immunomodulatory and anti-inflammatory effects and may also induce the recruitment of the adaptive immune response, which has been reported in several papers and reviews (Zasloff, 2002; Hirsch et al., 2008; Steinstraesser et al., 2009; Magliani et al., 2011; Hsieh and Hartshorn, 2016; Li et al., 2017).
Resistance to AFPs
Membrane remodeling of C. albicans has been associated with its resistance to current non-peptide antifungal drugs, mainly ergosterol-sphingolipid-rich lipid rafts containing multi-drug resistance (MDR) proteins attached to the membrane (Mukhopadhyay et al., 2004; Pasrija et al., 2005, 2008; Shahi and Moye-Rowley, 2009). In the case of resistance to AMPs and AFPs it is important to note, as discussed previously, that these peptides frequently function through membrane interaction, but that additional modes of action for microbial inhibition have also been demonstrated (Wu et al., 1999). Microorganisms, such as fungi, evolve rapidly, and can adapt quickly when exposed to antibiotics and antifungal drugs. However, it is important to note that cell membranes are slower to evolve. The rapid and potent effect on membrane, coupled with other inhibitory mechanisms exhibited by AFPs, de novo resistance is less likely to emerge in target microorganisms (Yeung et al., 2011).
As is the case for currently used antimicrobial drugs, the overuse of AFPs could accelerate the occurrence of fungal resistance. This issue complicates the application of antifungal therapeutics. Indeed, extensive echinocandin usage in hospitals has led to an increase in the number of strains with acquired (secondary) resistance to these first-line antifungals, especially among strains of C. glabrata (Sanguinetti et al., 2015; Pappas et al., 2018). The potential for the emergence of high level resistance to AMPs has been debated, but it is likely to occur at a reduced rate relative to that observed for other antifungals, though it will depend on how the antimicrobial peptide is administered. Besides, although a lower target concentration is required, the absence or change in the specific fungal target through spontaneous mutation can naturally lead to resistance. However, such modification of conserved molecules could, in turn, result in reduced pathogen virulence. In addition, combination therapy, involving the use of AMPs with antibiotics or with other peptides, will likely reduce the development of resistance markedly (Matejuk et al., 2010; Rautenbach et al., 2016).
Production of AFPs
Despite progress relating to the discovery and characterization of AMPs, their application remains challenging (Wimley and Hristova, 2011). There is a need for effective AMP production in sufficient amounts and purity to more extensively investigate their structure–function relationships, efficacy and safety, especially in clinical treatments. Efficient production is also required to serve market requirements should these investigations be successfully completed. There are three major strategies that one could employ to achieve this, i.e., direct isolation from natural producers, heterologous expression or chemical synthesis.
Natural Production
Currently, there are not many AMPs that are produced from their natural sources for clinical use. The purification of peptides from natural sources is laborious and expensive due to their low abundance and the multiplicity of compounds present in those sources (Vriens et al., 2014). Because of this, most industrial-scale productions of AMPs are performed by heterologous expression or chemical synthesis. Brief descriptions of two approaches for industrial-scale AMP production from natural sources are presented here: microbial fermentation and proteolysis of food proteins.
Echinocandins are exceptional examples of AMPs produced by microbial fermentation, and further chemical modification in case of the semisynthetic variants. Industrial-scale production of echinocandins is based on fermentation since they possess a high-complexity chemical structure. However, little information has been published about these processes. Echinocandin B, pneumocandin B0 and FR901379 are the natural echinocandins produced for commercial purposes from A. rugulosus, Glarea lozoyensis and Coleophoma empetri, respectively. The optimization of the fermentation process is essential to obtain a competitive product, since the fermentation and purification costs of natural echinocandins are the main variables that influence the overall production costs of semisynthetic derivatives. The clinical applications of natural echinocandins is limited by certain undesirable properties, such as a strong hemolytic activity, as discussed previously (Emri et al., 2013).
Industrial scale AMPs can also be obtained by proteolysis of food proteins and the release of encrypted peptides. Agyei and Danquah (2011) offered a brief description of the process for manufacturing pharmaceutical-grade peptides by this approach. The process involves, firstly, the acquisition of raw materials: food protein and proteolytic enzymes/or microorganisms. By-products from dairy, fish and meat industries are suitable cheap sources for proteins (Sibel Akalin, 2014; Ryder et al., 2016). The second step involves protein hydrolysis. The use of enzymatic hydrolysis is preferred over in situ microbial fermentation, particularly in food and pharmaceutical industries due to the lower or absent output of organic solvents and toxic chemical in the process and product (Sibel Akalin, 2014; Ryder et al., 2016). Under industrial-scale conditions, the use of immobilized enzymes offers several advantages over the conventional soluble enzymes, such as milder and controlled conditions and recycling of enzymes used (Sewczyk et al., 2018). The final step is fractionation and isolation of the bioactive peptides. Ultrafiltration, precipitation with solvents and liquid chromatography techniques (e.g., ion exchange, gel filtration) have been proposed for purification of peptides, however, their current implicit high costs make them prohibitive for large scale applications. It is estimated that up to 70% of the capital and operating costs in industrial biotechnology processes may correspond to the separation and purification stages (Brady et al., 2008). Electro-membrane filtration (EMF) is being established as an alternative method for the purification of bioactive peptides. It combines electrophoresis with conventional membrane filtration, being more cost-effective than chromatographic techniques (Bazinet and Firdaous, 2013). AMPs with antifungal activity, such as casocidin-I, kapacin A and lactoferrin-derived peptides can be produced by these methods as well.
Recombinant Production of AFPs
Considering the often low amounts of AMPs obtained by purification from natural sources and the high costs and difficulties that may arise from chemical synthesis (Li et al., 2010; Hou et al., 2017), recombinant production of AMPs provides a solid option to make these peptides accessible at low cost and high efficiency.
Genetically modified microorganisms facilitate the production and functional expression of any bioactive molecule but, importantly, also allow the production of bioengineered and encrypted peptides that would not be achievable otherwise (Wibowo and Zhao, 2019). Moreover, new sequencing technologies have made available a vast amount of genomic, transcriptomic and metabolic data, providing the means to further explore known and novel AMPs and the rational design of new antifungal peptides (Amaral et al., 2012; Porto et al., 2012; Tracanna et al., 2017).
However, the success of recombinant production can be highly variable. Understanding the composition and physicochemical properties of AMPs influences the selection and design of hosts and expression system. The choice of host, codon bias, protein expression vector, number of copies of the plasmid and fusion proteins can influence the correct synthesis, folding, and secretion of the recombinant peptide through the cell machinery (Deng et al., 2017). There are two other aspects to bear in mind: the toxicity of the AMP for the host and the high instability and susceptibility of peptides to degradation by proteases. To overcome this, AMPs can be synthesized in a form that is fused to another protein (fusion proteins) or in inactive forms (Kosobokova et al., 2016) initially.
Escherichia coli, yeasts (mainly Pichia pastoris) and plants are the most common recombinant expression platforms for biopharmaceutical proteins. Comprehensive reviews have previously reviewed the different heterologous production platforms available for AMPs (Sanchez-Garcia et al., 2016; Deng et al., 2017). Most AFPs produced by recombinant platforms target plant phytopathogens rather than human fungal pathogens. However, many of these AFPs may represent underutilized resources whose antifungal activity against human fungal pathogens is waiting to be discovered. In the next subsection, a brief overview of the production of AFPs in different hosts with clinical applications is provided.
Production of AFPs in Bacteria
E. coli is by far the bacterial species that is used most widely as a host for heterologous production of peptides and proteins (Li, 2011). Its genetic configuration is well-known, it is easy to manipulate and there is a broad variety of protein expression vectors and host strains available. The pET vectors (Novagen) are the most commonly used. Among the expression strains, the most popular are E. coli BL21 (DE3), deficient in proteases that may lead to protein degradation, or pLysS Origami and Rosetta and C41 (DE3) (Novagen), employed when disulfide bond formation is needed (Rosano and Ceccarelli, 2014). Many E. coli strains are unable to export proteins across their outer membrane, and proteins are secreted into the cytoplasm or periplasm generating inclusion bodies (Singh et al., 2015). Therefore, AFPs produced by E. coli are usually purified by sonication methods followed by reversed-phase chromatographies, giving relatively low yields. Several examples of AFPs production in E. coli are shown in Table 3.
Table 3.
Expression of antifungal peptides in E. coli.
| Type of AFP | Name | Source | Host | Expression vector | Fusion partner | Antifungal spectrum | Yield | References |
|---|---|---|---|---|---|---|---|---|
| Defensin | PvD1 | Phaseolus vulgaris L. (common bean) | Escherichia coli Rosetta Gami DE3 | pET-32 EK/LIC | Thioredoxin | Candida albicans | Unknown | de O Mello et al., 2014 |
| Hybrid peptide (lactoferricin+cecropin) | LF15-CA8 |
Hyalophora cecropia (giant silk moth) Bovine lactoferricin (encrypted peptide) |
E. coli BL21 (DE3) | pGEX-4T-2 | GST | Lactoferricin: C. tropicalis, C. krusei, C. albicans, C. glabrata, Aspergillus spp., Cryptococcus spp. | 10 mg/mL | Feng et al., 2014; Fernandes and Carter, 2017 |
| Lactoferricin | Lactoferricin B | Mammals | E. coli BL21 (DE3) | pET21d | MMIS | Lactoferricin:C. tropicalis, C. krusei, C. albicans, C. glabrata, Aspergillus spp., Cryptococcus spp. | Unkown | Kim et al., 2006; Fernandes and Carter, 2017 |
| Cecropin | CeA | Hyalophora cecropia (giant silk moth) | E. coli BL21 (DE3) | pET-30a | ELK16 self-assembly peptide (GyrA intein) | C. tropicalis, C. krusei, C. albicans, C. glabrata, Aspergillus spp., Cryptococcus spp. | 6.2 mg/mg wet cell | Wang et al., 2018 |
| Peptidyl nucleoside antibiotics | Nikkomycin | Streptomyces ansochromogenes | E. coli BL21 (DE3) | pET23b | His tag | C. albicans | 800 mg/L | Li et al., 2005 |
| Echinocandins | PH HtyE | A. pachycristatus | E. coli BL21 Gold | pET-28b(+) | His tag | Candida spp., Aspergillus spp. | 75 mg/L | Mattay et al., 2018 |
| Magainins | Magainin-2 | Xenopus laevis (African frog) | E. coli BL21 (DE3) | pET-21a | Carbohydrate-binding module, His tag |
C. albicans, C. neoformans S. cerevisiae |
Unkown | Zasloff, 2002; Ramos et al., 2013 |
| Dermaseptin | Dermaseptin S4 | Phyllomedusinae frogs (amphibian skin) | E. coli strain BL21 (DE3) | pGEX-4T-1 | Glutathione S-transferase (GST) | C. neoformans and A. fumigatus | Unknown | Belaid and Hani, 2011; Song et al., 2014 |
| Chitin-binding cysteine rich | Tachycitin | horseshoe crab hemocyte (Argopecten irradians) | Escherichia coli BL21 (DE3)/pLysS | pET-22b() | None | Paecilomyces variotii, Aspergillus spp., F. oxysporum, Neurospora crassa, B. cinerea, and Alternaria brassicola | 1 mg/L | Kawabata et al., 1996; Suetake et al., 2002 |
| Defensin | HBD5/ HBD6, HBD26, HBD27 |
Mammal | E. coli BL21 (DE3) | pET-32a (+) | Thioredoxin AHis6 | C. albicans | Unknown | Huang et al., 2008, 2009 |
Bacillus subtilis has also been explored as host for AFPs production. This includes plectasin (Zhang et al., 2015), cathelicidin (Luan et al., 2014), and the hybrid cecropin A– melittin (Ji et al., 2017) (Table 4). B. subtilis is a well-studied species, is non-pathogenic, has been approved by the Food and Drug Administration as a Generally Regarded As Safe (GRAS) microorganism and does not exhibit codon bias. It also reaches high cell density and releases proteins directly to the extracellular medium, simplifying the purification process. However, B. subtilis secretes proteolytic enzymes that can degrade the secreted recombinant proteins. Luckily, optimization of the cloning strategies and construction of protease-negative mutants are fostering its wider use (Cui et al., 2018).
Table 4.
Expression of antifungal peptides in other bacteria and yeasts.
| Type of AFP | Name | Source | Host | Expression vector | Fusion partner | Antifungal spectrum | Yield | References |
|---|---|---|---|---|---|---|---|---|
| BACTERIA | ||||||||
| Cyclic lipopeptide | Iturin A | Bacillus amyloliquefaciens and B. subtilis | B. amyloliquefaciens | Genome shuffling | Protoplast fusion technology | Saccharomyces cerevisiae, plant pathogens | 172.22 mg/L | Shi et al., 2018 |
| Defensin | Plectasin | Pseudoplectania nigrella | B. subtilis | pGJ148 | Small Ubiquitine-like modifier (SUMO) | Candida albicans | 5.5 mg/L | Zhang et al., 2015 |
| Polyoxins | Polyoxin P Polyoxin O |
Streptomyces ansochromogenes, S. cacaoi | S. ansochromogenes | pPOL | None | Alternaria kikuchiana, Aspergillus fumigates, Rhizoctonia solani, Botrytis cinerea and Trichoderma viride | Unknown | Li et al., 2012 |
| Human beta defensin | HBD-1 | Humans | Lactococcus lactis A164 | pOED1 | DsbC-Tag | C. albicans | Unknown | Choi et al., 2005 |
| YEASTS | ||||||||
| De novo designed peptide | PAF102 | Combinatorial screen against the phytopathogen Penicillium digitatum | Pichia pastoris | pGAPHA | Plant oleosin | P. digitatum, Magnaporthe oryzae, Fusarium oxysporum and B. cinerea | 180 mg/L | López-García et al., 2015; Popa et al., 2019 |
| Big defensin | AiBD | Argopecten irradians (mollusk) | P. pastoris GS115 | pPIC-9K | None | C. albicans | Unknown | Saito et al., 1995; Zhao et al., 2007 |
| Cathelicidin | Protegrin 1 (PG1) | Mammals | P. pastoris X-33 | pJ912 | His tag | C. albicans | 104 mg/mL | Huynh et al., 2018 |
| Cathelicidin | Protegrin 1 (PG1) | Mammals | P. pastoris X-33 | pPICZα-A | His tag | C. albicans | 15.6/100 mL | Niu et al., 2015 |
| Transferrin family | pLF (Porcine lactoferrin) | Sow's milk | P. pastoris GS115 (his4) | pPIC9 | None | C. tropicalis, C. krusei, and C. albicans | Unknown | Pecorini et al., 2005; Fernandes and Carter, 2017 |
| Plant defensin | HsAFP1 | Heuchera sanguinea (coral bells) | P. pastoris X-33 | pPICZαA | None |
S. cerevisiae, C. albicans, and F. culmorum |
40 mg/L | Aerts et al., 2011; Vriens et al., 2015 |
| Radish defensin | RsAFP2 | Raphanus sativus L. (radish seeds) | P. pastoris strain GS115 | Unknown | None | Alternaria spp., Fusarium spp., Trichoderma spp. | 100 mg/L | Terras et al., 1992; Vriens et al., 2016 |
| Chitinase | VuChiI | Vigna unguiculata (L.) (walp.Cow pea) | P. pastoris KM71H | pPICZαA | Histag | P. herquei | 18 mg/L | Landim et al., 2017 |
Lactic acid bacteria (LAB) have been extensively used for the heterologous production of bacteriocins, antibacterial peptides secreted by bacteria (Rodríguez et al., 2003; García-Fruitós, 2012). Genera such as Lactobacillus, Leuconostoc, Pediococcus, Lactococcus, Streptococcus, and Enterococcus (König and Fröhlich, 2017) can be part of the human microbiota at different body sites (George et al., 2018) and, for some of these genera, specific strains have been used as probiotics (Harzallah and Belhadj, 2013). However, the production of AFPs in LAB is challenging due to the antimicrobial sensitivity of the host. Choi et al. (2005) attempted the heterologous production of HBD-1 in a nisin Z L. lactis producer, but toxicity to the host was apparent (Choi et al., 2005) (Table 4).
In addition to being outstanding secondary metabolite producers, including of antibacterial and antifungal peptides (Harir et al., 2018), in their own right, Streptomyces species also offer many potential advantages as hosts for the expression and secretion of proteins (Baltz, 2010). In a small number of cases they have also been used for the heterologous production of AFPs (Li et al., 2012; Roldán-Tapia et al., 2017) (Table 4).
Several fusion partners have been used to facilitate the production of AFPs in bacteria (Li, 2009; Costa et al., 2014). Examples of AFPs produced by E. coli, Bacillus, Streptomomyces and L. lactis using different fusion partners are shown in Table 4. The His-tag, widely used beyond E. coli and consists of a short chain of six histidine residues. The His-tag is often combined with other, fusion tags to improve the production, solubility and recovery of the recombinant protein. Thioredoxin (TRX) and glutathione S-transferase (GST) are ubiquitous enzymes involved in redox and detoxification processes, respectively, which are often used as N-terminal fusion tagsTRX helps with the formation of disulphide bridges of the target protein, especially in strains unable to do so. On the other hand, GST functions as a chaperone that enhances the expression and solubility of recombinant proteins. In addition, GST-tagged fusion proteins can be purified by glutathione affinity chromatography, which facilitates the purification process. MMIS is a modified form of the magainin intervening sequence (MIS) that prevents the antimicrobial activity of the fused peptides until they are released. ELK16 is a self-assembling peptide that induces the formation of cytoplasmic inclusion bodies in E. coli. Carbohydrate-binding modules have also been used to enhance purification of AMPs though a cellulose matrix. More innovative fusion partners include protoplast for iturin production (Shi et al., 2018). Overall, fusion partners increase the solubility of the target peptides and protect them from degradation, but there is no evidence of higher yields.
Production of AFPs by Yeasts
With respect to heterologous expression, yeasts are fast growing and easy to manipulate genetically. Moreover, they are capable to perform correct protein processing and post-translational modifications. The methylotrophic yeast Pichia pastoris has been the preferred yeast for AMP production and for recombinant expression of AFPs (Table 4). Pichia protein expression vectors contain the alcohol oxidase gene promoter (AOX 1), inducible by the addition of methanol, which allows the overexpression of the gene introduced downstream. Three strains have been most widely used to produce AFPs: P. pastoris X-33, P. pastoris GS115, and P. pastoris KM71H. The strains differ in their genotypes, which affects the selection of selectable markers, typically antibiotic resistance genes or auxotrophic markers. His-tags are commonly used here also to facilitate the purification of recombinant proteinsaffinity metal-chelating chromatography (Niu et al., 2015; Landim et al., 2017). Plant oleosin fusion technology (Ling, 2007; Bhatla et al., 2010) has also been used for the production of iturin A (Popa et al., 2019). Table 4 shows some examples of AFPs produced by P. pastoris for clinical use.
Production in Fungi
Filamentous fungi are a well-known source of metabolites and enzymes (Hoffmeister and Keller, 2007), e.g., they naturally produce a wide range of primary metabolites such as organic and fatty acids, and important secondary metabolites, including the antibiotics penicillin, cephalosporin and griseofulvin, or the cholesterol lowering agent lovastatin (Alberti et al., 2017). The attraction of filamentous fungi as hosts for protein recombinant production relates to their relatively inexpensive growing requirements and their ability to naturally secrete large amounts of proteins into the growth medium. They can also perform complex posttranslational modifications including glycosylation, proteolytic cleavage and multiple disulphide bond formation. Moreover, they are very useful when whole synthetic pathways need to be recreated. Species such as Aureobasidium pullulans, Penicillium chrysogenum, and P. digitatum have been used to produce AFPs. A. pullulans has been used to produce several bioactive molecules, such as pullulan, a polysaccharide with numerous applications in health and the food industry, β glucan, and a wide variety of extracellular enzymes. It has also been reported to produce the antibacterial exophilin A, liamocins and heavy oils (Chi et al., 2009). Notably, A. pullulans has also been used to produce aureobasidin A as a consequence of homologous recombination (Table 5) (Slightom et al., 2009). P. chrysogenum and P. digitatum have also been successfully used to produce NFAP2 and AfpB, respectively. P. chrysogenum is known to produce a cationic antifungal protein which inhibits zoopathogens and plant-pathogenic fungi. This host has undergone many improvements to optimize fermentation conditions. Finally, P. digitatum was originally known as a fruit pathogen but has been used for the homologous production of AfpB.
Table 5.
Expression of antifungal peptides in fungi and plants.
| Type of AFP | Name | Source | Host | Expression vector | Fusion partner | Antifungal spectrum | Yield | Reference |
|---|---|---|---|---|---|---|---|---|
| FUNGI | ||||||||
| Aureobasidins | Aureobasidin A (BP-1938) | Aureobasidium pullulans | A. pullulans | pCR2.1 TOPO | None | Candida spp. | Unknown | Slightom et al., 2009 |
| Cysteine rich | Neosartorya fischeri antifungal protein 2 (NFAP2) | N. fischeri |
Penicillium chrysogenum Q176 GRAS-FDA |
pSK275nfap | None | Candida spp. | 15 mg/L | Tóth et al., 2018 |
| Cysteine rich | AfpB | P. digitatum CECT 20796 (PHI26) |
P. digitatum
P. pastoris X-33 |
pBHt2 pPICZαA |
None |
Saccharomyces cerevisiae, P. italicum, P. expansum, Botrytis cinerea, Magnaporthe oryzae, Fusarium oxysporum, P. digitatum |
12–20 mg protein/l (P. digitatum) 1.2–1.4 mg/l (P. pastoris) |
Garrigues et al., 2018 |
| PLANTS | ||||||||
| Lactoferrin-derived peptides | Lactoferrin+lactoferrampin chimera | Bovine milk | Nicotiana tabacum | pBI121 | His tag | C. tropicalis, C. krusei, C. albicans, C. glabrata, Aspergillus spp., Cryptococcus spp. | 4.8 μg/g fresh weight | Chahardoli et al., 2018 |
| Dermaseptin | Dermaseptin B1 | Skin glands of the South American hylid frog, Phyllomedusa bicolor | Nicotiana tabacum | pGSA1285 | Tandem repeat of Cladosporium fulvum Avr4 effector protein CBD | Agrobacterium tumefaciens (PTCC 1654), Pectobacterium carotovorum (PTCC 1675), Pseudomonas aeruginosa (PTCC 1558), Xanthomonas campestris (PTCC 1473), and Ralstonia solanacearum (ATCC 11696) bacteria, Alternaria alternata (PTCC 5224) and Pythium spp. | Unknown | Shams et al., 2019 |
Production in Plants
Plant-based expression systems have been explored as production hosts for recombinant expression of AMPs due to their capacity for large scale production and their cost-effectiveness. Advantages of plants are their capability to perform appropriate glycosylation, folding, and disulphide bond formation of recombinant AMPs. There are different genetic approaches to produce AMPs in plants: using whole plants, tissue specific expression, tissue culture, or transient expression (Holaskova et al., 2015). Nuclear transformation has been the preferred technique for plant-derived therapeutic proteins followed by purification from transgenic plants. The tobacco plant (Nicotiana tabacum) is the most commonly used transgenic expression system and strains of the bacterial species Agrobacterium tumefaciens are the most popular intermediate hosts (Desai et al., 2010). Setting up a higher plant production platform is more expensive than using bacteria, yeast or fungi. However, once the system is established, it is easier to handle and provides high capacity for scale-up. Moreover, plant-based systems do not generally need control of production. Chahardoli et al. used tobacco whole plants as a platform to produce a lactoferrin and lactoferrampin 34 amino acid chimera. To our knowledge this is the first study to produce an AFP in plants (Table 5) (Chahardoli et al., 2018).
Chemical Synthesis
Chemical synthesis of peptides can be divided in two types: solid- (SPPS) or liquid (solution) phase peptide synthesis (LPPS). In general terms, LPPS is suitable for large-scale manufacture of short peptides or structures that are not easily prepared by SPPS. SPPS is generally used for lower scales or to provide mechanistic insights about peptides and offers the potential for the creation and production of more cost-effective antifungal therapies (Matejuk et al., 2010). Currently, Fmoc SPPS is the preferred method for peptide chemical synthesis due to the versatility and low cost of very high-quality building blocks. The diversity of synthetic peptides entering clinical trials has increased over the last 13 years, stimulating advances in Fmoc SPPS technologies in response to the growing demand for medicinal chemistry and pharmacology (Behrendt et al., 2016). Several groups have synthesized linear (Tran et al., 2008; Magliani et al., 2011; Konno et al., 2015; Cools et al., 2017; Park et al., 2018) and cyclic peptides (Mosca et al., 2000; Schaaper et al., 2001; Konno et al., 2015; Ng-Choi et al., 2019) using Fmoc SPPS.
The long process of isolation and characterization of new natural AMPs delays their clinical use. In this regard, Fmoc SPPS is at the forefront in the design of therapeutic peptides since it permits the easy alteration of features such as hydrophobicity, polarity, charge, structure, and it may also enhance activity and overcome the limitations of natural peptides (Freitas and Franco, 2016). The rational design of synthetic sequences is a new approach of relevance, and results from optimizing the sequence and chemical characteristics shared by different AMPs (pharmacophoric patron) (Freitas and Franco, 2016). Ideally, an antifungal peptide agent should be short, as mentioned previously. De novo peptide design may help reducing production costs, potential toxicity and lability, as well as increasing the in vivo activity (Steckbeck et al., 2014).
Unfortunately, Fmoc SPPS is currently far from meeting its potential and still cannot compete with the template-based process of expression for the large-scale demand of therapeutics (Behrendt et al., 2016). However, it should be noted that companies specialized in the large-scale manufacture of peptides are currently being established, claiming productions from multi-10 kg/lot (SPPS) to multi-100 kg/lot (LPPS)1. This suggests that the versatility of chemical synthesis is on its way to reach the cost-efficiency and scale of peptide expression.
Current and Potential Applications in Human Medicine
Combined Therapy With Other Drugs
A recurrence of disease and establishment of chronic fungal infections may result when antifungal treatments are not sufficiently effective. Thevissen (2016) proposed that more efforts should focus on combination therapy in addition to screening for novel antifungal compounds. Synergy between the combined compounds is the main objective of combination therapy, thereby increasing their activity and diminishing their toxicity on the host. Another option is combining an antimycotic (either currently used or novel AMP) with an enhancer molecule that, for example, weakens one or multiple pathogen tolerance mechanisms, such as biofilms, without direct antifungal activity. Either way, it is essential to demonstrate efficacy and safety of the combination before proceeding to clinical trials.
Limited data from clinical trials are available in this regard. A study performed by Candoni et al. showed for the first time that early mortality of patients with invasive aspergillosis was reduced by combined treatment with two antifungal agents (Candoni et al., 2014). In a retrospective study published in 2017, Lee et al. analyzed records from a pediatric department in South Korea and described how the combined therapy of voriconazole and caspofungin was an effective and safe treatment for children with leukemia (Lee et al., 2017). In vitro and in vivo studies suggests that AFPs are excellent candidates for this type of approaches and have a great potential for clinical success. Lactoferrin-derived peptides such as Lf(1-11) and bLfcin have shown synergy with azole antifungal drugs or amphotericin B, greatly reducing the minimal inhibitory concentrations (MICs) against C. albicans, C. glabrata, C. krusei, C. parapsilosis, C. tropicalis, and fluconazole-resistant strains of C. albicans (Wakabayashi et al., 1996, 1998; Lupetti et al., 2003; Fernandes and Carter, 2017). Some other examples of these studies are summarized in Table 6.
Table 6.
Combination of antifungal compounds against pathogenic fungi.
| Compound A | Compound B | Type of combination | Fungus treated | Type of study | Outcome | References |
|---|---|---|---|---|---|---|
| AmB | bacillomycin D | A - A | C. albicans | in vitro (keratinocytes) | Synergy; Anti-biofilm and wound-healing activities | Tabbene et al., 2016 |
| Ds7 | C. tropicalis | in vitro | Synergy; Anti-biofilm and membrane lytic activities | Singh et al., 2017 | ||
| Crotalicidin | Candida spp. | in vitro (HK-2 cells) | Synergy; MIC reduction; less cytotoxic and hemolytic than Amb | Cavalcante et al., 2017 | ||
| AmB or VCZ | bLfcin | C. albicans | in vitro | Synergy; 4-16 fold MIC reduction, reduced formation of biofilms | Fernandes and Carter, 2017 | |
| FCZ | hLf(1-11) | C. glabrata, C. krusei, C. parapsilosis, C. tropicalis, C. albicans | in vitro | Synergy; fungicidal effect | Lupetti et al., 2003 | |
| CaThi | F. solani | in vitro | Synergy; 100% fungicidal effect | Taveira et al., 2017 | ||
| Af or Cf | DermaseptinS3(1-16) | C. glabrata, C. albicans | in vitro | Synergy; MIC reduction | Harris and Coote, 2010 | |
| Renalexin | C. glabrata, C. albicans | in vivo (mice) | Synergy; MIC reduction; no effect in in vivo tests | Harris and Coote, 2010 | ||
| Magainin2 | C. glabrata, C. albicans | in vitro | Synergy; MIC reduction | Harris and Coote, 2010 | ||
| 6752 | C. glabrata, C. albicans | in vitro | Synergy; MIC reduction | Harris and Coote, 2010 | ||
| GS14K4 | C. glabrata, C. albicans | in vitro | Synergy; MIC reduction | Harris and Coote, 2010 | ||
| MUC7 12-mer | Hsn5 12-mer | C. albicans, C. neoformans | in vitro | Synergy; MIC reduction, low hemolytic activity | Wei and Bobek, 2004 | |
| Amb | C. albicans, C. neoformans | in vitro | Synergy; MIC reduction | Wei and Bobek, 2004 | ||
| Miconazole | C. albicans, C. neoformans | in vitro | Synergy; MIC reduction | Wei and Bobek, 2004 | ||
| Cf | Hepcidin 20 | C. glabrata | in vitro | Synergy; MIC reduction | Tavanti et al., 2011 | |
| VCZ | Pneumocystis jirovecii, Aspergillus spp. | in vivo (children with leukemia) | Overall response 90% after combination treatment; 10% mild liver side effect | Lee et al., 2017 | ||
| VCZ | Candida spp. , Aspergillus spp., Fusarium spp. | in vivo (humans) | Reduction in early mortality of patients with invasive aspergillosis | Candoni et al., 2014 | ||
| LAmB | Candida spp., Aspergillus spp., Fusarium spp. | in vivo (humans) | Reduction in early mortality of patients with invasive aspergillosis | Candoni et al., 2014 | ||
| FCZ | Retigeric acid B (RAB) | A - P | C. albicans | in vitro - in vivo (mice) | Synergy; inhibition of hyphal formation and adherence to host cells | Chang et al., 2012 |
| 2-adamantanamine (AC17) | C. albicans | in vivo (guinea pigs) | Synergy; reduction in fungal tissue burden (cutaneous candidiasis) | Lafleur et al., 2013 | ||
| FK506 | C. albicans | in vivo (rats) | Synergy; inhibition of biofilm formation in catheter model | Uppuluri et al., 2008 | ||
| Cephalosporin A (CsA) | C. albicans | in vivo (rats) | Synergy; inhibition of biofilm formation in catheter model | Uppuluri et al., 2008 | ||
| Cf | diclofenac | C. albicans | in vivo (rats) | Synergy; inhibition of biofilm formation in catheter model | Bink et al., 2012 | |
| AmB | bLf peptide 2 - GM-CSF | A - A - P | C. albicans | in vivo (mice) | Synergy; upregulation of phagocytes; extended survival of mice up | Tanida et al., 2001 |
Af, anidulafungin; Cf, caspofungin; FCZ, fluconazole; VCZ, voriconazole; AmB, amphotericin B; MUC7, human mucin-derived peptide; CaThi, Thionine-like peptide from Capsicum anuum; DS7; synthetic peptide derived from Aspergillus giganteous antifungal protein. Hsn5, histatin 5; LAmB, liposomal AmB; hLf(1-11), human lactoferrin peptide 1-11; bLf, bovine lactoferrin; bLfcin, bovine lactoferricin; GM-CSF, granulocyte-macrophage colony-stimulating factor; A, Antifungal; P, Potentiator.
Commercial Products and Formulations
P113 is a 12-mer peptide developed by the company PacGen Life Science (Vancouver, Canada) as a mouth rinse formulation for the topical treatment of oral candidiasis. Clinical trials from PacGen demonstrated that oral candidiasis was effectively treated by P113, which compared favorably to the efficacy of nystatin, a standard treatments for oral candidiasis. Vaginal, dermatological and ophthalmic applications are on the list of P113 therapeutic potential (Duncan and O'Neil, 2013). Currently, the P113-containing line of products includes oral rinse solution and spray, feminine soothing spray and cleansing wash, and antibacterial hand cream, among others2. NP213/Novexatin was the lead product of NovaBiotics of Aberdeen, UK. It is an arginine-rich cyclic cationic peptide based on human α and β defensins (among others). It was used for treatment of toenails stubborn fungal infections such as onychomycosis (patents PCT/GB2006/004890 and PCT/GB2005/003245). Indeed, independent podiatrist analysis determined the treatment against mild to moderate onychomycosis as being 80% clinically effective. This effectiveness rate is significantly higher than that provided by existing topical treatments for onychomycosis in different territories, including United States and Europe (Duncan and O'Neil, 2013; Fox, 2013)3. However, the clinical study did not acomplish the main goal of a Phase IIb study by not showing differences over the placebo treatment under FDA current guidelines4.
Potential Applications
Despite their promising properties, only a few AFPs have reached the clinical phase. One example is hLF(1-11), a peptide that was developed for the systemic treatment of bloodstream and deep tissue infections produced by fungi and bacteria in severely immunocompromised transplant recipients. However, after favorable safety and tolerability clinical trials of hLF(1–11), no more studies have taken place putting on hold the commercialization of this peptide (van der Velden et al., 2009; Martin et al., 2015; Bruni et al., 2016). Another example is CZEN-002, a synthetic octapeptide derived from alpha-Melanocyte-Stimulating Hormone (a-MSH). CZEN-002 modulates immune and inflammatory responses, and has been shown to kill C. albicans as well (Fjell et al., 2011; Mahlapuu et al., 2016). This AMP had been in phase II clinical trials for the treatment of vulvovaginal candidiasis. However, no recent development has been reported. It was developed by Zengen, Abiogen Pharma and Lee's pharmaceuticals (Duncan and O'Neil, 2013)5.
In addition, there are many cases of peptides with promising properties currently being evaluated pre-clinically (Koo and Seo, 2019). Other pre-clinical examples are described in the review by Duncan and O'Neill (Duncan and O'Neil, 2013) and in the database DRAMP 2.0 (Kang et al., 2019).
Delivery and Formulations
As discussed, different formulations and delivery strategies for AFPs might be explored depending on the peptide properties, the potential toxicity (suitable for topical and/or systemic use) and the marketing strategies of the companies (e.g., mouth rinse, toothpaste, spray, dermal cream, etc.). Different carriers can enhance the pharmacodynamics and stability, and reduce toxicity of the active peptide, such as liposome encapsulation or the use of peptoids, the D-conformation-based peptide and β-peptides (Sajjan et al., 2001; Porter et al., 2002; da Silva Malheiros et al., 2010; Chongsiriwatana et al., 2011). Diverse pre-clinical delivery tools and formulations are being studied. In one case, Park et al. developed a pH-responsive and redox-sensitive polymer-based AmB-delivery carrier system (Park et al., 2017), by conjugation with histatin 5 acting both as a synergistic antifungal molecule and a targeting ligand against C. albicans. Other authors describe that some properties of the peptides could make them suitable for self-delivery systems. Examples include the synthetic killer peptide (KP) and the ultrashort peptide NapFFKK-OH, which, at certain pHs and concentration conditions, undergo a self-assembly process. The results are hydrogel-like aggregates that could slowly release the peptides in physiological conditions, as well as reducing the proteolytic susceptibility and increasing the storage stability of the active compound (Magliani et al., 2011; Albadr et al., 2018). Future clinical studies of these hydrogels and other delivery technologies will determine their safety and efficiency. Additional drug delivery strategies include carbon nanotubes and magnetic nanoparticles (López-Abarrategui et al., 2013; Chaudhari et al., 2016).
Conclusions
Mycoses are a serious and rising threat to humans. Survival rates remain unacceptably low and no new antifungals have been introduced in more than 13 years since echinochandins and pneumocandins. AFPs have obvious potential as more efficient and safer therapeutic agents than conventional antifungal drugs. Research on AFPs has been highly active and over one thousand peptides have been described. Nevertheless, few molecules have reached late clinical stage studies or have entered the market. Major challenges for AFPs commercialization relate to their specificity and safety. Moreover, stability of the formulations, delivery strategies and the overall therapeutic efficiency together with production costs at industrial scale and regulatory barriers remain to be resolved. The mode of action of AFPs is not fully understood which also raises safety concerns.
Regarding exploitation, AFP production yields from natural sources are very low and requires complex and costly procedures for extraction and purification. Their peptidic nature enables production through recombinant platforms, but scaling-up procedures are not always successful and require extensive optimization. Currently, chemical synthesis is economically viable only for short peptides and high value applications, but novel synthesis and purification technologies such as EMF are on the way to meet these requirements for such short peptides and high value applications.
Notably, indications from the FDA and European regulatory entities indicate that regulatory laws are changing, encouraging companies to invest in antimicrobial discovery and development. Among other measures, new guidance documents have been released, including plainspoken clinical criteria for evaluating antimicrobials and broadening the spectrum of volunteers for clinical trials. Other steps will help in trials designed for the evaluation of drug-resistant pathogens (Fox, 2013).
Considering that the global market for antifungals is set to be worth $12.2 billion6, AFPs are commercially attractive candidates in terms of manufacturing costs, options, increasing regulatory acceptance of peptide therapeutics, etc. (Duncan and O'Neil, 2013). Thus, an integral action plan on this field needs to be driven by academy, biotechnological and pharmaceutical companies and regulatory entities in order to enhance and thrust the development of novel antifungal therapies. Nonetheless, the path has been paved for these promising molecules.
Author Contributions
MF, SA, and EG-G designed and wrote the manuscript. PC critically revised the manuscript. All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1Synthesis—PolyPeptide. Available online at: https://www.polypeptide.com/commercial-manufacturing/ (accessed August 12, 2019).
2P113 | Pacgen Life Science. Available online at: http://www.pacgenlife.com/node/171 (accessed August 15, 2019).
3Elewski, B. E. Dermatologists and Podiatrists Interviewed, 14.
5CZEN-002 Available online at: https://www.pharmacodia.com/yaodu/html/v1/chemicals/f84aa65357bec670cbba3ae77711c233.html (accessed August 15, 2019).
Funding. This publication has emanated from research conducted with the financial support of Science Foundation Ireland (SFI) under grant number SFI/12/RC/2273. MF and SA have received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement number 754535. EG-G was supported by Walsh Fellowship Project 2015066.
References
- Abe H., Kawada M., Sakashita C., Watanabe T., Shibasaki M. (2018). Structure-activity relationship study of leucinostatin A, a modulator of tumor–stroma interaction. Tetrahedron 74, 5129–5137. 10.1016/j.tet.2018.05.064 [DOI] [Google Scholar]
- Aerts A. M., Bammens L., Govaert G., Carmona-Gutierrez D., Madeo F., Cammue B. P. A., et al. (2011). The antifungal plant defensin HsAFP1 from Heuchera sanguinea induces apoptosis in Candida albicans. Front. Microbiol. 2:47. 10.3389/fmicb.2011.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal P., Bhalla S., Chaudhary K., Kumar R., Sharma M., Raghava G. P. S. (2018). In silico approach for prediction of antifungal peptides. Front. Microbiol. 9:323. 10.3389/fmicb.2018.00323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agyei D., Danquah M. K. (2011). Industrial-scale manufacturing of pharmaceutical-grade bioactive peptides. Biotechnol. Adv. 29, 272–277. 10.1016/j.biotechadv.2011.01.001 [DOI] [PubMed] [Google Scholar]
- Akkam Y. (2016). A review of antifungal peptides: basis to new era of antifungal drugs. Jordan J. Pharm. Sci. 9, 51–75. [Google Scholar]
- Albadr A. A., Coulter S. M., Porter S. L., Thakur R. R. S., Laverty G. (2018). Ultrashort self-assembling peptide hydrogel for the treatment of fungal infections. Gels 4:48. 10.3390/gels4020048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti F., Foster G. D., Bailey A. M. (2017). Natural products from filamentous fungi and production by heterologous expression. Appl. Microbiol. Biotechnol. 101, 493–500. 10.1007/s00253-016-8034-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral A. C., Silva O. N., Mundim N. C. C. R., de Carvalho M. J. A., Migliolo L., Leite J. R. S. A., et al. (2012). Predicting antimicrobial peptides from eukaryotic genomes: in silico strategies to develop antibiotics. Peptides 37, 301–308. 10.1016/j.peptides.2012.07.021 [DOI] [PubMed] [Google Scholar]
- Andreu D., Rivas L. (1998). Animal antimicrobial peptides: an overview. J. Pept. Sci. 47, 415–433. [DOI] [PubMed] [Google Scholar]
- Auchtung T. A., Fofanova T. Y., Stewart C. J., Nash A. K., Wong M. C., Gesell J. R., et al. (2018). Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere 3:e00092–18. 10.1128/mSphere.00092-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar A. A., Ren D. (2013). Antimicrobial peptides. Pharmaceuticals 6, 1543–1575. 10.3390/ph6121543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkovec J. M., Black R. M., Hammond M. L., Heck J. V., Zambias R. A., Abruzzo G., et al. (1992). Synthesis, stability, and biological evaluation of water-soluble prodrugs of a new echinocandin lipopeptide. Discovery of a potential clinical agent for the treatment of systemic candidiasis and Pneumocystis carinii pneumonia (PCP). J. Med. Chem. 35, 194–198. 10.1021/jm00079a027 [DOI] [PubMed] [Google Scholar]
- Baltz R. H. (2010). Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J. Ind. Microbiol. Biotechnol. 37, 759–772. 10.1007/s10295-010-0730-9 [DOI] [PubMed] [Google Scholar]
- Bazinet L., Firdaous L. (2013). Separation of bioactive peptides by membrane processes: technologies and devices. Recent. Pat. Biotechnol. 7, 9–27. 10.2174/1872208311307010003 [DOI] [PubMed] [Google Scholar]
- Bechinger B., Lohner K. (2006). Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Biophys. Acta 1758, 1529–1539. 10.1016/j.bbamem.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Bednarska N. G., Wren B. W., Willcocks S. J. (2017). The importance of the glycosylation of antimicrobial peptides: natural and synthetic approaches. Drug Discov. Today 22, 919–926. 10.1016/j.drudis.2017.02.001 [DOI] [PubMed] [Google Scholar]
- Behrendt R., White P., Offer J. (2016). Advances in Fmoc solid-phase peptide synthesis. J. Pept. Sci. 22, 4–27. 10.1002/psc.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaid A., Hani K. (2011). Antiviral and antifungal activity of some dermaseptin S4 analogues. Afr. J. Biotechnol. 10, 14962–14967. 10.5897/AJB11.1108 [DOI] [Google Scholar]
- Besson F., Michel G. (1984). Action of the antibiotics of the iturin group on artificial membranes. J. Antibiot. 37, 646–651. 10.7164/antibiotics.37.646 [DOI] [PubMed] [Google Scholar]
- Bewley C. A., Faulkner D. J. (1994). Theonegramide, an antifungal glycopeptide from the Philippine lithistid sponge Theonella swinhoei. J. Org. Chem. 59, 4849–4852. 10.1021/jo00096a028 [DOI] [Google Scholar]
- Beyda N. D., Lewis R. E., Garey K. W. (2012). Echinocandin resistance in Candida species: mechanisms of reduced susceptibility and therapeutic approaches. Ann. Pharmacother. 46, 1086–1096. 10.1345/aph.1R020 [DOI] [PubMed] [Google Scholar]
- Bhatla S. C., Kaushik V., Yadav M. K. (2010). Use of oil bodies and oleosins in recombinant protein production and other biotechnological applications. Biotechnol. Adv. 28, 293–300. 10.1016/j.biotechadv.2010.01.001 [DOI] [PubMed] [Google Scholar]
- Bink A., Kucharíková S., Neirinck B., Vleugels J., van Dijck P., Cammue B. P. A., et al. (2012). The nonsteroidal antiinflammatory drug diclofenac potentiates the in vivo activity of caspofungin against Candida albicans biofilms. J. Infect. Dis. 206, 1790–1797. 10.1093/infdis/jis594 [DOI] [PubMed] [Google Scholar]
- Blondelle S. E., Lohner K. (2000). Combinatorial libraries: a tool to design antimicrobial and antifungal peptide analogues having lytic specificities for structure–activity relationship studies. J. Pept. Sci. 55, 74–87. [DOI] [PubMed] [Google Scholar]
- Bondaryk M., Staniszewska M., Zielinska P., Urbanczyk-Lipkowska Z. (2017). Natural antimicrobial peptides as inspiration for design of a new generation antifungal compounds. J. Fungi 3:46. 10.3390/jof3030046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongomin F., Gago S., Oladele R. O., Denning D. W. (2017). Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi 3:E57. 10.3390/jof3040057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady R., Woonton B., Gee M. L., O'Connor A. J. (2008). Hierarchical mesoporous silica materials for separation of functional food ingredients — a review. Innov. Food Sci. Emerg. Technol. 9, 243–248. 10.1016/j.ifset.2007.10.002 [DOI] [Google Scholar]
- Bruni N., Capucchio M. T., Biasibetti E., Pessione E., Cirrincione S., Giraudo L., et al. (2016). Antimicrobial activity of lactoferrin-related peptides and applications in human and veterinary medicine. Molecules 21:752. 10.3390/molecules21060752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows L. L., Stark M., Chan C., Glukhov E., Sinnadurai S., Deber C. M. (2006). Activity of novel non-amphipathic cationic antimicrobial peptides against Candida species. J. Antimicrob. Chemother. 57, 899–907. 10.1093/jac/dkl056 [DOI] [PubMed] [Google Scholar]
- Campbell-Platt G., Cook P. E. (2008). Fungi in the production of foods and food ingredients. J. Appl. Microbiol. 67, 117s−131s. 10.1111/j.1365-2672.1989.tb03776.x [DOI] [Google Scholar]
- Candoni A., Caira M., Cesaro S., Busca A., Giacchino M., Fanci R., et al. (2014). Multicentre surveillance study on feasibility, safety and efficacy of antifungal combination therapy for proven or probable invasive fungal diseases in haematological patients: the SEIFEM real-life combo study. Mycoses 57, 342–350. 10.1111/myc.12161 [DOI] [PubMed] [Google Scholar]
- Cavalcante C. S. P., Falcão C. B., Fontenelle R. O., Andreu D., Rádis-Baptista G. (2017). Anti-fungal activity of Ctn[15–34], the C-terminal peptide fragment of crotalicidin, a rattlesnake venom gland cathelicidin. J. Antibiot. Res. 70, 231–237. 10.1038/ja.2016.135 [DOI] [PubMed] [Google Scholar]
- Chahardoli M., Fazeli A., Ghabooli M. (2018). Recombinant production of bovine lactoferrin-derived antimicrobial peptide in tobacco hairy roots expression system. Plant Physiol. Biochem. 123, 414–421. 10.1016/j.plaphy.2017.12.037 [DOI] [PubMed] [Google Scholar]
- Chang W., Li Y., Zhang L., Cheng A., Liu Y., Lou H. (2012). Retigeric acid B enhances the efficacy of azoles combating the virulence and biofilm formation of Candida albicans. Biol. Pharm. Bull. 35, 1794–1801. 10.1248/bpb.b12-00511 [DOI] [PubMed] [Google Scholar]
- Chaudhari A. A., Ashmore D., Nath S., deb Kate K., Dennis V., Singh S. R., et al. (2016). A novel covalent approach to bio-conjugate silver coated single walled carbon nanotubes with antimicrobial peptide. J. Nanobiotechnology 14:58. 10.1186/s12951-016-0211-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. C.-A., Slavin M. A., Sorrell T. C. (2011). Echinocandin antifungal drugs in fungal infections: a comparison. Drugs 71, 11–41. 10.2165/11585270-000000000-00000 [DOI] [PubMed] [Google Scholar]
- Chi Z., Wang F., Chi Z., Yue L., Liu G., Zhang T. (2009). Bioproducts from aureobasidium pullulans, a biotechnologically important yeast. Appl. Microbiol. Biotechnol. 82, 793–804. 10.1007/s00253-009-1882-2 [DOI] [PubMed] [Google Scholar]
- Choi H. J., Seo M. J., Lee J. C., Cheigh C. I., Park H., Ahn C., et al. (2005). Heterologous expression of human β-defensin-1 in bacteriocin-producing Lactococcus lactis. J. Microbiol. Biotechnol. 15, 330–336. [Google Scholar]
- Chongsiriwatana N. P., Miller T. M., Wetzler M., Vakulenko S., Karlsson A. J., Palecek S. P., et al. (2011). Short alkylated peptoid mimics of antimicrobial lipopeptides. Antimicrob. Agents Chemother. 55, 417–420. 10.1128/AAC.01080-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons K. V., Stevens D. A. (1997). Efficacy of nikkomycin Z against experimental pulmonary blastomycosis. Antimicrob. Agents Chemother. 41, 2026–2028. 10.1128/AAC.41.9.2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools T. L., Struyfs C., Drijfhout J. W., Kucharíková S., Lobo Romero C., van Dijck P., et al. (2017). A linear 19-Mer Plant Defensin-Derived peptide acts synergistically with Caspofungin against Candida albicans biofilms. Front. Microbiol. 8:2051. 10.3389/fmicb.2017.02051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa S., Almeida A., Castro A., Domingues L. (2014). Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: the novel Fh8 system. Front. Microbiol. 5:63. 10.3389/fmicb.2014.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W., Han L., Suo F., Liu Z., Zhou L., Zhou Z. (2018). Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond. World J. Microbiol. Biotechnol. 34:145. 10.1007/s11274-018-2531-7 [DOI] [PubMed] [Google Scholar]
- da Silva Malheiros P., Daroit D. J., Brandelli A. (2010). Food applications of liposome-encapsulated antimicrobial peptides. Trends Food Sci. Technol. 21, 284–292. 10.1016/j.tifs.2010.03.003 [DOI] [Google Scholar]
- De Lucca A. J. (2000). Antifungal peptides: potential candidates for the treatment of fungal infections. Expert Opin. Investig. Drugs 9, 273–299. 10.1517/13543784.9.2.273 [DOI] [PubMed] [Google Scholar]
- De Lucca A. J., Bland J. M., Jacks T. J., Grimm C., Walsh T. J. (1998). Fungicidal and binding properties of the natural peptides cecropin B and dermaseptin. Med. Mycol. 36, 291–298. 10.1080/02681219880000461 [DOI] [PubMed] [Google Scholar]
- De Lucca A. J., Walsh T. J. (1999). Antifungal peptides: novel therapeutic compounds against emerging pathogens. Antimicrob. Agents Chemother. 43, 1–11. 10.1128/AAC.43.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de O Mello E., dos Santos I. S., de O Carvalho A., de Souza L. S., de Souza-Filho G. A., do Nascimento VV., et al. (2014). Functional expression and activity of the recombinant antifungal defensin PvD1r from Phaseolus vulgaris L. (common bean) seeds. BMC Biochem. 15:7 10.1186/1471-2091-15-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza M. C. P., Santos A. G. D., Reis A. M. M. (2016). Adverse drug reactions in patients receiving systemic antifungal therapy at a high-complexity hospital. J. Clin. Pharmacol. 56, 1507–1515. 10.1002/jcph.772 [DOI] [PubMed] [Google Scholar]
- Deng T., Ge H., He H., Liu Y., Zhai C., Feng L., et al. (2017). The heterologous expression strategies of antimicrobial peptides in microbial systems. Protein Expr. Purif. 140, 52–59. 10.1016/j.pep.2017.08.003 [DOI] [PubMed] [Google Scholar]
- Desai P. N., Shrivastava N., Padh H. (2010). Production of heterologous proteins in plants: Strategies for optimal expression. Biotechnol. Adv. 28, 427–435. 10.1016/j.biotechadv.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Dimarcq J.-L., Bulet P., Hetru C., Hoffmann J. (1998). Cysteine-rich antimicrobial peptides in invertebrates. J. Pept. Sci. 47, 465–477. [DOI] [PubMed] [Google Scholar]
- Duncan V. M. S., O'Neil D. A. (2013). Commercialization of antifungal peptides. Fungal Biol. Rev. 26, 156–165. 10.1016/j.fbr.2012.11.001 [DOI] [Google Scholar]
- Emri T., Majoros L., Tóth V., Pócsi I. (2013). Echinocandins: production and applications. Appl. Microbiol. Biotechnol. 97, 3267–3284. 10.1007/s00253-013-4761-9 [DOI] [PubMed] [Google Scholar]
- Enaud R., Vandenborght L.-E., Coron N., Bazin T., Prevel R., Schaeverbeke T., et al. (2018). The mycobiome: a neglected component in the microbiota-gut-brain axis. Microorganisms 6:22. 10.3390/microorganisms6010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M., Takesako K., Kato I., Yamaguchi H. (1997). Fungicidal action of aureobasidin A, a cyclic depsipeptide antifungal antibiotic, against Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 41, 672–676. 10.1128/AAC.41.3.672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenauer G., Depestel D. D., Carver P. L. (2007). Comparison of echinocandin antifungals. Ther. Clin. Risk Manag. 3, 71–97. 10.2147/tcrm.2007.3.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfand R., Tomalia D. A. (2001). Poly(amidoamine) (PAMAM) dendrimers: from biomimicry to drug delivery and biomedical applications. Drug Discov. Today 6, 427–436. 10.1016/S1359-6446(01)01757-3 [DOI] [PubMed] [Google Scholar]
- Essig A., Hofmann D., Münch D., Gayathri S., Künzler M., Kallio P. T., et al. (2014). Copsin, a novel peptide-based fungal antibiotic interfering with the peptidoglycan synthesis. J. Biol. Chem. 289, 34953–34964. 10.1074/jbc.M114.599878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X.-J., Xing L.-W., Liu D., Song X.-Y., Liu C.-L., Li J., et al. (2014). Design and high-level expression of a hybrid antimicrobial peptide LF15-CA8 in Escherichia coli. J. Ind. Microbiol. Biotechnol. 41, 527–534. 10.1007/s10295-013-1382-3 [DOI] [PubMed] [Google Scholar]
- Fernandes K. E., Carter D. A. (2017). The antifungal activity of lactoferrin and its derived peptides: mechanisms of action and synergy with drugs against fungal pathogens. Front. Microbiol. 8:2. 10.3389/fmicb.2017.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell C. D., Hancock R. E. W., Cherkasov A. (2007). AMPer: a database and an automated discovery tool for antimicrobial peptides. Bioinformatics 23, 1148–1155. 10.1093/bioinformatics/btm068 [DOI] [PubMed] [Google Scholar]
- Fjell C. D., Hiss J. A., Hancock R. E. W., Schneider G. (2011). Designing antimicrobial peptides: form follows function. Nat. Rev. Drug Discov. 11, 37–51. 10.1038/nrd3591 [DOI] [PubMed] [Google Scholar]
- Fjell C. D., Jenssen H., Fries P., Aich P., Griebel P., Hilpert K., et al. (2008). Identification of novel host defense peptides and the absence of α-defensins in the bovine genome. Proteins 73, 420–430. 10.1002/prot.22059 [DOI] [PubMed] [Google Scholar]
- Fosgerau K., Hoffmann T. (2015). Peptide therapeutics: current status and future directions. Drug Discov. Today 20, 122–128. 10.1016/j.drudis.2014.10.003 [DOI] [PubMed] [Google Scholar]
- Fox J. L. (2013). Antimicrobial peptides stage a comeback. Nat. Biotechnol. 31, 379–382. 10.1038/nbt.2572 [DOI] [PubMed] [Google Scholar]
- Freitas C. G., Franco O. L. (2016). “Antifungal peptides with potential against pathogenicf fungi” in Recent Trends in Antifungal Agents and Antifungal Therapy, eds A. Basak, R. Chakraborty, and S. M. Mandal (New Delhi: Springer India; ), 75–95. 10.1007/978-81-322-2782-3_3 [DOI] [Google Scholar]
- García-Fruitós E. (2012). Lactic acid bacteria: a promising alternative for recombinant protein production. Microb. Cell Fact. 11:157. 10.1186/1475-2859-11-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigues S., Gandía M., Borics A., Marx F., Manzanares P., Marcos J. F. (2017). Mapping and identification of antifungal peptides in the putative antifungal protein AfpB from the filamentous fungus Penicillium digitatum. Front. Microbiol. 8:592. 10.3389/fmicb.2017.00592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigues S., Gandía M., Castillo L., Coca M., Marx F., Marcos J. F., et al. (2018). Three antifungal proteins from Penicillium expansum: different patterns of production and antifungal activity. Front. Microbiol. 9:2370. 10.3389/fmicb.2018.02370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George F., Daniel C., Thomas M., Singer E., Guilbaud A., Tessier F. J., et al. (2018). Occurrence and dynamism of lactic acid bacteria in distinct ecological niches: a multifaceted functional health perspective. Front. Microbiol. 9:2899. 10.3389/fmicb.2018.02899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Hu H., Zhao Q., Wang T., Zou Y., Yu S., et al. (2012). Synthesis and antifungal activities of glycosylated derivatives of the cyclic peptide fungicide caspofungin. ChemMedChem 7, 1496–1503. 10.1002/cmdc.201200214 [DOI] [PubMed] [Google Scholar]
- Guyard C., Dehecq E., Tissier J.-P., Polonelli L., Dei-Cas E., Cailliez J.-C., et al. (2002). Involvement of [beta]-glucans in the wide-spectrum antimicrobial activity of Williopsis saturnus var. mrakii MUCL 41968 killer toxin. Mol. Med. 8, 686–694. 10.1007/BF03402032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Chapple D. S. (1999). Peptide antibiotics. Antimicrob. Agents Chemother. 43, 1317–1323. 10.1128/AAC.43.6.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harir M., Bendif H., Bellahcene M., Fortas Z., Pogni R. (2018). Streptomyces Secondary Metabolites, Basic Biology and Applications of Actinobacteria, Shymaa Enany. IntechOpen. Available online at: https://www.intechopen.com/books/basic-biology-and-applications-of-actinobacteria/streptomyces-secondary-metabolites
- Harris M. R., Coote P. J. (2010). Combination of caspofungin or anidulafungin with antimicrobial peptides results in potent synergistic killing of Candida albicans and Candida glabrata in vitro. Int. J. Antimicrob. Agents 35, 347–356. 10.1016/j.ijantimicag.2009.11.021 [DOI] [PubMed] [Google Scholar]
- Harzallah D., Belhadj H. (2013). “Lactic acid bacteria as probiotics: characteristics, selection criteria and role in immunomodulation of human GI muccosal barrier,” in Lactic Acid Bacteria - R & D for Food, Health and Livestock Purposes. Marcelino Kongo (IntechOpen) Available online at: https://www.intechopen.com/books/lactic-acid-bacteria-r-d-for-food-health-and-livestock-purposes/lactic-acid-bacteria-as-probiotics-characteristics-selection-criteria-and-role-in-immunomodulation-o [Google Scholar]
- Hector R. F., Zimmer B. L., Pappagianis D. (1990). Evaluation of nikkomycins X and Z in murine models of coccidioidomycosis, histoplasmosis, and blastomycosis. Antimicrob. Agents Chemother. 34, 587–593. 10.1128/aac.34.4.587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmerhorst E. J., Breeuwer P., van't Hof W., Walgreen-Weterings E., Oomen L. C., Veerman E. C., et al. (1999). The cellular target of histatin 5 on Candida albicans is the energized mitochondrion. J. Biol. Chem. 274, 7286–7291. 10.1074/jbc.274.11.7286 [DOI] [PubMed] [Google Scholar]
- Hirsch T., Metzig M., Niederbichler A., Steinau H.-U., Eriksson E., Steinstraesser L. (2008). Role of host defense peptides of the innate immune response in sepsis. Shock 30, 117–126. 10.1097/shk.0b013e318160de11 [DOI] [PubMed] [Google Scholar]
- Hoffmeister D., Keller N. P. (2007). Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep. 24, 393–416. 10.1039/b603084j [DOI] [PubMed] [Google Scholar]
- Holaskova E., Galuszka P., Frebort I., Oz M. T. (2015). Antimicrobial peptide production and plant-based expression systems for medical and agricultural biotechnology. Biotechnol. Adv. 33, 1005–1023. 10.1016/j.biotechadv.2015.03.007 [DOI] [PubMed] [Google Scholar]
- Hori M., Kakiki K., Misato T. (1974). Interaction between polyoxin and active center of chitin synthetase. Agric. Biol. Chem. 38, 699–705. 10.1080/00021369.1974.10861226 [DOI] [Google Scholar]
- Hou W., Zhang X., Liu C.-F. (2017). Progress in chemical synthesis of peptides and proteins. Trans. Tianjin Univ. 23, 401–419. 10.1007/s12209-017-0068-8 [DOI] [Google Scholar]
- Houwaart S., Youssar L., Hüttel W. (2014). Pneumocandin biosynthesis: involvement of a trans-selective proline hydroxylase. ChemBioChem 15, 2365–2369. 10.1002/cbic.201402175 [DOI] [PubMed] [Google Scholar]
- Hsieh I.-N., Hartshorn K. L. (2016). The role of antimicrobial peptides in influenza virus infection and their potential as antiviral and immunomodulatory therapy. Pharmaceuticals 9:53. 10.3390/ph9030053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. W. (2006). Molecular mechanism of antimicrobial peptides: the origin of cooperativity. Biochim. Biophys. Acta 1758, 1292–1302. 10.1016/j.bbamem.2006.02.001 [DOI] [PubMed] [Google Scholar]
- Huang L., Ching C. B., Jiang R., Leong S. S. J. (2008). Production of bioactive human beta-defensin 5 and 6 in Escherichia coli by soluble fusion expression. Protein Expr. Purif. 61, 168–174. 10.1016/j.pep.2008.05.016 [DOI] [PubMed] [Google Scholar]
- Huang L., Leong S. S. J., Jiang R. (2009). Soluble fusion expression and characterization of bioactive human beta-defensin 26 and 27. Appl. Microbiol. Biotechnol. 84, 301–308. 10.1007/s00253-009-1982-z [DOI] [PubMed] [Google Scholar]
- Huffnagle G. B., Noverr M. C. (2013). The emerging world of the fungal microbiome. Trends. Microbiol 21, 334–341. 10.1016/j.tim.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh E., Akhtar N., Li J. (2018). Efficient production of recombinant protegrin-1 from Pichia pastoris, and its antimicrobial and in vitro cell migration activity. Front. Microbiol. 9:12. 10.3389/fmicb.2018.02300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K., Asahi K., Suzuki S. (1969). Polyoxins, antifungal antibiotics. XIII. Structure of polyoxins. J. Am. Chem. Soc. 91, 7490–7505. 10.1021/ja01054a045 [DOI] [PubMed] [Google Scholar]
- Jang W. S., Bajwa J. S., Sun J. N., Edgerton M. (2010). Salivary histatin 5 internalization by translocation, but not endocytosis, is required for fungicidal activity in Candida albicans. Mol. Microbiol. 77, 354–370. 10.1111/j.1365-2958.2010.07210.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S., Li W., Baloch A. R., Wang M., Li H., Cao B., et al. (2017). Efficient biosynthesis of a Cecropin A-melittin mutant in Bacillus subtilis WB700. Sci. Rep. 7:40587 10.1038/srep40587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Dong F., Shi C., Liu S., Sun J., Chen J., et al. (2019). DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data 6:148. 10.1038/s41597-019-0154-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitan M., Niemiec M. J., Steimle A., Frick J. S., Jacobsen I. D. (2018). Fungi as part of the microbiota and interactions with intestinal bacteria. Curr. Top. Microbiol. Immunol. 422, 265–301. 10.1007/82_2018_117 [DOI] [PubMed] [Google Scholar]
- Karlowsky J. A., Harding G. A., Zelenitsky S. A., Hoban D. J., Kabani A., Balko T. V., et al. (1997). In vitro kill curves of a new semisynthetic echinocandin, LY-303366, against fluconazole-sensitive and -resistant Candida species. Antimicrob. Agents Chemother. 41, 2576–2578. 10.1128/AAC.41.11.2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik N., Pujalte G. G. A., Reese S. T. (2015). Superficial fungal infections. Prim. Care 42, 501–516. 10.1016/j.pop.2015.08.004 [DOI] [PubMed] [Google Scholar]
- Kawabata S., Nagayama R., Hirata M., Shigenaga T., Agarwala K. L., Saito T., et al. (1996). Tachycitin, a small granular component in horseshoe crab hemocytes, is an antimicrobial protein with chitin-binding activity. J. Biochem. 120, 1253–1260. 10.1093/oxfordjournals.jbchem.a021549 [DOI] [PubMed] [Google Scholar]
- Kim H.-K., Chun D.-S., Kim J.-S., Yun C.-H., Lee J.-H., Hong S.-K., et al. (2006). Expression of the cationic antimicrobial peptide lactoferricin fused with the anionic peptide in Escherichia coli. Appl. Microbiol. Biotechnol. 72, 330–338. 10.1007/s00253-005-0266-5 [DOI] [PubMed] [Google Scholar]
- König H., Fröhlich J. (2017). “Lactic Acid Bacteria,” in Biology of Microorganisms on Grapes, in Must and in Wine, eds H. König, G. Unden, and J. Fröhlich (Cham: Springer International Publishing; ), 3–41. 10.1007/978-3-319-60021-5_1 [DOI] [Google Scholar]
- Konno H., Abumi K., Sasaki Y., Yano S., Nosaka K. (2015). Structure activity relationship study of burkholdine analogues toward simple antifungal agents. Bioorg. Med. Chem. Lett. 25, 3199–3202. 10.1016/j.bmcl.2015.05.088 [DOI] [PubMed] [Google Scholar]
- Koo H. B., Seo J. (2019). Antimicrobial peptides under clinical investigation. J. Pept. Sci. 111:e24122 10.1002/pep2.24122 [DOI] [Google Scholar]
- Koshlukova S. E., Lloyd T. L., Araujo M. W., Edgerton M. (1999). Salivary histatin 5 induces non-lytic release of ATP from Candida albicans leading to cell death. J. Biol. Chem. 274, 18872–18879. 10.1074/jbc.274.27.18872 [DOI] [PubMed] [Google Scholar]
- Kosobokova E. N., Skrypnik K. A., Kosorukov V. S. (2016). Overview of fusion tags for recombinant proteins. Biochem. Mosc. 81, 187–200. 10.1134/S0006297916030019 [DOI] [PubMed] [Google Scholar]
- Kumar R., Chadha S., Saraswat D., Bajwa J. S., Li R. A., Conti H. R., et al. (2011). Histatin 5 uptake by Candida albicans utilizes polyamine transporters Dur3 and Dur31 proteins. J. Biol. Chem. 286, 43748–43758. 10.1074/jbc.M111.311175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakidis I., Tragiannidis A., Munchen S., Groll A. H. (2017). Clinical hepatotoxicity associated with antifungal agents. Expert Opin. Drug Saf. 16, 149–165. 10.1080/14740338.2017.1270264 [DOI] [PubMed] [Google Scholar]
- Lafleur M. D., Sun L., Lister I., Keating J., Nantel A., Long L., et al. (2013). Potentiation of azole antifungals by 2-adamantanamine. Antimicrob. Agents Chemother. 57, 3585–3592. 10.1128/AAC.00294-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshminarayanan R., Liu S., Li J., Nandhakumar M., Aung T. T., Goh E., et al. (2014). Synthetic multivalent antifungal peptides effective against fungi. PLoS ONE 9:e87730. 10.1371/journal.pone.0087730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landim P. G. C., Correia T. O., Silva F. D. A., Nepomuceno D. R., Costa H. P. S., Pereira H. M., et al. (2017). Production in Pichia pastoris, antifungal activity and crystal structure of a class I chitinase from cowpea (Vigna unguiculata): Insights into sugar binding mode and hydrolytic action. Biochimie 135, 89–103. 10.1016/j.biochi.2017.01.014 [DOI] [PubMed] [Google Scholar]
- Landy M., Warren G. H., Rosenman M. S. B., Colio L. G. (1948). Bacillomycin: an antibiotic from Bacillus subtilis active against pathogenic fungi. Proc. Soc. Exp. Biol. Med. 67, 539–541. 10.3181/00379727-67-16367 [DOI] [PubMed] [Google Scholar]
- Latoud C., Peypoux F., Michel G., Genet R., Morgat J. L. (1986). Interactions of antibiotics of the iturin group with human erythrocytes. Biochim. Biophys. Acta 856, 526–535. 10.1016/0005-2736(86)90144-6 [DOI] [PubMed] [Google Scholar]
- Lee D. G., Kim P. I., Park Y., Woo E.-R., Choi J. S., Choi C.-H., et al. (2002). Design of novel peptide analogs with potent fungicidal activity, based on PMAP-23 antimicrobial peptide isolated from porcine myeloid. Biochem. Biophys. Res. 293, 231–238. 10.1016/S0006-291X(02)00222-X [DOI] [PubMed] [Google Scholar]
- Lee K. H., Lim Y. T., Hah J. O., Kim Y. K., Lee C. H., Lee J. M. (2017). Voriconazole plus caspofungin for treatment of invasive fungal infection in children with acute leukemia. Blood Res. 52, 167–173. 10.5045/br.2017.52.3.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Blencke H.-M., Paulsen V., Haug T., Stensvåg K. (2010). Powerful workhorses for antimicrobial peptide expression and characterization. Bioeng. Bugs 1, 217–220. 10.4161/bbug.1.3.11721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li L., Feng C., Chen Y., Tan H. (2012). Novel polyoxins generated by heterologously expressing polyoxin biosynthetic gene cluster in the sanN inactivated mutant of Streptomyces ansochromogenes. Microb. Cell Fact. 11:135. 10.1186/1475-2859-11-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. S., Sun J. N., Okamoto-Shibayama K., Edgerton M. (2006). Candida albicans cell wall ssa proteins bind and facilitate import of salivary histatin 5 required for toxicity. J. Biol. Chem. 281, 22453–22463. 10.1074/jbc.M604064200 [DOI] [PubMed] [Google Scholar]
- Li Y. (2009). Carrier proteins for fusion expression of antimicrobial peptides in Escherichia coli. Biotechnol. Appl. Biochem. 54, 1–9. 10.1042/BA20090087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. (2011). Recombinant production of antimicrobial peptides in Escherichia coli: a review. Protein Expr. Purif. 80, 260–267. 10.1016/j.pep.2011.08.001 [DOI] [PubMed] [Google Scholar]
- Li Y., Ling H., Li W., Tan H. (2005). Improvement of nikkomycin production by enhanced copy of sanU and sanV in Streptomyces ansochromogenes and characterization of a novel glutamate mutase encoded by sanU and sanV. Metab. Eng. 7, 165–173. 10.1016/j.ymben.2005.01.002 [DOI] [PubMed] [Google Scholar]
- Li Z., Mao R., Teng D., Hao Y., Chen H., Wang X., et al. (2017). Antibacterial and immunomodulatory activities of insect defensins-DLP2 and DLP4 against multidrug-resistant Staphylococcus aureus. Sci. Rep. 7, 1–16. 10.1038/s41598-017-10839-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H. (2007). Oleosin fusion expression systems for the production of recombinant proteins. Biologia 62, 119–123. 10.2478/s11756-007-0041-4 [DOI] [Google Scholar]
- López-Abarrategui C., Figueroa-Espí V., Reyes-Acosta O., Reguera E., Otero-Gonzalez A. J. (2013). Magnetic nanoparticles: new players in antimicrobial peptide therapeutics. Curr. Protein Pept. Sci. 14, 595–606. 10.2174/1389203711209070682 [DOI] [PubMed] [Google Scholar]
- López-García B., Harries E., Carmona L., Campos-Soriano L., López J. J., Manzanares P., et al. (2015). Concatemerization increases the inhibitory activity of short, cell-penetrating, cationic and tryptophan-rich antifungal peptides. Appl. Microbiol. Biotechnol. 99, 8011–8021. 10.1007/s00253-015-6541-1 [DOI] [PubMed] [Google Scholar]
- Luan C., Zhang H. W., Song D. G., Xie Y. G., Feng J., Wang Y. Z. (2014). Expressing antimicrobial peptide cathelicidin-BF in Bacillus subtilis using SUMO technology. Appl. Microbiol. Biotechnol. 98, 3651–3658. 10.1007/s00253-013-5246-6 [DOI] [PubMed] [Google Scholar]
- Lum K. Y., Tay S. T., Le C. F., Lee V. S., Sabri N. H., Velayuthan R. D., et al. (2015). Activity of novel synthetic peptides against Candida albicans. Sci. Rep. 5, 9657–9657. 10.1038/srep09657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupetti A., Paulusma-Annema A., Welling M. M., Dogterom-Ballering H., Brouwer C. P. J. M., Senesi S., et al. (2003). Synergistic activity of the N-terminal peptide of human lactoferrin and fluconazole against Candida species. Antimicrob. Agents Chemother. 47, 262–267. 10.1128/aac.47.1.262-267.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliani W., Conti S., Ciociola T., Giovati L., Zanello P. P., Pertinhez T., et al. (2011). Killer peptide: a novel paradigm of antimicrobial, antiviral and immunomodulatory auto-delivering drugs. Future Med. Chem. 3, 1209–1231. 10.4155/fmc.11.71 [DOI] [PubMed] [Google Scholar]
- Mahlapuu M., Håkansson J., Ringstad L., Björn C. (2016). Antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 6:194. 10.3389/fcimb.2016.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mania D., Hilpert K., Ruden S., Fischer R., Takeshita N. (2010). Screening for antifungal peptides and their modes of action in Aspergillus nidulans. Appl. Environ. Microbiol. 76, 7102–7108. 10.1128/AEM.01560-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L., van Meegern A., Doemming S., Schuerholz T. (2015). Antimicrobial peptides in human sepsis. Front. Immunol. 6:404. 10.3389/fimmu.2015.00404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matejuk A., Leng Q., Begum M. D., Woodle M. C., Scaria P., Chou S.-T., et al. (2010). Peptide-based antifungal therapies against emerging infections. Drugs Future 35:197. 10.1358/dof.2010.035.03.1452077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattay J., Houwaart S., Hüttel W. (2018). Cryptic production of trans-3-hydroxyproline in echinocandin b biosynthesis. Appl. Environ. Microbiol. 84:e02370-17. 10.1128/AEM.02370-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy P. J., Troke P. F., Gull K. (1985). Mechanism of action of nikkomycin and the peptide transport system of Candida albicans. J. Gen. Microbiol. 131, 775–780. 10.1099/00221287-131-4-775 [DOI] [PubMed] [Google Scholar]
- McDonald E. E., Goldberg H. A., Tabbara N., Mendes F. M., Siqueira W. L. (2011). Histatin 1 resists proteolytic degradation when adsorbed to hydroxyapatite. J. Dent. Res. 90, 268–272. 10.1177/0022034510388653 [DOI] [PubMed] [Google Scholar]
- McNair L. K. F., Siedler S., Vinther J. M. O., Hansen A. M., Neves A. R., Garrigues C., et al. (2018). Identification and characterization of a new antifungal peptide in fermented milk product containing bioprotective Lactobacillus cultures. FEMS Yeast Res. 18:foy094. 10.1093/femsyr/foy094 [DOI] [PubMed] [Google Scholar]
- Melo M. N., Ferre R., Castanho M. A. R. B. (2009). Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 7, 245–250. 10.1038/nrmicro2095 [DOI] [PubMed] [Google Scholar]
- Mikamo H., Sato Y., Tamaya T. (2000). In vitro antifungal activity of FK463, a new water-soluble echinocandin-like lipopeptide. J. Antimicrob. Chemother. 46, 485–487. 10.1093/jac/46.3.485 [DOI] [PubMed] [Google Scholar]
- Money N. P. (2016). Fungi and biotechnology. J. Fungi. 2016, 401–424. 10.1016/B978-0-12-382034-1.00012-8 [DOI] [Google Scholar]
- Mor A., Nguyen V. H., Delfour A., Migliore-Samour D., Nicolas P. (1991). Isolation, amino acid sequence and synthesis of dermaseptin, a novel antimicrobial peptide of amphibian skin. Biochemistry 30, 8824–8830. 10.1021/bi00100a014 [DOI] [PubMed] [Google Scholar]
- Mosca D. A., Hurst M. A., So W., Viajar B. S., Fujii C. A., Falla T. J. (2000). IB-367, a protegrin peptide with in vitro and in vivo activities against the microflora associated with oral mucositis. Antimicrob. Agents Chemother. 44, 1803–1808. 10.1128/aac.44.7.1803-1808.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D., Singh S., Kumar M., Kumar V., Datta S., Dhanjal D. S. (2018). “Fungal biotechnology: role and aspects,” in Fungi and their Role in Sustainable Development: Current Perspectives, eds P. Gehlot and J. Singh (Singapore: Springer Singapore; ), 91–103. 10.1007/978-981-13-0393-7_6 [DOI] [Google Scholar]
- Mukhopadhyay K., Prasad T., Saini P., Pucadyil T. J., Chattopadhyay A., Prasad R. (2004). Membrane sphingolipid-ergosterol interactions are important determinants of multidrug resistance in Candida albicans. Antimicrob. Agents Chemother. 48, 1778–1787. 10.1128/aac.48.5.1778-1787.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagiec M. M., Nagiec E. E., Baltisberger J. A., Wells G. B., Lester R. L., Dickson R. C. (1997). Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J. Biol. Chem. 272, 9809–9817. 10.1074/jbc.272.15.9809 [DOI] [PubMed] [Google Scholar]
- Ng-Choi I., Oliveras À., Planas M., Feliu L. (2019). Solid-phase synthesis of biaryl cyclic peptides containing a histidine-tyrosine linkage. Tetrahedron 75, 2625–2636. 10.1016/j.tet.2019.03.014 [DOI] [Google Scholar]
- Nguyen L. T., Haney E. F., Vogel H. J. (2011). The expanding scope of antimicrobial peptide structures and their modes of action. Trends. Biotechnol. 29, 464–472. 10.1016/j.tibtech.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Nicola A. M., Albuquerque P., Paes H. C., Fernandes L., Costa F. F., Kioshima E. S., et al. (2019). Antifungal drugs: new insights in research & development. Pharmacol. Ther. 195, 21–38. 10.1016/j.pharmthera.2018.10.008 [DOI] [PubMed] [Google Scholar]
- Niu M., Chai S., You X., Wang W., Qin C., Gong Q., et al. (2015). Expression of porcine protegrin-1 in Pichia pastoris and its anticancer activity in vitro. Exp. Ther. Med. 9, 1075–1079. 10.3892/etm.2015.2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani M. F., Yordanova S., Cangiotti M., Vasileva-Tonkova E., Coppola C., Stoyanov S., et al. (2016). Spectral characterization and in vitro microbiological activity of new bis-1,8-naphthalimides and their Cu(II) complexes. J. Mol. Struct. 1110, 72–82. 10.1016/j.molstruc.2016.01.033 [DOI] [Google Scholar]
- Pappas P. G., Lionakis M. S., Arendrup M. C., Ostrosky-Zeichner L., Kullberg B. J. (2018). Invasive candidiasis. Nat. Rev. Dis. Primers 4:18026. 10.1038/nrdp.2018.26 [DOI] [PubMed] [Google Scholar]
- Park S.-C., Kim J.-Y., Kim E.-J., Cheong G.-W., Lee Y., Choi W., et al. (2018). Hydrophilic linear peptide with histidine and lysine residues as a key factor affecting antifungal activity. Int. J. Mol. Sci. 19:3781. 10.3390/ijms19123781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-C., Kim Y.-M., Lee J.-K., Kim N.-H., Kim E.-J., Heo H., et al. (2017). Targeting and synergistic action of an antifungal peptide in an antibiotic drug-delivery system. J. Control. Release 256, 46–55. 10.1016/j.jconrel.2017.04.023 [DOI] [PubMed] [Google Scholar]
- Pasrija R., Panwar S. L., Prasad R. (2008). Multidrug transporters CaCdr1p and CaMdr1p of Candida albicans display different lipid specificities: both ergosterol and sphingolipids are essential for targeting of CaCdr1p to membrane rafts. Antimicrob. Agents Chemother. 52, 694–704. 10.1128/AAC.00861-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasrija R., Prasad T., Prasad R. (2005). Membrane raft lipid constituents affect drug susceptibilities of Candida albicans. Biochem. Soc. Trans. 33, 1219–1223. 10.1042/BST0331219 [DOI] [PubMed] [Google Scholar]
- Pecorini C., Savazzini F., Martino P. A., Fusi E., Fogher C., Baldi A. (2005). Heterologous expression of biologically active porcine lactoferrin in Pichia pastoris yeast. Vet. Res. Commun. 29 (Suppl. 2), 379–382. 10.1007/s11259-005-0086-1 [DOI] [PubMed] [Google Scholar]
- Pfaller M., Gordee R., Gerarden T., Yu M., Wenzel R. (1989). Fungicidal activity of cilofungin (LY121019) alone and in combination with anticapsin or other antifungal agents. Eur. J. Clin. Microbiol. Infect. Dis. 8, 564–567. 10.1007/BF01967483 [DOI] [PubMed] [Google Scholar]
- Popa C., Shi X., Ruiz T., Ferrer P., Coca M. (2019). Biotechnological production of the cell penetrating antifungal PAF102 Peptide in Pichia pastoris. Front. Microbiol. 10:1472. 10.3389/fmicb.2019.01472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter E. A., Weisblum B., Gellman S. H. (2002). Mimicry of host-defense peptides by unnatural oligomers: antimicrobial β-peptides. J. Am. Chem. Soc. 124, 7324–7330. 10.1021/ja0260871 [DOI] [PubMed] [Google Scholar]
- Porto W. F., Silva O. N., Franco O. L. (2012). “Prediction and rational design of antimicrobial Peptides,” in Protein Structure Eshel Faraggi (IntechOpen). Available online at: https://www.intechopen.com/books/protein-structure/prediction-and-rational-design-of-antimicrobial-peptides
- Pound M. W., Townsend M. L., Dimondi V., Wilson D., Drew R. H. (2011). Overview of treatment options for invasive fungal infections. Med. Mycol. 49, 561–580. 10.3109/13693786.2011.560197 [DOI] [PubMed] [Google Scholar]
- Raj P. A., Marcus E., Sukumaran D. K. (1998). Structure of human salivary histatin 5 in aqueous and nonaqueous solutions. Biopolymers 45, 51–67. [DOI] [PubMed] [Google Scholar]
- Ramos R., Moreira S., Rodrigues A., Gama M., Domingues L. (2013). Recombinant expression and purification of the antimicrobial peptide magainin-2. Biotechnol. Prog. 29, 17–22. 10.1002/btpr.1650 [DOI] [PubMed] [Google Scholar]
- Rautemaa-Richardson R., Richardson M. D. (2017). Systemic fungal infections. Medicine 45, 757–762. 10.1016/j.mpmed.2017.09.007 [DOI] [Google Scholar]
- Rautenbach M., Troskie A. M., Vosloo J. A. (2016). Antifungal peptides: to be or not to be membrane active. Biochimie 130, 132–145. 10.1016/j.biochi.2016.05.013 [DOI] [PubMed] [Google Scholar]
- Robinson J. A. (2011). Protein epitope mimetics as anti-infectives. Curr. Opin. Chem. Biol. 15, 379–386. 10.1016/j.cbpa.2011.02.015 [DOI] [PubMed] [Google Scholar]
- Rodríguez J. M., Martínez M. I., Horn N., Dodd H. M. (2003). Heterologous production of bacteriocins by lactic acid bacteria. Int. J. Food Microbiol. 80, 101–116. 10.1016/S0168-1605(02)00153-8 [DOI] [PubMed] [Google Scholar]
- Roemer T., Krysan D. J. (2014). Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harb. Perspect. Med. 4:a019703. 10.1101/cshperspect.a019703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldán-Tapia M., Anné J., Reyes A. G., Carrasco U., Millán-Pacheco C., Barrios-González J., et al. (2017). Streptomyces as overexpression system for heterologous production of an antimicrobial peptide. Protein Pept. Lett. 24, 483–488. 10.2174/0929866524666170208154327 [DOI] [PubMed] [Google Scholar]
- Rosano G. L., Ceccarelli E. A. (2014). Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol. 5:172. 10.3389/fmicb.2014.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscetto E., Contursi P., Vollaro A., Fusco S., Notomista E., Catania M. R. (2018). Antifungal and anti-biofilm activity of the first cryptic antimicrobial peptide from an archaeal protein against Candida spp. clinical isolates. Sci. Rep. 8:17570. 10.1038/s41598-018-35530-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein D. M., Spacciapoli P., Tran L. T., Xu T., Roberts F. D., Dalla Serra M., et al. (2001). Anticandida activity is retained in p-113, a 12-amino-acid fragment of histatin 5. Antimicrob. Agents Chemother. 45, 1367–1373. 10.1128/AAC.45.5.1367-1373.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder K., Bekhit A. E.-D., McConnell M., Carne A. (2016). Towards generation of bioactive peptides from meat industry waste proteins: generation of peptides using commercial microbial proteases. Food Chem. 208, 42–50. 10.1016/j.foodchem.2016.03.121 [DOI] [PubMed] [Google Scholar]
- Saito T., Kawabata S., Shigenaga T., Takayenoki Y., Cho J., Nakajima H., et al. (1995). A novel big defensin identified in horseshoe crab hemocytes: isolation, amino acid sequence, and antibacterial activity. J. Biochem. 117, 1131–1137. 10.1093/oxfordjournals.jbchem.a124818 [DOI] [PubMed] [Google Scholar]
- Sajjan U. S., Tran L. T., Sole N., Rovaldi C., Akiyama A., Friden P. M., et al. (2001). P-113D, an antimicrobial peptide active against Pseudomonas aeruginosa, retains activity in the presence of sputum from cystic fibrosis patients. Antimicrob. Agents Chemother. 45, 3437–3444. 10.1128/AAC.45.12.3437-3444.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garcia L., Martín L., Mangues R., Ferrer-Miralles N., Vázquez E., Villaverde A. (2016). Recombinant pharmaceuticals from microbial cells: a 2015 update. Microb. Cell Fact. 15:33. 10.1186/s12934-016-0437-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti M., Posteraro B., Lass-Flörl C. (2015). Antifungal drug resistance among Candida species: mechanisms and clinical impact. Mycoses 58, 2–13. 10.1111/myc.12330 [DOI] [PubMed] [Google Scholar]
- Schaaper W. M., Posthuma G. A., Plasman H. H., Sijtsma L., Fant F., Borremans F. A., et al. (2001). Synthetic peptides derived from the beta2-beta3 loop of Raphanus sativus antifungal protein 2 that mimic the active site. J. Pept. Res. 57, 409–418. 10.1034/j.1399-3011.2001.00842.x [DOI] [PubMed] [Google Scholar]
- Schibli D. J., Epand R. F., Vogel H. J., Epand R. M. (2002). Tryptophan-rich antimicrobial peptides: comparative properties and membrane interactions. Biochem. Cell Biol. 80, 667–677. 10.1139/o02-147 [DOI] [PubMed] [Google Scholar]
- Schneider G., Fechner U. (2005). Computer-based de novo design of drug-like molecules. Nat. Rev. Drug Discov. 4, 649–663. 10.1038/nrd1799 [DOI] [PubMed] [Google Scholar]
- Semlali A., Leung K. P., Curt S., Rouabhia M. (2011). Antimicrobial decapeptide KSL-W attenuates Candida albicans virulence by modulating its effects on toll-like receptor, human β-defensin, and cytokine expression by engineered human oral mucosa. Peptides 32, 859–867. 10.1016/j.peptides.2011.01.020 [DOI] [PubMed] [Google Scholar]
- Sewczyk T., Hoog Antink M., Maas M., Kroll S., Beutel S. (2018). Flow rate dependent continuous hydrolysis of protein isolates. AMB Express 8:18. 10.1186/s13568-018-0548-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahi P., Moye-Rowley W. S. (2009). Coordinate control of lipid composition and drug transport activities is required for normal multidrug resistance in fungi. Biochim. Biophys. Acta 1794, 852–859. 10.1016/j.bbapap.2008.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai Y. (2002). Mode of action of membrane active antimicrobial peptides. Biopolymers 66, 236–248. 10.1002/bip.10260 [DOI] [PubMed] [Google Scholar]
- Shams M. V., Nazarian-Firouzabadi F., Ismaili A., Shirzadian-Khorramabad R. (2019). Production of a recombinant dermaseptin peptide in nicotiana tabacum hairy roots with enhanced antimicrobial activity. Mol. Biotechnol. 61, 241–252. 10.1007/s12033-019-00153-x [DOI] [PubMed] [Google Scholar]
- Sher Khan R., Iqbal A., Malak R., Shehryar K., Attia S., Ahmed T., et al. (2019). Plant defensins: types, mechanism of action and prospects of genetic engineering for enhanced disease resistance in plants. Biotechnology 9:192. 10.1007/s13205-019-1725-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zhu X., Lu Y., Zhao H., Lu F., Lu Z. (2018). Improving iturin a production of bacillus amyloliquefaciens by genome shuffling and its inhibition against Saccharomyces cerevisiae in orange juice. Front. Microbiol. 9:2683. 10.3389/fmicb.2018.02683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnar A. E., Butler K. L., Park H. J. (2003). Cathelicidin family of antimicrobial peptides: proteolytic processing and protease resistance. Bioorg. Chem. 31, 425–436. 10.1016/S0045-2068(03)00080-4 [DOI] [PubMed] [Google Scholar]
- Sibel Akalin A. (2014). Dairy-derived antimicrobial peptides: action mechanisms, pharmaceutical uses and production proposals. Trends Food Sci. Technol. 36, 79–95. 10.1016/j.tifs.2014.01.002 [DOI] [Google Scholar]
- Sinden S. L., DeVay J. E., Backman P. A. (1971). Properties of syringomycin, a wide spectrum antibiotic and phytotoxin produced by Pseudomonas syringae, and its role in the bacterial canker disease of peach trees. Physiol. Plant Pathol. 1, 199–213. 10.1016/0048-4059(71)90029-4 [DOI] [Google Scholar]
- Singh A., Upadhyay V., Upadhyay A. K., Singh S. M., Panda A. K. (2015). Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Fact. 14:41. 10.1186/s12934-015-0222-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Shekhar S., Yadav Y., Xess I., Dey S. (2017). DS6: anticandidal, antibiofilm peptide against Candida tropicalis and exhibit synergy with commercial drug. J. Pept. Sci. 23, 228–235. 10.1002/psc.2973 [DOI] [PubMed] [Google Scholar]
- Slightom J. L., Metzger B. P., Luu H. T., Elhammer A. P. (2009). Cloning and molecular characterization of the gene encoding the aureobasidin A biosynthesis complex in Aureobasidium pullulans BP-1938. Gene 431, 67–79. 10.1016/j.gene.2008.11.011 [DOI] [PubMed] [Google Scholar]
- Song D., Chen Y., Li X., Zhu M., Gu Q. (2014). Heterologous expression and purification of dermaseptin s4 fusion in Escherichia coli and recovery of biological activity. Prep. Biochem. Biotech. 44, 598–607. 10.1080/10826068.2013.835735 [DOI] [PubMed] [Google Scholar]
- Steckbeck J. D., Deslouches B., Montelaro R. C. (2014). Antimicrobial peptides: new drugs for bad bugs? Expert Opin. Biol. Ther. 14, 11–14. 10.1517/14712598.2013.844227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinstraesser L., Kraneburg U. M., Hirsch T., Kesting M., Steinau H.-U., Jacobsen F., et al. (2009). Host defense peptides as effector molecules of the innate immune response: a sledgehammer for drug resistance? Int. J. Mol. Sci. 10, 3951–3970. 10.3390/ijms10093951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetake T., Aizawa T., Koganesawa N., Osaki T., Kobashigawa Y., Demura M., et al. (2002). Production and characterization of recombinant tachycitin, the Cys-rich chitin-binding protein. Protein Eng. 15, 763–769. 10.1093/protein/15.9.763 [DOI] [PubMed] [Google Scholar]
- Suzuki S., Isono K., Nagatsu J., Mizutani T., Kawashima Y., Mizuno T. (1965). A new antibiotic, polyoxin A. J. Antibiot. 18:131. [PubMed] [Google Scholar]
- Tabbene O., Azaiez S., Grazia A. D., Karkouch I., Slimene I. B., Elkahoui S., et al. (2016). Bacillomycin D and its combination with amphotericin B: promising antifungal compounds with powerful antibiofilm activity and wound-healing potency. J. Appl. Microb. 120, 289–300. 10.1111/jam.13030 [DOI] [PubMed] [Google Scholar]
- Tanida T., Rao F., Hamada T., Ueta E., Osaki T. (2001). Lactoferrin peptide increases the survival of Candida albicans- inoculated mice by upregulating neutrophil and macrophage functions, especially in combination with amphotericin B and granulocyte-macrophage colony-stimulating factor. Infect. Immun. 69, 3883–3890. 10.1128/IAI.69.6.3883-3890.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavanti A., Maisetta G., Del Gaudio G., Petruzzelli R., Sanguinetti M., Batoni G., et al. (2011). Fungicidal activity of the human peptide hepcidin 20 alone or in combination with other antifungals against Candida glabrata isolates. Peptides 32, 2484–2487. 10.1016/j.peptides.2011.10.012 [DOI] [PubMed] [Google Scholar]
- Taveira G. B., Mello É. O., Carvalho A. O., Regente M., Pinedo M., Canal L., et al. (2017). Antimicrobial activity and mechanism of action of a thionin-like peptide from Capsicum annuum fruits and combinatorial treatment with fluconazole against Fusarium solani. Pept. Sci. 108:e23008. 10.1002/bip.23008 [DOI] [PubMed] [Google Scholar]
- Terras F. R., Schoofs H. M., de Bolle M. F., van Leuven F., Rees S. B., Vanderleyden J., et al. (1992). Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J. Biol. Chem. 267, 15301–15309. [PubMed] [Google Scholar]
- Thevissen K. (2016). How promising are combinatorial drug strategies in combating Candida albicans biofilms? Future Med. Chem. 8, 1383–1385. 10.4155/fmc-2016-0127 [DOI] [PubMed] [Google Scholar]
- Tóth L., Váradi G., Borics A., Batta G., Kele Z., Vendrinszky Á., et al. (2018). Anti-candidal activity and functional mapping of recombinant and synthetic Neosartorya fischeri antifungal protein 2 (NFAP2). Front. Microbiol. 9:393. 10.3389/fmicb.2018.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracanna V., de Jong A., Medema M. H., Kuipers O. P. (2017). Mining prokaryotes for antimicrobial compounds: from diversity to function. FEMS Microbiol. Rev. 41, 417–429. 10.1093/femsre/fux014 [DOI] [PubMed] [Google Scholar]
- Tran D., Tran P., Roberts K., Osapay G., Schaal J., Ouellette A., et al. (2008). Microbicidal properties and cytocidal selectivity of rhesus macaque theta defensins. Antimicrob. Agents Chemother. 52, 944–953. 10.1128/AAC.01090-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppuluri P., Nett J., Heitman J., Andes D. (2008). Synergistic effect of calcineurin inhibitors and fluconazole against Candida albicans biofilms. Antimicrob. Agents Chemother. 52, 1127–1132. 10.1128/AAC.01397-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden W. J., van Iersel T. M., Blijlevens N. M., Donnelly J. P. (2009). Safety and tolerability of the antimicrobial peptide human lactoferrin 1-11 (hLF1-11). BMC Med. 7:44 10.1186/1741-7015-7-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weerden N. L., Bleackley M. R., Anderson M. A. (2013). Properties and mechanisms of action of naturally occurring antifungal peptides. Cell. Mol. Life Sci. 70, 3545–3570. 10.1007/s00018-013-1260-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vouldoukis I., Shai Y., Nicolas P., Mor A. (1996). Broad spectrum antibiotic activity of skin-PYY. FEBS Lett. 380, 237–240. 10.1016/0014-5793(96)00050-6 [DOI] [PubMed] [Google Scholar]
- Vriens K., Cammue B. P. A., Thevissen K. (2014). Antifungal plant defensins: mechanisms of action and production. Molecules 19, 12280–12303. 10.3390/molecules190812280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriens K., Cools T. L., Harvey P. J., Craik D. J., Braem A., Vleugels J., et al. (2016). The radish defensins RsAFP1 and RsAFP2 act synergistically with caspofungin against Candida albicans biofilms. Peptides 75, 71–79. 10.1016/j.peptides.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Vriens K., Cools T. L., Harvey P. J., Craik D. J., Spincemaille P., Cassiman D., et al. (2015). Synergistic activity of the plant defensin hsafp1 and caspofungin against Candida albicans biofilms and planktonic cultures. PLoS ONE 10:e0132701. 10.1371/journal.pone.0132701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi H., Abe S., Okutomi T., Tansho S., Kawase K., Yamaguchi H. (1996). Cooperative anti-Candida effects of lactoferrin or its peptides in combination with azole antifungal agents. Microbiol. Immunol. 40, 821–825. 10.1111/j.1348-0421.1996.tb01147.x [DOI] [PubMed] [Google Scholar]
- Wakabayashi H., Abe S., Teraguchi S., Hayasawa H., Yamaguchi H. (1998). Inhibition of hyphal growth of azole-resistant strains of Candida albicans by triazole antifungal agents in the presence of lactoferrin-related compounds. Antimicrob. Agents Chemother. 42, 1587–1591. 10.1128/AAC.42.7.1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Li X., Wang Z. (2016). APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–D1093. 10.1093/nar/gkv1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Zheng K., Lin J., Huang M., Ma Y., Li S., et al. (2018). Rapid and efficient production of cecropin A antibacterial peptide in Escherichia coli by fusion with a self-aggregating protein. BMC Biotechnol. 18:62. 10.1186/s12896-018-0473-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G.-X., Bobek L. A. (2004). In vitro synergic antifungal effect of MUC7 12-mer with histatin-5 12-mer or miconazole. J. Antimicrob. Chemother. 53, 750–758. 10.1093/jac/dkh181 [DOI] [PubMed] [Google Scholar]
- Wibowo D., Zhao C.-X. (2019). Recent achievements and perspectives for large-scale recombinant production of antimicrobial peptides. Appl. Microbiol. Biotechnol. 103, 659–671. 10.1007/s00253-018-9524-1 [DOI] [PubMed] [Google Scholar]
- Wimley W. C., Hristova K. (2011). Antimicrobial Peptides: successes, challenges and unanswered questions. J. Membr. Biol. 239, 27–34. 10.1007/s00232-011-9343-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Maier E., Benz R., Hancock R. E. (1999). Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry 38, 7235–7242. 10.1021/bi9826299 [DOI] [PubMed] [Google Scholar]
- Yeaman M. R., Yount N. Y. (2003). Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55, 27–55. 10.1124/pr.55.1.2 [DOI] [PubMed] [Google Scholar]
- Yeung A. T. Y., Gellatly S. L., Hancock R. E. W. (2011). Multifunctional cationic host defence peptides and their clinical applications. Cell. Mol. Life Sci. 68, 2161–2176. 10.1007/s00018-011-0710-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M. (2002). Antimicrobial peptides of multicellular organisms. Nature 415, 389–395. 10.1038/415389a [DOI] [PubMed] [Google Scholar]
- Zhang L., Gallo R. L. (2016). Antimicrobial peptides. Curr. Biol. 26, R14–R19. 10.1016/j.cub.2015.11.017 [DOI] [PubMed] [Google Scholar]
- Zhang L., Li X., Wei D., Wang J., Shan A., Li Z. (2015). Expression of plectasin in Bacillus subtilis using SUMO technology by a maltose-inducible vector. J. Ind. Microbiol. Biotechnol. 42, 1369–1376. 10.1007/s10295-015-1673-y [DOI] [PubMed] [Google Scholar]
- Zhao J., Song L., Li C., Ni D., Wu L., Zhu L., et al. (2007). Molecular cloning, expression of a big defensin gene from bay scallop Argopecten irradians and the antimicrobial activity of its recombinant protein. Mol. Immunol. 44, 360–368. 10.1016/j.molimm.2006.02.025 [DOI] [PubMed] [Google Scholar]