Abstract
Multidrug-resistant hypervirulent Klebsiella pneumoniae (MDR-hvKP) has been increasingly reported and is now recognized as a significant threat to public health; however, characterization of MDR-hvKP has not been systematically investigated. In the present study, 124 of 428 (28.92%) K. pneumoniae clinical isolates collected from January 2010 to December 2016 were identified with aerobactin and defined as hvKP; these included 94 non-MDR-KP, 20 extended-spectrum β-lactamase-producing K. pneumoniae (ESBL-KP), and 10 carbapenem-resistant K. pneumoniae (CR-KP) isolates. The remaining 304 isolates without presence of virulence factor aerobactin were defined as classic K. pneumoniae (cKP). The antimicrobial resistance rate of cKP was significantly higher than that of the hvKP isolates in the non-MDR-KP group, but showed no significant differences in the ESBL-KP and CR-KP groups. The detection frequencies of capsular serotype K1 (magA), hypermucoviscosity, sequence type 23 (ST23), and the virulence gene rmpA were significantly higher in the hvKP than cKP isolates in all three groups (P < 0.05). Most of the hypervirulent ESBL-KP and CR-KP isolates were K non-typeable (16/30) and harbored at least one gene for virulence (26/30). The hypervirulent ESBL-KP isolates primarily carried blaCTX–M (12/20, 60%) genes, and the hypervirulent CR-KP isolates mainly carried blaNDM–1 (8/10, 80%) genes. Moreover, three hypervirulent ESBL-KP and two hypervirulent CR-KP isolates showed resistance to tigecycline but were sensitive to colistin. The transcriptional levels of rmpA in cKP were much lower than that in hvKP isolates in all three groups. Furthermore, overexpression of rmpA in the rmpA-low-expression cKP isolates could enhance bacterial virulence in the mouse infection experiment. In conclusion, our data suggest that the capsular serotype K1 (magA), rmpA, hypermucoviscosity, and ST23 were strongly associated with hvKP in non-MDR-KP, ESBL-KP, and CR-KP groups, and low rmpA expression levels contributed to the absence of hypervirulent phenotype.
Keywords: Klebsiella pneumoniae, hypervirulence, extended-spectrum β-lactamases, carbapenem resistance, rmpA, aerobactin
Introduction
The hypervirulent Klebsiella pneumoniae (hvKP) was a new type of K. pneumoniae that caused several serious infections, such as pyogenic liver abscesses, meningitis, and endophthalmitis in young and healthy individuals (Struve et al., 2015). Since it was first reported in Taiwan in the mid-1980s (Liu et al., 1986), the hvKP infections have been reported around the world over the past few years (Decre et al., 2011; Siu et al., 2011; Chang et al., 2013). The hvKP and classic K. pneumoniae (cKP) strains can be distinguished by a combination of phenotypic and genotypic characteristics. Hypermucoviscosity, which is determined by a string test on agar plates, is a typical feature of hvKP strains and has been commonly used to define hvKP in previous studies (Li et al., 2014; Zhan et al., 2017). Most of the hvKP isolates belong to K1 and K2 capsular serotypes (Yeh et al., 2007). Several virulence genes, including regulator of mucoid phenotype A (rmpA and rmpA2), mucoviscosity-associated gene A (magA) and wcaG (encoding GDP-fucose synthetase) have been found to contribute to the hypervirulent phenotype (Lai et al., 2003; Yu et al., 2006; Hunt et al., 2011; Zhang R. et al., 2016). A recent study showed that aerobactin was a major virulence factor for the increased siderophore production in hvKP isolates and used for the definition of hvKP (Russo et al., 2014).
In recent years several studies have reported the existence and increasing incidence of multidrug-resistant hvKP (MDR-hvKP) isolates, especially extended-spectrum β-lactamase-producing hvKP (ESBL-hvKP) and carbapenem-resistant hvKP (CR-hvKP) (Yu et al., 2015b; Zhang et al., 2015; Zhang Y. et al., 2016; Zhan et al., 2017), which is now recognized as a serious threat to public health. However, the clinical and molecular characteristics of these MDR-hvKP isolates are not yet well understood. In this study, K. pneumoniae isolates carrying aerobactin were designated as hvKP and the differences in the distribution, molecular epidemiology, and clinical characteristics of hypervirulent determinants were investigated and compared among the non-MDR-KP, ESBL-KP, and CR-KP isolates. We also assessed the impact of rmpA expression level on virulence of cKP isolates and hypothesized that low levels of rmpA expression in cKP strains contributed to the absence of hypervirulent phenotype.
Materials and Methods
Bacterial Strains and Growth Conditions
A total of 428 non-duplicate K. pneumoniae strains were collected from inpatients at the Affiliated Shenzhen Sixth Hospital of Guangdong Medical University in China between January 2010 and December 2016. Bacterial species were identified by standard methods with a VITEK 2 compact system (Biomérieux, Marcy l’Etoile, France). All procedures performed were approved by the Ethical Committee of Shenzhen Nanshan People’s Hospital and were in accordance with the tenets of the 1964 Helsinki declaration and its later amendments. All strains were cultured in Luria-Bertani (LB; Oxoid, Basingstoke, United Kingdom) medium at 37°C with shaking at 220 rpm.
Antimicrobial Susceptibility Testing and Phenotypic Confirmation of ESBL-KP and CR-KP
Antimicrobial susceptibility to several commonly used antibiotics such as amoxicillin/clavulanate, piperacillin/tazobactam, cefotaxime, ceftazidime, cefepime, ciprofloxacin, and amikacin were detected by broth microdilution in the VITEK 2 compact system. The minimal inhibitory concentrations (MICs) of tigecycline, colistin, imipenem, and meropenem were determined by the agar dilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI-M100-S27). The sensitivity results of antimicrobials were confirmed based on CLSI-M100-S27. ESBL production was detected by the agar dilution test with cefotaxime and ceftazidime, and CR-KP was confirmed by the modified Hodge test, both in accordance with the CLSI guidelines (CLSI-M100-S27). Escherichia coli ATCC 25922 was used as the quality control for all antimicrobial susceptibility tests.
PCR Detection of Virulence-Associated Factors, β-Lactamase, and Carbapenemase Genes
Aerobactin positivity was determined by polymerase chain reaction (PCR) using the primers aerobactin-F/aerobactin-R and aerobactin-positive strains were designated as hvKP (Zhang Y. et al., 2016). K. pneumoniae virulence genes (rmpA, rmpA2, magA, and wcaG) and capsular serotypes (K1, K2, K5, K20, K54, and K57) were detected by PCR as previously described (Li et al., 2014; Candan and Aksoz, 2015; Yu et al., 2017). The extracted DNA of K. pneumoniae isolates served as templates for the PCR amplification. The rmpA amplicons were recovered for further sequencing. Mutations of rmpA in K. pneumoniae isolates were identified by comparing with the reference sequence of the K. pneumoniae NTUH-K2044 genotype (GenBank accession number: AP006725) (Wu et al., 2009). The β-lactamase genes blaTEM, blaSHV, blaCTX–M, and blaOXA–1, and carbapenemase genes blaKPC, blaNDM–1, blaOXA–48, blaIMP, and blaVIM were determined by PCR as previously described (Lewis et al., 2007; Candan and Aksoz, 2015). The primers used in this study are listed in Supplementary Table S1.
String Test
The hypermucoviscous phenotype of K. pneumoniae isolates was confirmed by string test. The bacterial strains were grown on an agar plate at 37°C overnight and the formation of a mucoviscous string measuring >5 mm on a bacteriology inoculation loop was defined as a positive string test (Zheng et al., 2018a).
Multilocus Sequence Typing (MLST) and Clonal Complex (CC)
Multilocus sequence typing (MLST) was conducted as previously described (Zheng et al., 2018a). Briefly, the housekeeping genes gapA, infB, mdh, pgi, phoE, rpoB, and tonB were amplified by PCR and sequenced. The sequence types (STs) were identified by the MLST database1. Klebsiella pneumoniae CCs were assigned using the phyloviz-2.0a program2. All primer sequences used in MLST are listed in Supplementary Table S1.
Quantitative Real-Time (qRT)-PCR Analysis
The transcriptional levels of the virulence gene rmpA in hvKP and cKP isolates were analyzed by qRT-PCR. Sixteen hvKP (aerobactin positive and hypermucoviscous) and 13 cKP (aerobactin negative and non-hypermucoviscous) isolates were selected randomly from the non-MDR-KP, ESBL-KP, or CR-KP groups and grown to mid-log phase (4 h) in LB medium at 37°C. Total bacterial RNA was extracted using the RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany). The extracted RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit (Takara Bio Inc., Shiga, Japan). The qRT-PCR was performed in a MasterCycler RealPlex System (Eppendorf, Hamburg, Germany) at the following amplification conditions: 95°C for 2 min, followed by 40 cycles of 5 s at 95°C and 34 s at 60°C. The housekeeping gene rrsE was used to normalize the expression of target genes. The transcriptional levels of rmpA were determined based on the 2–ΔΔCt method and compared with that of K. pneumoniae NTUH-K2044 genotype. Each sample was carried out in triplicate. The primers used for qRT-PCR were designed based on the genome of K. pneumoniae NTUH-K2044, by using a Beacon designer software (Premier Biosoft International Ltd., Palo Alto, CA, United States) (Supplementary Table S1).
The rmpA Overexpression Assay
The full-length of rmpA was amplified from the genomic DNA of NTUH-K2044. The PCR fragments were inserted into the plasmid pZP1137 with endonucleases NheI and BglII for gene overexpression (Zheng et al., 2018b). The positive clones were screened by kanamycin and verified by PCR and sequencing. The rmpA overexpression plasmid (prmpA) were then introduced separately into three cKP isolates (LBKP61, EKP190, and CRKP16) with low expression levels of rmpA in qRT-PCR. The strains, plasmids, and primers used for rmpA overexpression are listed in Supplementary Tables S2, S3. The transcriptional level of rmpA was measured by qRT-PCR as described above. The overexpression of rmpA was induced with 1 mM arabinose (Ara).
Intranasal Infection Model
The effect of rmpA overexpression on cKP isolates was determined by the mouse intranasal infection model as previously described (Palacios et al., 2018). Briefly, overnight bacterial cultures were diluted 1:200 in LB containing 1 mM arabinose and grown at 37°C until the optical density (OD) at 600 nm measured 1.0. Cells were harvested by centrifugation at 5,000 g for 5 min and resuspended in phosphate-buffered saline (PBS). Female C57BL/6 mice aged 5–7 weeks were anesthetized using a mixture containing ketamine (50 mg/kg) and xylazine (5 mg/kg) and were inoculated intranasally with 5 × 104 colony forming unit (CFU)/mouse of each strain (n = 8 mice in each group). At 72 h post-inoculation, the mice were euthanized with pentobarbital (40 mg/kg). Lungs were removed, homogenized in PBS, serially diluted, and plated onto LB agar to quantify the CFU level. Results are reported as log10 CFU per gram of lung. Inoculation with PBS served as a negative control.
Statistical Analysis
Experimental data were analyzed with Student’s t-test or one-way analysis of variance in IBM SPSS Statistics (version 20.0, Chicago, IBM, United States). Differences with a P-value < 0.05 were regarded statistically significant.
Results
Detection Frequency of hvKP in Non-MDR-KP, ESBL-KP, and CR-KP Isolates
A total of 428 non-repetitive K. pneumoniae clinical isolates were categorized into three groups: non-MDR-KP (205), ESBL-KP (181), and CR-KP (42) (Table 1). The detection frequency of hvKP (defined by aerobactin positivity) in all clinical isolates, non-MDR-KP, ESBL-KP, and CR-KP were 28.92% (124/428), 45.85% (94/205), 11.05% (20/181), and 23.81% (10/42), respectively. This indicates that the frequency of hvKP in ESBL-KP and CR-KP groups is significantly lower than that in non-MDR KP group (Table 1). Characteristics and comparison of the sample specimens’ origins among non-MDR-KP, ESBL-KP, and CR-KP groups are shown in Table 1.
TABLE 1.
Distribution of hypervirulent (aerobactin positive) phenotype among non-MDR-KP, ESBL-KP, and CR-KP.
| Isolates (n) | hvKPb (n = 124) | cKP (n = 304) | P-valuec |
| Non-MDR-KP (205) | 94(45.85%) | 111(54.15%) | < 0.0001(vs.ESBL-KP) |
| Sputum (66) | 35(53.03%) | 31(46.97%) | < 0.0001(vs.sputumESBL-KP) |
| Urine (23) | 7(30.43%) | 16(69.57%) | 0.09(vs.urineESBL-KP) |
| Blood (94) | 35(37.23%) | 59(62.77%) | 0.879(vs.bloodESBL-KP) |
| Abscess (15) | 14(93.33%) | 1(6.67%) | < 0.0001(vs.othersESBL-KP) |
| Othersa (7) | 3(42.86%) | 4(57.14%) | |
| ESBL-KP (181) | 20(11.05%) | 161(88.95%) | < 0.0001(vs.non-MDR-KP) |
| Sputum (110) | 8(7.27%) | 102(92.73%) | < 0.0001(vs.sputumnon-MDR-KP) |
| Urine (39) | 5(12.82%) | 34(87.18%) | 0.09(vs.urinenon-MDR-KP) |
| Blood (17) | 6(35.29%) | 11(64.71%) | 0.879(vs.bloodnon-MDR-KP) |
| Others (15) | 1(6.67%) | 14(93.33%) | |
| CR-KP (42) | 10(23.81%) | 32(76.19%) | 0.008(vs.non-MDR-KP) |
| 0.29(vs.ESBL-KP) | |||
| Sputum (22) | 5(22.73%) | 17(77.27%) | 0.013(vs.sputumnon-MDR-KP) |
| 0.026(vs.sputumESBL-KP) | |||
| Urine (4) | 1(25.00%) | 3(75.00%) | 0.826(vs.urinenon-MDR-KP) |
| 0.503(vs.urineESBL-KP) | |||
| Blood (16) | 4(25.00%) | 12(75.00%) | 0.344(vs.bloodnon-MDR-KP) |
| 0.52(vs.bloodESBL-KP) | |||
| Others (1) | 0(0.00%) | 1(100.00%) |
aOthers contain ascites, bile, pleural effusion, bronchoalveolar lavage fluid, and pericardial effusion. bThe hvKP were defined by aerobactin positivity. cP value for hvKP among K. pneumoniae isolates.
Comparison of the Antimicrobial Susceptibility of hvKP and cKP Isolates in Non-MDR-KP, ESBL-KP, and CR-KP Groups
The antimicrobial susceptibilities of K. pneumoniae isolates are shown in Supplementary Table S4. Overall, the antimicrobial resistance rate of cKP was significantly higher than that of hvKP isolates in non-MDR-KP group but showed no differences in ESBL-KP and CR-KP groups. For example, 9.57% (9/94) of hvKP and 27.93% (31/111) of cKP isolates were resistant to ciprofloxacin in non-MDR-KP group while the hvKP and cKP isolates in ESBL-KP and CR-KP groups exhibited high-frequency resistance (resistance rate > 50%) to this antibiotic. The resistance rates of amikacin in hvKP and cKP isolates were less than 15% in non-MDR-KP and ESBL-KP groups but more than 30% in CR-KP group. Tigecycline susceptibility among non-MDR-KP, ESBL-KP, and CR-KP showed no significant difference; these isolates remained highly susceptible to tigecycline. Notably, colistin exhibited excellent antimicrobial activity against hvKP and cKP isolates in all three groups.
Distribution of Virulence-Associated Factors in hvKP and cKP Isolates Among Non-MDR-KP, ESBL-KP, and CR-KP Groups
The distribution of virulence-associated factors of K. pneumoniae isolates are shown in Table 2. The capsular serotype K1 (magA) and virulence gene rmpA were more common in hvKP than cKP isolates in all three groups (P < 0.05). Further, K2, K5, K20, K54, and K57 were not significantly different between the hvKP and cKP isolates. The presence of wcaG and rmpA2 were more common in hvKP isolates than cKP isolates in non-MDR-KP and ESBL-KP groups (P < 0.05), whereas they showed no difference between hvKP and cKP isolates in CR-KP group. The majority of hvKP isolates (97/124; 78.23%) while only 3.95% of the cKP (12/304) were hypermucoviscous (Table 2).
TABLE 2.
Distribution of virulence factors among non-MDR-KP, ESBL-KP, and CR-KP isolates.
| Characteristics | Non-MDR-KP (n = 205) |
ESBL-KP (n = 181) |
CR-KP (n = 42) |
||||||
| hvKP (n = 94) | cKP (n = 111) | P-value | hvKP (n = 20) | cKP (n = 161) | P-value | hvKP (n = 10) | cKP (n = 32) | P-value | |
| K serotype | |||||||||
| K1 | 37(39.36%) | 5(4.50%) | <0.001 | 6(30.00%) | 12(7.45%) | 0.001 | 3(30.00%) | 2(6.25%) | 0.043 |
| K2 | 14(14.89%) | 4(3.60%) | 0.004 | 2(10.00%) | 6(3.73%) | 0.198 | 0(0.00%) | 2(6.25%) | 0.418 |
| K5 | 1(1.06%) | 0(0.00%) | 0.276 | 0(0.00%) | 0(0.00%) | / | 0(0.00%) | 0(0.00%) | / |
| K20 | 1(1.06%) | 1(0.90%) | 0.906 | 0(0.00%) | 3(1.86%) | 0.538 | 0(0.00%) | 0(0.00%) | / |
| K54 | 6(6.38%) | 6(5.41%) | 0.766 | 1(5.00%) | 0(0.00%) | 0.004 | 0(0.00%) | 3(9.38%) | 0.315 |
| K57 | 2(2.13%) | 3(2.70%) | 0.790 | 1(5.00%) | 0(0.00%) | 0.004 | 1(10.00%) | 0(0.00%) | 0.07 |
| Virulence gene | |||||||||
| wcaG | 12(12.77%) | 6(5.41%) | 0.064 | 3(15.00%) | 5(3.11%) | 0.015 | 0(0.00%) | 0(0.00%) | / |
| rmpA | 75(79.79%) | 8(7.21%) | <0.001 | 18(90.00%) | 4(2.48%) | <0.001 | 7(70.00%) | 1(3.13%) | <0.001 |
| rmpA2 | 26(27.66%) | 5(4.50%) | <0.001 | 2(10.00%) | 3(1.86%) | 0.036 | 0(0.00%) | 0(0.00%) | / |
| magA | 37(39.36%) | 5(4.50%) | <0.001 | 6(30.00%) | 12(7.45%) | 0.001 | 3(30.00%) | 2(6.25%) | 0.043 |
| MLST genotype | |||||||||
| ST23 | 18(19.15%) | 2(1.80%) | <0.001 | 5(25.00%) | 1(0.62%) | <0.001 | 3(30.00%) | 0(0.00%) | 0.001 |
| ST11 | 2(2.13%) | 8(7.21%) | 0.093 | 6(30.00%) | 12(7.45%) | 0.001 | 2(20.00%) | 3(9.38%) | 0.365 |
| Hypermucoviscosity | 73(77.66%) | 7(6.31%) | <0.001 | 17(85.00%) | 3(1.86%) | <0.001 | 7(70.00%) | 2(6.25%) | <0.001 |
Comparison of MLST Genotyping Between hvKP and cKP Isolates
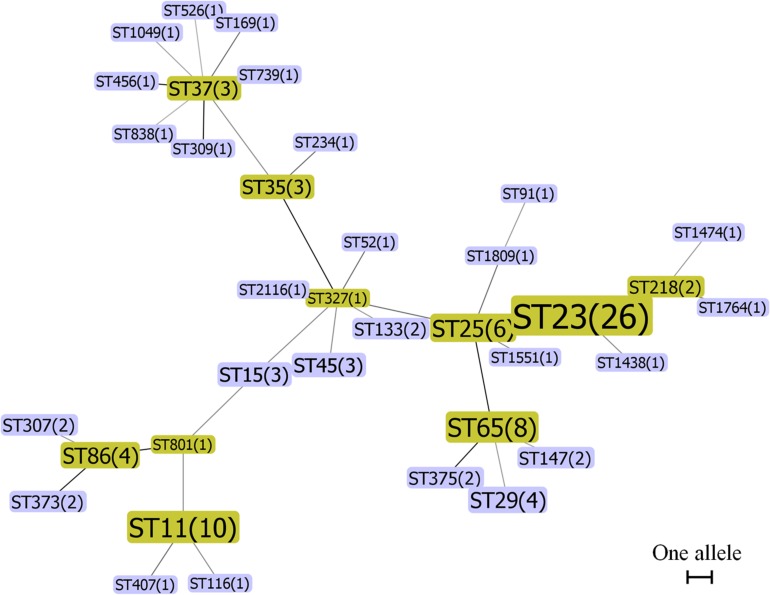
To compare the genetic relationship of hvKP and cKP isolates, the MLST genotyping and minimum spanning tree analysis were conducted. MLST enabled a clear sequence type (ST) assignment for 102 of the 124 hvKP isolates, in total 36 different STs. The most prevalent ST in the hvKP isolates was ST23 (n = 26, 25.49%), followed by ST11 (n = 10, 9.8%), ST65 (n = 8, 7.84%), and ST25 (n = 6, 5.88%). Two major MLST groups, ST23-like and ST317-like, were obtained based on minimum spanning tree analysis (Figure 1). The STs of 257 of 304 cKP isolates could be determined and were assigned to 88 different STs. ST37 was the most prevalent ST (n = 35, 13.62%), followed by ST15 (n = 31, 12.06%), ST11 (n = 23, 8.95%), and ST133 (n = 15, 5.84%). Minimum spanning tree analysis showed that all of the cKP isolates belonged to three major MLST groups, ST15-like, ST17-like, and ST25-like (Supplementary Figure S1). Sixteen K. pneumoniae isolates, including 6 hvKP and 10 cKP isolates, were identified with new allele code combinations (Supplementary Table S5). The STs of the remaining 53 isolates could not be determined due to the presence of one or more incomplete alleles. Moreover, the ST23 was more common in hvKP than cKP isolates in all three groups (P < 0.05, Table 2). These results indicated that ST23 was strongly associated with hvKP, while ST37 and ST15 were more common in the cKP isolates.
FIGURE 1.
Minimum spanning tree of 102 hvKP isolates by MLST type and gene allele profile. Seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) were PCR-amplified and sequenced from all isolates according to the Klebsiella pneumoniae MLST protocol. Alleles and sequence types (STs) were assigned by the MLST database. K. pneumoniae CCs were identified by the phyloviz-2.0a program. Each node within the tree represented a single ST and the number of isolates. The size of the nodes was proportional to the number of isolates. Lines connecting each node indicate CCs. Length of lines between the node was proportional to the number of different alleles.
Characteristics of the Hypervirulent ESBL-KP and CR-KP Isolates
Characteristics of the hypervirulent ESBL-KP and hypervirulent CR-KP isolates are shown in Table 3 (data of the hypervirulent non-MDR-KP isolates are listed in Supplementary Table S6). Among 20 hypervirulent ESBL-KP isolates, six were K1 capsular serotype and 10 were K non-typeable. Eighteen of these isolates harbored at least one virulence associated gene (rmpA, rmpA2, magA, or wcaG) in addition to aerobactin, and 17 isolates were hypermucoviscous. Six of them were ST11 and five, ST23. Twelve, seven, and two of the isolates carried blaCTX–M, blaTEM, and blaSHV genes, respectively, encoding different ESBLs. Of the 10 hypervirulent CR-KP isolates, three were K1 and one was of K57 capsular serotype. Eight isolates harbored at least one gene for virulence (rmpA or magA). Seven isolates were hypermucoviscous. Three were ST23, two were ST11, two were ST25, and two were ST133. Eight isolates were New Delhi metallo-beta-lactamase 1 (NDM-1) positive and three were K. pneumoniae carbapenemase-2 (KPC-2) positive. Notably, three hypervirulent ESBL-KP and two hypervirulent CR-KP isolates were resistant to tigecycline but all of them were sensitive to colistin (Table 3).
TABLE 3.
Molecular characteristics of the hypervirulent (aerobactin positive) ESBL-KP and CR-KP isolates.
| Strain | Source | Capsule | Virulence genes | String-test | MLST | ESBL or carbapenemase types | Tigecycline susceptibility | Colistin susceptibility |
| ESBL-KP (n = 20) | ||||||||
| EKP3 | Blood | K non-typeable | rmpA | + | ST15 | CTX-M-14, | S | S |
| EKP16 | Urine | K2 | rmpA, rmpA2 | + | ST65 | CTX-M-14 | S | S |
| EKP19 | Sputum | K non-typeable | + | NTa | TEM-15 | S | S | |
| EKP29 | Urine | K1 | rmpA, magA, wcaG | + | ST23 | CTX-M-15 | S | S |
| EKP60 | Urine | K non-typeable | rmpA | − | ST11 | TEM-15, CTX-M-6 | R | S |
| EKP85 | Sputum | K non-typeable | rmpA | − | ST526 | TEM-15 | R | S |
| EKP91 | Sputum | K1 | rmpA, magA | + | ST23 | CTX-M-15 | S | S |
| EKP109 | Urine | K1 | rmpA. magA | + | ST23 | CTX-M-14 | S | S |
| EKP110 | Blood | K54 | rmpA | + | ST11 | TEM-104 | S | S |
| EKP120 | Ascites | K1 | rmpA, magA | + | ST11 | TEM-15 | S | S |
| EKP130 | Sputum | K1 | rmpA, magA | + | ST23 | CTX-M-14 | S | S |
| EKP138 | Blood | K1 | rmpA, magA | + | ST25 | CTX-M-15 | S | S |
| EKP141 | Sputum | K57 | rmpA, wcaG | + | ST11 | SHV-12 | S | S |
| EKP146 | Blood | K non-typeable | + | NT | CTX-M-15 | S | S | |
| EKP162 | Sputum | K non-typeable | rmpA | + | ST11 | TEM-15 | S | S |
| EKP169 | Blood | K non-typeable | rmpA | − | ST1438 | CTX-M-14 | S | S |
| EKP170 | Urine | K non-typeable | rmpA | + | ST11 | CTX-M-18 | R | S |
| EKP181 | Sputum | K2 | rmpA | + | ST37 | CTX-M-15 | S | S |
| EKP198 | Blood | K non-typeable | rmpA, rmpA2 | + | ST327 | SHV-12 | S | S |
| EKP204 | Sputum | K non-typeable | rmpA, wcaG | + | ST23 | TEM-48 | S | S |
| CR-KP (n = 10) | ||||||||
| CRKP6 | Blood | K non-typeable | rmpA | + | ST25 | NDM-1 | S | S |
| CRKP7 | Blood | K1 | rmpA, magA | + | ST23 | NDM-1, KPC-2 | S | S |
| CRKP10 | Blood | K non-typeable | rmpA | + | ST11 | NDM-1 | R | S |
| CRKP14 | Blood | K1 | rmpA, magA | + | ST23 | NDM-1 | S | S |
| CRKP17 | Sputum | K non-typeable | − | ST133 | KPC-2 | S | S | |
| CRKP34 | Sputum | K non-typeable | rmpA | + | ST23 | NDM-1 | S | S |
| CRKP36 | Sputum | K1 | magA | + | ST25 | NDM-1 | S | S |
| CRKP39 | Sputum | K non-typeable | rmpA | + | ST11 | NDM-1 | R | S |
| CRKP40 | Sputum | K non-typeable | rmpA | − | ST133 | KPC-2 | S | S |
| CRKP42 | Urine | K57 | − | NT | NDM-1 | S | S | |
aNT, not typeable.
Characteristics of rmpA-Positive cKP Isolates
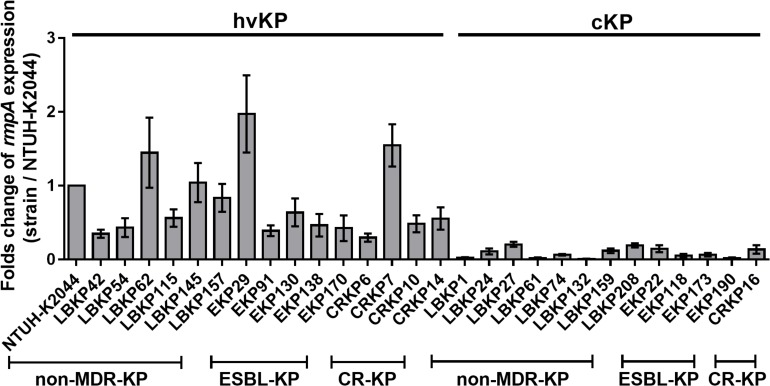
The rmpA gene was more frequently detected in hvKP than cKP isolates in all three groups (79.79% vs. 7.21%, P < 0.001, 90.00% vs. 2.48%, P < 0.001, 70% vs. 3.14%, P < 0.001, Table 2). This indicates an association of rmpA with hypervirulent phenotype (aerobactin positive) in K. pneumoniae. However, there were 13 rmpA-positive K. pneumoniae isolates (eight non-MDR-KP, four ESBL-KP, and one CR-KP) that did not show hypervirulence (aerobactin negative and non-hypermucoviscous). Owing to the high frequency of rmpA in hvKP isolates, the potential impact of rmpA on the virulence of cKP was investigated. First, genetic mutations in rmpA were determined. Among these 13 rmpA-positive strains, only two mutation sites of rmpA were found (LBKP27-K69I and EKP22-C102R), suggesting that gene polymorphism cannot entirely explain the phenotypic difference (hypervirulent or classic phenotype) caused by rmpA (Supplementary Table S7). Further, the difference in expression of rmpA between rmpA-positive hvKP (aerobactin positive, hypermucoviscous) and cKP (aerobactin negative, non-hypermucoviscous) isolates were compared by qRT-PCR. The transcriptional levels of rmpA in cKP isolates were much lower than those in hvKP isolates in all three groups (Figure 2), indicating that low expression of rmpA may lead to the absence of hypervirulent phenotype in cKP isolates.
FIGURE 2.
Relative gene expression of rmpA in 29 clinical isolates of K. pneumoniae. The hvKP and cKP isolates were cultured in LB for 4 h. Total RNA was extracted and transcriptional levels of rmpA were examined by qRT-PCR. The housekeeping gene rrsE was used as the endogenous reference gene. The K. pneumoniae NTUH-K2044 was used as the reference strain (transcriptional level = 1.0). All qRT-PCRs were carried out in triplicate. The hvKP isolates used in this assay were aerobactin positive and hypermucoviscous.
Overexpression of rmpA Enhances the Virulence of rmpA-Positive cKP Isolates
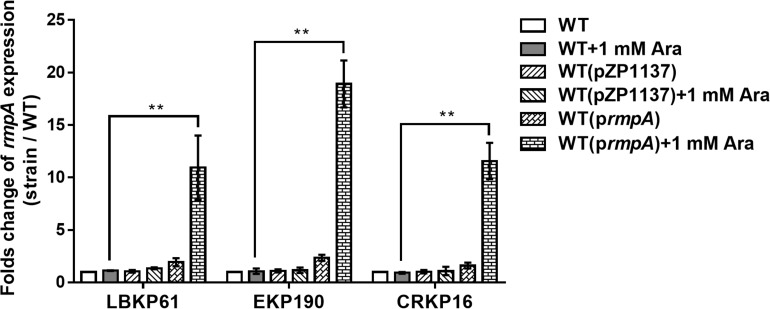
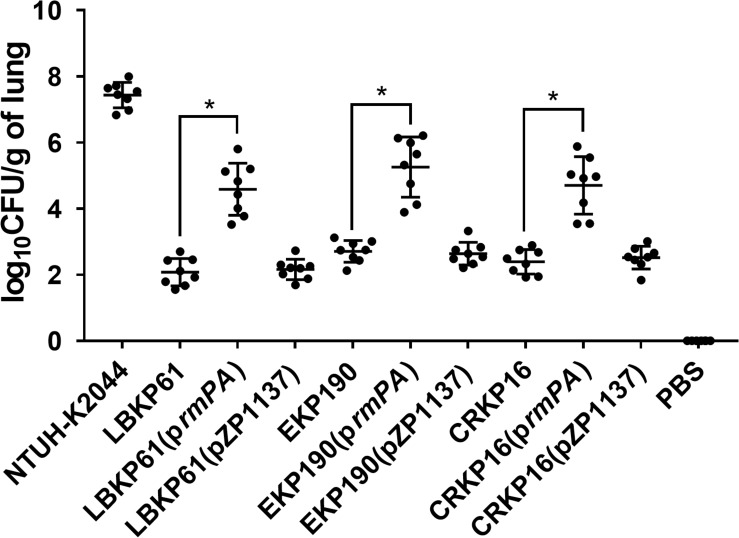
To confirm the impact of rmpA expression level on virulence in rmpA-positive cKP isolates, rmpA overexpressing plasmid was constructed and transformed into the rmpA-low-expression cKP isolates (LBKP61, EKP190, and CRKP16). The stable overexpression of rmpA in the transformed isolates was confirmed by qRT-PCR. The transcriptional level of rmpA in the transformed strains with 1 mM arabinose (Ara) induction increased 10.68–17.67-fold compared with that in the wild-type isolates (Figure 3). Further, the virulence of rmpA overexpression strains was determined by the mouse intranasal infection model. As showed in Figure 4, the colonized level of all three rmpA overexpression strains in the lung increased by more than two logs than that of the wild-type strains, indicating that overexpression of rmpA could enhance the virulence of the rmpA-low-expression cKP isolates.
FIGURE 3.
Relative transcriptional analysis of rmpA used in overexpression experiments. The K. pneumoniae strains were grown in LB for 4 h induced with or without 1 mM arabinose. Total RNA was extracted and transcriptional levels of rmpA were examined by qRT-PCR. The housekeeping gene rrsE was used as the endogenous reference gene. The clinical wild-type strains were used as the reference strain (transcriptional level = 1.0). All qRT-PCRs were carried out in triplicate. **P < 0.01.
FIGURE 4.
Effect of rmpA overexpression in rmpA-low-expression cKP isolates on virulence. Mice were inoculated intranasally with K. pneumoniae isolates. At 72 h post-inoculation, mice were euthanized, and lungs were homogenized and plated onto LB agar to quantify the CFU level. Each symbol represents one mouse. Mann–Whitney tests were performed for statistical analyses. *P < 0.05.
Discussion
Despite on-going research, there is still no consensus about the definition of hvKP. It is traditionally defined by hypermucoviscosity with a positive string test, but recent research has indicated that hypermucoviscosity and hypervirulence are two different phenotypes (Catalan-Najera et al., 2017). K. pneumoniae clinical isolates causing pyogenic liver abscesses without the characteristic hypermucoviscous phenotype have been found in an increasing number of studies (Luo et al., 2014; Cubero et al., 2016; Wu et al., 2017). These results suggested that the hypervirulent phenotype of K. pneumoniae is not dependent on hypermucoviscosity. Therefore, using hypermucoviscosity to define hvKP may not be accurate. The siderophore aerobactin is a major virulence factor in the progression of hvKP infection and survival in a human host (Russo et al., 2014, 2015). It is located on the large virulence plasmid (pLVPK) that is present in most hvKP isolates, but rarely present in cKP strains and considered as a genetic factor for both hypermucoviscous and hypervirulent phenotypes (Nassif and Sansonetti, 1986; Paczosa and Mecsas, 2016; Catalan-Najera et al., 2017). Because of the crucial role in hvKP, aerobactin positivity was considered a defining genetic trait for hvKP (Zhang Y. et al., 2016). In the present study, 45.85% of non-MDR-KP, 11.05% of ESBL-KP, and 23.81% of CR-KP isolates were defined as hvKP based on aerobactin positivity. Regarding hypermucoviscosity, 39.02, 11.05, and 21.43% of those isolates were identified as string test positivity in our study (Supplementary Table S8). Compared with the traditional designation criteria for hvKP by hypermucoviscosity, using aerobactin to define hvKP was more sensitive but less specific, which was similar to previous research findings (Zhang Y. et al., 2016). Considering that these two definitions of hvKP have a high degree of overlap (Supplementary Table S8), it may be more appropriate to combine aerobactin positivity and hypermucoviscosity when defining hvKP. Notably, hvKP isolates from abscesses contained more hypervirulent-associated factors compared to the hvKP isolates from other sources. All the 14 hvKP isolates from abscesses were rmpA-positive and hypermucoviscous. Eleven of them were K1 capsular serotype and nine were ST23 genotype (Supplementary Table S6).
Previous studies have shown that the rmpA gene, which is located on the large virulence plasmid pLVPK and participates in the enhancement of capsular production (Cheng et al., 2010), is a major virulence-associated factor in hvKP isolates (Yu et al., 2006; Zhang Y. et al., 2016). In our study, 80.65% (100/124) of hvKP isolates harbored rmpA gene while only 13 of 304 cKP isolates were rmpA-positive, indicating a strong association between rmpA and hypervirulence. Moreover, the potential impact of rmpA on virulence in those rmpA-positive cKP isolates was investigated. It has been reported that high tendency of rmpA mutations contributes to low virulence of rmpA-positive K. pneumoniae isolates (Yu et al., 2015a, b). We screened for rmpA genetic mutations in our rmpA-positive cKP strains, but did not identify any important mutation site. We further compared the expression levels of rmpA between hvKP and cKP isolates and found that the rmpA expression levels in cKP isolates were significantly lower than that in hvKP isolates in all three groups. Therefore, low expression of rmpA may lead to absence of the hypervirulent phenotype in cKP isolates. To confirm this assumption, a rmpA overexpressing plasmid was constructed and transformed into the rmpA-low-expression cKP isolates. We found that overexpression of rmpA could enhance the virulence of cKP isolates in the mouse model, which supported our hypothesis. However, the reason for the low expression of rmpA in cKP isolates is still unclear. A previous study showed that the expression of rmpA is regulated by the iron-responsive regulator Fur (Cheng et al., 2010), indicating that iron availability contributes to the expression of rmpA. Considering that aerobactin is one of the most important siderophores and all cKP isolates in our study were aerobactin-negative, the rmpA expression may depend on aerobactin, which needs to be further studied.
Besides rmpA, the capsular serotypes K1 and K2 have shown a strong correlation with hvKP in many reports (Zhao et al., 2016; Catalan-Najera et al., 2017; Guo et al., 2017). A previous study in Taiwan found that K. pneumoniae isolates with K1 or K2 capsular serotypes showed significantly higher virulence in the mouse model than isolates with other capsular serotypes (Yeh et al., 2007). In this study, the detection frequencies of hvKP isolates with K1 capsular serotype were 39.36% (37/94), 30% (6/20), and 30% (3/10) in non-MDR-KP, ESBL-KP, and CR-KP groups, respectively, which were significantly higher than that of cKP isolates. The proportion of K2 capsular serotype in hvKP and cKP isolates did not show a significant difference in our study. The virulence gene magA belongs to the K1 capsular operon and shows the same distribution pattern with K1 capsular serotype. ST23 was found to be the most prevalent ST in hvKP isolates and strongly correlated with the K1 capsular serotype in several previous studies (Siu et al., 2011; Qu et al., 2015; Guo et al., 2017). In this study, the ST23 genotype was more common in hvKP than cKP isolates in all three groups, suggesting the strong correlation between ST23 and hvKP. The proportion of ST23 in K1 capsular serotype hvKP isolates was 43.48% (20/46), which was lower than that reported in previous studies (Guo et al., 2017; Lee et al., 2017).
The hvKP strains are generally susceptible to commonly used clinical antimicrobial drugs (Paczosa and Mecsas, 2016). However, with the dissemination of genetic elements encoding ESBLs and carbapenemases, hypervirulent ESBL-KP and CR-KP isolates have been increasingly reported in the past few years (Yu et al., 2015b; Zhan et al., 2017; Liu and Guo, 2019). Previous studies showed that the identified CR-KP strains in China were mostly KPC-2-producer with ST11 or ST23 (Lee et al., 2017). In this study, 20 hypervirulent ESBL-KP and 10 hypervirulent CR-KP isolates were identified. Eight of them were ST11 and eight were ST23, implying the transmission of virulence genes to clonal lineages of cKP, e.g., ST11. The hypervirulent ESBL-KP isolates mainly carried blaCTX–M (12/20) genes, which have been reported as the most common ESBL genotype in China (Yu et al., 2007). Notably, eight of 10 hypervirulent CR-KP isolates were NDM-1 type, which has rarely been reported in China before (Lee et al., 2016). Three hypervirulent ESBL-KP and two hypervirulent CR-KP isolates were resistant to tigecycline, which further reduced the therapeutic options for successful treatment of infection with these hypervirulent MDK-KP strains.
Conclusion
Using the aerobactin positivity for defining hvKP in the present study revealed that the ESBL-KP and CR-KP isolates had significantly lower prevalence of hypervirulent phenotype but higher antibiotic resistance rates than the non-MDR-KP isolates. We also found that the capsular serotype K1 (magA), rmpA, hypermucoviscosity, and ST23 were strongly associated with hvKP in non-MDR-KP, ESBL-KP, and CR-KP groups. We showed that in addition to genetic mutations, low expression levels of rmpA also contributed to the hypervirulence-negative phenotype.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
All procedures performed were approved by the Ethical Committee of Shenzhen Nanshan People’s Hospital and were in accordance with the tenets of the 1964 Helsinki declaration and its later amendments.
Author Contributions
ZL participated in the design of the study and drafted the manuscript. ZL, JZ, BB, and GX participated in antibiotic susceptibility test, string test, and detection of virulence factors, β-lactamase, and carbapenemase genes and participated in data analysis. FL and ZC performed the MLST and qRT-PCR assay. ZL and XS conducted the rmpA overexpression assay and mouse intranasal infection assay. DQ, ZY, and QD designed the study and assisted in revisions of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Yun-song Yu (Department of Infectious Diseases, College of Medicine, Zhejiang University, Zhejiang, China) for providing the K. pneumoniae NTUH-K2044 strain.
Funding. This study was supported by the National Natural Science Foundation of China (81902033 and 81170370), the Science, Technology and Innovation Commission of Shenzhen Municipality of basic research funds (JCYJ20180302144403714, JCYJ20180302144721183, and JCYJ20180302144340004) and key funds (JCYJ20170412143551332 and JCYJ20180508162403996), the Sanming Project of Medicine in Shenzhen (SMGC201705029), and the Shenzhen Nanshan District Scientific Research Program of China (2018021).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00436/full#supplementary-material
Minimum spanning tree of 257 cKP isolates by MLST type and gene allele profile. Seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) were PCR-amplified and sequenced from all isolates according to the K. pneumoniae MLST protocol. Alleles and sequence types (STs) were assigned by the MLST database. K. pneumoniae CCs were identified by the phyloviz-2.0a program. Each node within the tree represented a single ST and the number of isolates. The size of the nodes was proportional to the number of isolates. Lines connecting each node indicated CCs. Length of lines between the node was proportional to the number of different alleles.
Primers used in this study.
Strains and plasmids used for the overexpression of rmpA in Klebsiella pneumoniae.
Primers used for the overexpression of rmpA in K. pneumoniae.
Distribution of antimicrobial resistance among non-MDR-KP, ESBL-KP, and CR-KP isolates.
The allele information of 6 hvKP and 10 cKP isolates with new allele code combinations.
Molecular characteristics of the hypervirulent non-MDR-KP isolates.
The genotypes of rmpA among K. pneumoniae without hypervirulent phenotype.
Comparison of the sensitivity and specificity of hvKP defined by aerobactin or hypermucoviscosity.
References
- Candan E. D., Aksoz N. (2015). Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim. Pol. 62 867–874. 10.18388/abp.2015_1148 [DOI] [PubMed] [Google Scholar]
- Catalan-Najera J. C., Garza-Ramos U., Barrios-Camacho H. (2017). Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence 8 1111–1123. 10.1080/21505594.2017.1317412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Bastian I., Warner M. (2013). Survey of Klebsiella pneumoniae bacteraemia in two South Australian hospitals and detection of hypermucoviscous phenotype and magA/rmpA genotypes in K. pneumoniae isolates. Infection 41 559–563. 10.1007/s15010-012-0374-y [DOI] [PubMed] [Google Scholar]
- Cheng H. Y., Chen Y. S., Wu C. Y., Chang H. Y., Lai Y. C., Peng H. L. (2010). RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J. Bacteriol. 192 3144–3158. 10.1128/JB.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubero M., Grau I., Tubau F., Pallares R., Dominguez M. A., Linares J., et al. (2016). Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007-2013). Clin. Microbiol. Infect. 22 154–160. 10.1016/j.cmi.2015.09.025 [DOI] [PubMed] [Google Scholar]
- Decre D., Verdet C., Emirian A., Le Gourrierec T., Petit J. C., Offenstadt G., et al. (2011). Emerging severe and fatal infections due to Klebsiella pneumoniae in two university hospitals in France. J. Clin. Microbiol. 49 3012–3014. 10.1128/JCM.00676-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Wang S., Zhan L., Jin Y., Duan J., Hao Z., et al. (2017). Microbiological and clinical characteristics of hypermucoviscous Klebsiella pneumoniae isolates associated with invasive infections in China. Front. Cell. Infect. Microbiol. 7:24. 10.3389/fcimb.2017.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt J. J., Wang J. T., Callegan M. C. (2011). Contribution of mucoviscosity-associated gene A (magA) to virulence in experimental Klebsiella pneumoniae endophthalmitis. Invest. Ophthalmol. Vis. Sci. 52 6860–6866. 10.1167/iovs.11-7798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y. C., Peng H. L., Chang H. Y. (2003). RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 gene expression at the transcriptional level. J. Bacteriol. 185 788–800. 10.1128/jb.185.3.788-800.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. R., Lee J. H., Park K. S., Jeon J. H., Kim Y. B., Cha C. J., et al. (2017). Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front. Cell. Infect. Microbiol. 7:483. 10.3389/fcimb.2017.00483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. R., Lee J. H., Park K. S., Kim Y. B., Jeong B. C., Lee S. H. (2016). Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7:895. 10.3389/fmicb.2016.00895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. S., II, Herrera M., Wickes B., Patterson J. E., Jorgensen J. H. (2007). First report of the emergence of CTX-M-type extended-spectrum beta-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob. Agents Chemother. 51 4015–4021. 10.1128/aac.00576-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Sun G., Yu Y., Li N., Chen M., Jin R., et al. (2014). Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin. Infect. Dis. 58 225–232. 10.1093/cid/cit675 [DOI] [PubMed] [Google Scholar]
- Liu C., Guo J. (2019). Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann. Clin. Microbiol. Antimicrob. 18:4. 10.1186/s12941-018-0302-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. C., Cheng D. L., Lin C. L. (1986). Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 146 1913–1916. 10.1001/archinte.146.10.1913 [DOI] [PubMed] [Google Scholar]
- Luo Y., Wang Y., Ye L., Yang J. (2014). Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin. Microbiol. Infect. 20 O818–O824. 10.1111/1469-0691.12664 [DOI] [PubMed] [Google Scholar]
- Nassif X., Sansonetti P. J. (1986). Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 54 603–608. 10.1128/iai.54.3.603-608.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczosa M. K., Mecsas J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80 629–661. 10.1128/MMBR.00078-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios M., Miner T. A., Frederick D. R., Sepulveda V. E., Quinn J. D., Walker K. A., et al. (2018). Identification of two regulators of virulence that are conserved in Klebsiella pneumoniae classical and hypervirulent strains. mBio 9 e01443–18. 10.1128/mBio.01443-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu T. T., Zhou J. C., Jiang Y., Shi K. R., Li B., Shen P., et al. (2015). Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect. Dis. 15:161. 10.1186/s12879-015-0899-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo T. A., Olson R., Macdonald U., Beanan J., Davidson B. A. (2015). Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect. Immun. 83 3325–3333. 10.1128/IAI.00430-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo T. A., Olson R., Macdonald U., Metzger D., Maltese L. M., Drake E. J., et al. (2014). Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect. Immun. 82 2356–2367. 10.1128/IAI.01667-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu L. K., Fung C. P., Chang F. Y., Lee N., Yeh K. M., Koh T. H., et al. (2011). Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J. Clin. Microbiol. 49 3761–3765. 10.1128/JCM.00977-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struve C., Roe C. C., Stegger M., Stahlhut S. G., Hansen D. S., Engelthaler D. M., et al. (2015). Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 6:e00630. 10.1128/mBio.00630-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Li D., Zhou H., Sun Y., Guo L., Shen D. (2017). Bacteremia and other body site infection caused by hypervirulent and classic Klebsiella pneumoniae. Microb. Pathog. 104 254–262. 10.1016/j.micpath.2017.01.049 [DOI] [PubMed] [Google Scholar]
- Wu K. M., Li L. H., Yan J. J., Tsao N., Liao T. L., Tsai H. C., et al. (2009). Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J. Bacteriol. 191 4492–4501. 10.1128/JB.00315-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K. M., Kurup A., Siu L. K., Koh Y. L., Fung C. P., Lin J. C., et al. (2007). Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J. Clin. Microbiol. 45 466–471. 10.1128/jcm.01150-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W. L., Ko W. C., Cheng K. C., Lee H. C., Ke D. S., Lee C. C., et al. (2006). Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 42 1351–1358. 10.1086/503420 [DOI] [PubMed] [Google Scholar]
- Yu W. L., Lee M. F., Chang M. C., Chuang Y. C. (2015a). Intrapersonal mutation of rmpA and rmpA2: a reason for negative hypermucoviscosity phenotype and low virulence of rmpA-positive Klebsiella pneumoniae isolates. J. Glob. Antimicrob. Resist. 3 137–141. 10.1016/j.jgar.2015.03.008 [DOI] [PubMed] [Google Scholar]
- Yu W. L., Lee M. F., Chen C. C., Tang H. J., Ho C. H., Chuang Y. C. (2017). Impacts of hypervirulence determinants on clinical features and outcomes of bacteremia caused by extended-spectrum beta-lactamase-producing Klebsiella pneumoniae. Microb. Drug Resist. 23 376–383. 10.1089/mdr.2016.0018 [DOI] [PubMed] [Google Scholar]
- Yu W. L., Lee M. F., Tang H. J., Chang M. C., Chuang Y. C. (2015b). Low prevalence of rmpA and high tendency of rmpA mutation correspond to low virulence of extended spectrum beta-lactamase-producing Klebsiella pneumoniae isolates. Virulence 6 162–172. 10.1080/21505594.2015.1016703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Ji S., Chen Y., Zhou W., Wei Z., Li L., et al. (2007). Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J. Infect. 54 53–57. 10.1016/j.jinf.2006.01.014 [DOI] [PubMed] [Google Scholar]
- Zhan L., Wang S., Guo Y., Jin Y., Duan J., Hao Z., et al. (2017). Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a tertiary hospital in China. Front. Cell. Infect. Microbiol. 7:182. 10.3389/fcimb.2017.00182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Lin D., Chan E. W., Gu D., Chen G. X., Chen S. (2016). Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob. Agents Chemother. 60 709–711. 10.1128/AAC.02173-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zeng J., Liu W., Zhao F., Hu Z., Zhao C., et al. (2015). Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J. Infect. 71 553–560. 10.1016/j.jinf.2015.07.010 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhao C., Wang Q., Wang X., Chen H., Li H., et al. (2016). High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob. Agents Chemother. 60 6115–6120. 10.1128/AAC.01127-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Chen J., Zhao M., Qiu X., Chen X., Zhang W., et al. (2016). Multilocus sequence types and virulence determinants of hypermucoviscosity-positive Klebsiella pneumoniae isolated from community-acquired infection cases in Harbin, North China. Jpn. J. Infect. Dis. 69 357–360. 10.7883/yoken.JJID.2015.321 [DOI] [PubMed] [Google Scholar]
- Zheng J. X., Lin Z. W., Chen C., Chen Z., Lin F. J., Wu Y., et al. (2018a). Biofilm formation in Klebsiella pneumoniae bacteremia strains was found to be associated with CC23 and the presence of wcaG. Front. Cell. Infect. Microbiol. 8:21. 10.3389/fcimb.2018.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. X., Lin Z. W., Sun X., Lin W. H., Chen Z., Wu Y., et al. (2018b). Overexpression of OqxAB and MacAB efflux pumps contributes to eravacycline resistance and heteroresistance in clinical isolates of Klebsiella pneumoniae. Emerg. Microbes Infect. 7:139. 10.1038/s41426-018-0141-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Minimum spanning tree of 257 cKP isolates by MLST type and gene allele profile. Seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) were PCR-amplified and sequenced from all isolates according to the K. pneumoniae MLST protocol. Alleles and sequence types (STs) were assigned by the MLST database. K. pneumoniae CCs were identified by the phyloviz-2.0a program. Each node within the tree represented a single ST and the number of isolates. The size of the nodes was proportional to the number of isolates. Lines connecting each node indicated CCs. Length of lines between the node was proportional to the number of different alleles.
Primers used in this study.
Strains and plasmids used for the overexpression of rmpA in Klebsiella pneumoniae.
Primers used for the overexpression of rmpA in K. pneumoniae.
Distribution of antimicrobial resistance among non-MDR-KP, ESBL-KP, and CR-KP isolates.
The allele information of 6 hvKP and 10 cKP isolates with new allele code combinations.
Molecular characteristics of the hypervirulent non-MDR-KP isolates.
The genotypes of rmpA among K. pneumoniae without hypervirulent phenotype.
Comparison of the sensitivity and specificity of hvKP defined by aerobactin or hypermucoviscosity.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.