Abstract
The primary cilium is a cell surface organelle that is an important component of cellular biology. While it was once believed to be a vestigial structure without biologic function, it is now known to have essential roles in critical cellular signaling pathways such as Hedgehog (HH) and Wnt. The HH and Wnt pathways are involved in pathogenesis of basal cell carcinoma and melanoma, respectively, and this knowledge is now beginning to inform therapeutic and diagnostic options for patients. The purpose of this review is to familiarize clinicians with primary cilia biology and how this complex cellular organelle has started to translate into clinical care.
Keywords: Basal cell carcinoma, Diagnostic tool, Hedgehog, Melanoma, Primary cilia, Sonidegib, Vismodegib
Key Summary Points
| The primary cilium is a cell surface organelle that was discovered in the 1800s, but only in the last 20 years has been recognized for its importance in cellular biology. |
| The primary cilium has critical roles in signaling pathways such as Hedgehog (HH) and Wnt. |
| Basal cell carcinoma (BCC) depends on HH pathway activation through the primary cilium, which is the basis for the BCC therapies vismodegib and sonidegib. |
| Immunofluorescence staining of conventional melanocytic nevi, which retain primary cilia, and melanoma, which shows primary cilia loss, provides the basis for a novel diagnostic tool. |
Introduction
The primary cilium is an antenna-like structure that extends from the surface of nearly all cells in the human body [1]. Acting as the cellular “antenna,” primary cilia help to transmit, regulate, and decode extracellular signals [2]. They have distinct membrane receptors that act as the main signaling depots for common molecular signal pathways in cancer, including the Wnt [3–7], platelet-derived growth factor [5], Hedgehog (HH) signaling [6], among others.
Basal cell carcinoma (BCC) depends on HH pathway activation through the primary cilium. This dependence on the HH pathway is the basis for the BCC therapies vismodegib and sonidegib, which target and inhibit signaling through the primary cilium. Recent studies have shown the ability of BCC to phenotype switch to squamous differentiation in part by reducing dependence on HH signaling through the primary cilium [7].
Melanoma and non-melanocytic skin tumors are the most common cancers worldwide, and are projected to increase in incidence and mortality for the foreseeable future [8]. Given the wide range of clinical and histopathological appearances of melanocytic neoplasms, diagnostic consistency and accuracy can be particularly challenging for both dermatologists and dermatopathologists [9, 10]. Though there are a variety of diagnostic tools available to aid in achieving the most accurate assessment of malignancy [11, 12], diagnostic uncertainty can persist in ambiguous lesions. Multiple studies have now demonstrated that primary cilia are lost in melanoma and retained in nevi [13–17]. These studies suggest the potential for using primary cilia staining as an adjunct diagnostic tool for dermatopathologists.
In this review, we will give a brief history of the primary cilium in modern medicine and cancer, discuss the role of HH signaling in BCC, and explore the how primary cilia are giving new insights into melanoma.
Methods
A literature search using the PubMed database was conducted using the terms (“primary cilia”) AND (“history” OR “cancer”); (“basal cell carcinoma”) AND (“cilia” OR “treatment” OR “resistance” OR “hedgehog signaling”). For the melanoma component, the search terms included (“primary cilia” OR “primary cilium”) AND (“melanoma” OR “nevus”). Only articles focusing on histopathologic diagnosis of cutaneous melanocytic neoplasms were included.
The tissue samples used for generating Figs. 1 and 4 were procured from the archives of the University of California San Francisco (UCSF) Department of Dermatopathology, and all the tissues were collected in accordance with the institutional review board with regard to informed consent. Written patient consent was not required because only existing and de-identified specimens were used. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964, as revised in 2013.
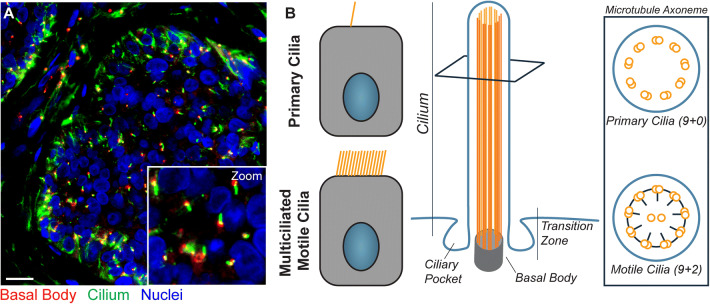
Fig. 1.
Immunofluorescence staining and structure of primary cilia. a Immunofluorescence staining of primary cilia on basal cell carcinoma is shown (scale bar 50 µm). Nuclei are highlighted in blue (DAPI), the ciliary axoneme is shown in green (acetylated alpha-tubulin), and centrioles are displayed in red (gamma-tubulin). b Unlike motile cilia, only one primary cilium is present per cell, and the primary cilium’s axoneme does not contain central microtubule singlets nor dynein arms
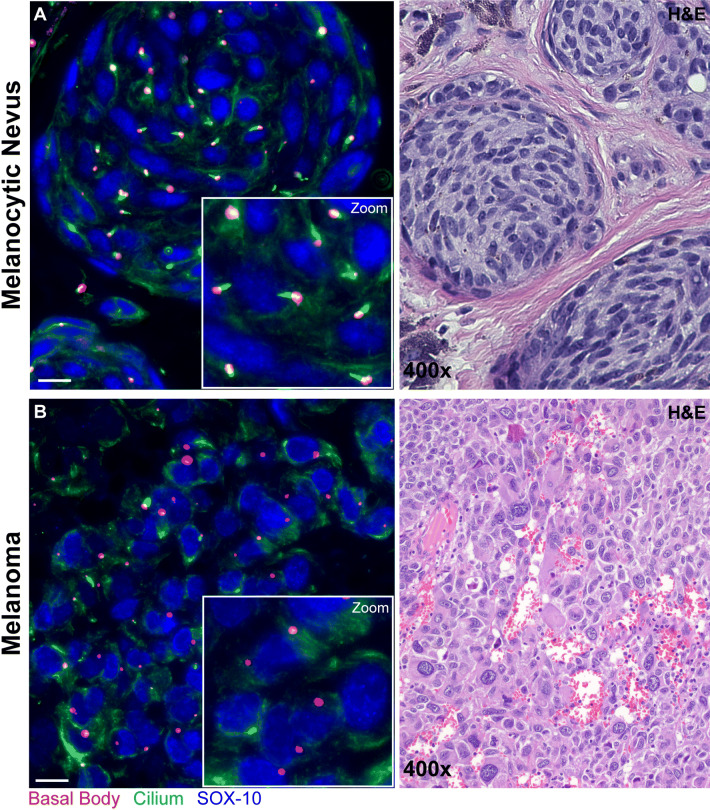
Fig. 4.
Primary cilia immunofluorescence staining in melanocytic nevus and melanoma. Immunofluorescence staining shows melanocyte nuclei highlighted blue (SOX-10), ciliary axoneme shown in green (acetylated alpha-tubulin), and centrioles in red (gamma-tubulin). For the purpose of quantification for calculation of the ciliation index (% ciliated lesional melanocytes), a cell is considered positive if it contains an elongated ciliary axoneme extending from a centriole and negative if only centrioles are identified. Only melanocytes with identifiable centrioles are counted in at least three high-power fields, with an overall minimum of 150 melanocytes. An example of the immunofluorescence and corresponding hematoxylin and eosin (H&E) stains are shown for a melanocytic nevus (a) and invasive melanoma (b) (scale bar 50 µm)
Results and Discussion
Primary Cilia Structure
The primary cilium is a microtubule-based organelle that projects from the surface of most vertebrate cells. Under most circumstances, each cell only has one primary cilium (Fig. 1a, b). Unlike motile cilia, such as those found on respiratory epithelial cells and eukaryotic flagella, primary cilia are not involved in motility. Rather, primary cilia are responsible for transmitting cellular signals, and have a specialized structure that facilitates signal transduction [18]. Vertebrate primary cilia project from the surface of the cell, emerging from a single centrosomal centriole known as the basal body, and are surrounded by the cell membrane. Within the membrane is the axoneme, which consists of a ring of nine microtubule doublets, known as a 9+0 configuration. In contrast, motile cilia additionally have two central microtubule singlets and dynein arms attached to the outer microtubule doublets, altogether known as the 9+2 configuration (Fig. 1b).
The transition zone at the base of the cilium acts as a gate that controls protein entry and exit [19]. Thus, primary cilia serve as a unique microenvironment where specific proteins and lipids can interact in a dynamic fashion to alter cell signaling, function, and fate. Consequently, dysfunctional primary cilia structure or signaling can lead to cancer and congenital conditions collectively known as “ciliopathies” [20]. Thus, understanding primary cilia is crucial to understanding the pathogenesis and clinical manifestations of many diseases.
History
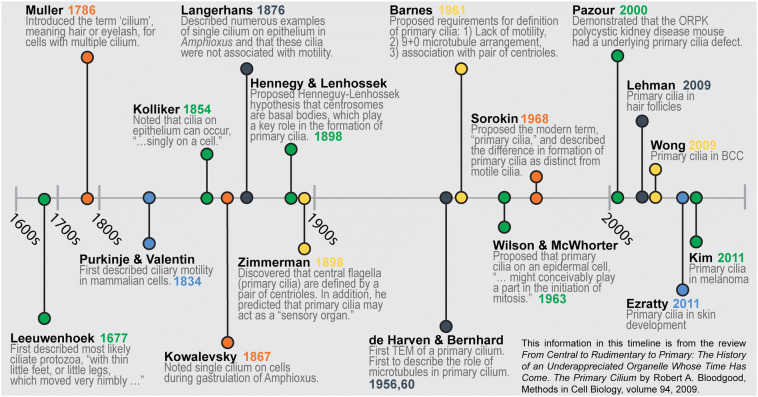
Cilia were initially observed in protozoa by Antony van Leeuwenhoek in 1677, and were defined by their motility as the first cellular organelle [21, 22]. Almost two centuries later, Alexander Kovalevsky perceived that many vertebrate cells had a single, immotile cilia [23]. This discovery was soon corroborated by other scientists, including Karl Wilhelm Zimmermann, who named them “centralgeissel” (central flagella) in mammalian cells and suggested that they may have sensory potential [24, 25]. Despite the short burst of scientific interest in centralgeissel in the late nineteenth century, the organelle was largely neglected and dismissed as vestigial because of lack of any obvious function. In 1968, immotile centralgeissel on mammalian cells were renamed primary cilia because their expression could be identified first during fetal development, before the emergence of motile cilia [26].
Interest in primary cilia smoldered to life in 1993 with the seemingly unrelated discovery of intraflagellar transport (IFT) in the unicellular green alga Chlamydomonas [27]. IFT is the method by which cells move cargo, such as proteins, along the microtubules within cilia and flagella. Specifically, scientists discovered that loss of the IFT88 gene in Chlamydomonas resulted in loss of flagella [28]. Interestingly, mice that lack Tg737 (the murine homolog of IFT88) have deformed primary cilia in their kidney cells and develop polycystic kidney disease [28]. This nascent connection between cilia and disease renewed scientific interest in the primary cilium and sparked an explosion of primary cilium investigation that has persisted to the present day. A more detailed timeline and summary of significant advances in the cilia literature can be found in Fig. 2 and Table 1, which are based on information from a review article by Bloodgood [29].
Fig. 2.
Timeline of major contributions to the cilia literature
Table 1.
Major contributions to the cilia literature
| Author(s) (reference) | Year | Discovery/contribution/advancement |
|---|---|---|
| von Leeuwenhoek [21] | 1677 | First described most likely ciliate protozoa, “with thin little feet, or little legs, which moved very nimbly…” |
| Muller [94] | 1786 | Introduced the term “cilium,” meaning hair or eyelash, for cells with multiple cilia |
| Purkinje and Valentin [95] | 1834 | First described ciliary motility in mammalian cells |
| DuJardin [96] | 1841 | Introduced the term “flagellum,” meaning whip, for cells with a single cilium |
| Ecker [97] | 1844 | Noted single cilium on the epithelium of the semicircular ear canals in sea lamprey |
| Kolliker [98] | 1854 | Noted that cilia on epithelium can occur “singly on a cell” |
| Kowalevsky [23] | 1867 | Noted single cilium on cells during gastrulation of Amphioxus |
| Flemming and van Beneden [99, 100] | 1875/76 | Discovered the centrosome (aka centriole, basal body) |
| Langerhans [24] | 1876 | Described numerous examples of single cilium on epithelium in Amphioxus and that these cilia were not associated with motility |
| Zimmermann [25] | 1898 | Discovered that central flagella (primary cilia) are defined, and distinguished from cilia, by a pair of centrioles. In addition, he predicted that primary cilia may act as a “sensory organ” for these cells |
| Henneguy and Lenhossek [101–103] | 1898 | Proposed the Henneguy–Lenhossek hypothesis that centrosomes are basal bodies, which play a key role in the formation of primary cilia |
| Alverdes [104] | 1927 | Proposed that primary cilium “may represent a cellular receptor, which communicates fluctuations…” |
| Sjostrand [105] | 1953 | First transmission electron micrograph (TEM) of a mature photoreceptor connecting epithelium (a sensory cilium) |
| de Harven and Bernhard [106] | 1956 | First TEM of a primary cilium |
| de Harven and Bernhard [107] | 1960 | First to describe the role of microtubules in primary cilium |
| Barnes [108] | 1961 | Proposed requirements for definition of primary cilia: (1) Lack of motility, (2) 9 + 0 microtubule arrangement, and (3) association with pair of centrioles |
| Wilson and McWhorter [109] | 1963 | Proposed that primary cilia on epidermal cell “might conceivably play a part in the initiation of mitosis” |
| Sorokin [26] | 1968 | Proposed the modern term “primary cilia” and described the difference in formation of primary cilia as distinct from motile cilia |
| Archer and Wheatley [110] | 1971 | First to demonstrate experimentally that primary cilia completely disappear during mitosis |
| Afzelius [111] | 1976 | Discovered the connection between Kartagener’s syndrome and motile cilia with a defect in the dynein arms |
| Kozminski [27] | 1993 | Discovered intraflagellar transport in Chlamydomonas |
| Pazour [28] | 2000 | Demonstrated that the ORPK (Oak Ridge polycystic kidney disease) mouse had an underlying primary cilia defect |
| Lehman [112] | 2009 | Described role of primary cilium in hair follicles |
| Wong [60] | 2009 | Described role of primary cilia in BCC |
| Ezratty [113] | 2011 | Described role of primary cilia in skin development |
| Kim [14] | 2011 | Described role of primary cilia in melanoma |
Since the connection to polycystic kidney disease, an entire class of disorders, known as “ciliopathies” and spanning almost every organ system, have been linked to dysfunction in the primary cilium [30]. Ciliopathies are inherited genetic disorders that are associated with mutations in genes that are important for the function of primary cilia. Outside of an inherited context, dysfunction of primary cilia has also been implicated in sporadic cancers [31, 32]. The mechanisms underlying both ciliopathies and the primary cilium’s role in cancer have been linked to more recent discoveries implicating the primary cilium as a central hub for many gene expression programs that are crucial for development and adult stem cell homeostasis, such as the HH [6], Wnt [33], and Notch [34] pathways.
BCC and HH Signaling
BCC affects approximately two million people per year and is the most common cancer in the USA [35]. BCC typically arises in adult patients as a result of ultraviolet radiation-induced genomic mutations. Indeed, BCCs are the most mutated human cancer, with about 65 mutations per megabase [36]. Although one might expect these numerous mutations to be random, BCCs are unified by misactivation of the HH pathway in both sporadic and syndromic cases. With respect to the latter, individuals with inherited heterozygous mutations in HH pathway inhibitor genes develop nevoid BCC syndrome (also known as Gorlin syndrome), which is characterized by numerous BCCs and other HH-associated cancers such as medulloblastoma and rhabdomyosarcoma [37–41]. Primary cilia play a crucial part in the pathogenesis of BCC because of the primary cilium’s essential role in the HH signaling pathway.
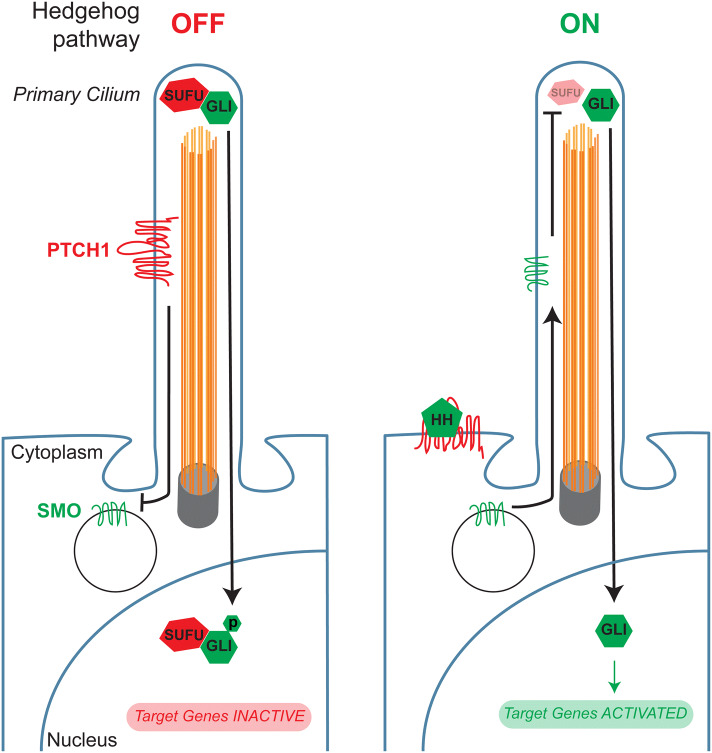
The HH pathway is activated by HH ligands, which are secreted lipoproteins [42]. The best studied HH ligand gene encodes Sonic hedgehog (SHH), a crucial regulator of embryonic development and adult tissue homeostasis [43, 44]. HH ligands stimulate the HH pathway, often in nearby cells, by binding to their transmembrane receptors, patched 1 (PTCH1) and patched 2 (PTCH2), which are present at the ciliary membrane [45–47] (Fig. 3). HH signaling is transduced by the primary cilium. When HH binds to PTCH proteins, another transmembrane protein, Smoothened (SMO), accumulates at the cilium to activate the downstream pathway [48, 49] (Fig. 3). SMO activates GLI2, the primary activator of the HH transcriptional program, which in turn regulates GLI1, a feed-forward amplifier of transcriptional activity [50, 51]. When the HH pathway is inactive, suppressor of fused (SUFU) binds to GLI family members and represses their transcriptional activity [52–54] (Fig. 3).
Fig. 3.
Hedgehog pathway activation through the primary cilium. When the Hedgehog pathway is off, patched (PTCH1) localizes to the primary cilium and prevents Smoothened (SMO) from entering the cilium. Simultaneously, suppressor of fused homolog (SUFU) binds to and inhibits the activity of glioma-associated oncogene homolog (GLI) transcription factors. Hedgehog ligands (HH) activate the pathway by binding to PTCH1, causing it to leave the cilium, and allowing SMO to enter. SMO in turn activates GLI proteins, allowing for the transcription of Hedgehog target genes
Primary cilia and the HH pathway play crucial roles in normal skin development and growth, thus their dysregulation in BCC is perhaps not surprising. In the developing skin, SHH secreted by epidermal placodes induces proliferation and hair follicle formation in the underlying dermal condensates [55]. Without SHH, epidermal placodes form but fail to grow down into the dermis [56]. Similarly, in postnatal skin, hair follicle growth also relies on signaling through the HH pathway. For example, SHH is expressed during anagen, the hair follicle phase during which there is downward growth of the follicle into the dermis, and is important for maintaining epidermal stem cells and regulating epidermal growth [57–59].
The most common HH pathway-activating mutations in BCC include biallelic loss-of-function mutations in pathway inhibitors (e.g., PTCH1, PTCH2, and SUFU); amplification of GLI2, the principal HH pathway transcriptional activator; and monoallelic activating mutations in pathway activators, such as SMO [36]. BCC cells possess cilia (Fig. 1a), which transduce HH signals from SMO to GLI2 [60, 61]. In fact, in mouse models of BCC, scientists have shown that both the ciliary gene Intu and HH signaling are necessary for BCC tumorigenesis and progression [61, 62]. Thus, the primary cilium, through HH signaling, acts a crucial nexus in the pathogenesis of BCC.
BCC Therapy
BCCs are typically slow-growing and are most often effectively treated with local excision. However, many factors can prevent complete excision, such as number or size of tumors, or proximity to critical structures, including the eye, lip, and nose. In these cases, nonsurgical local treatments, such as topical cytotoxic agents, radiotherapy, photodynamic therapy, and cryotherapy, can be used [63]. In the small subset of patients with locally advanced or metastatic BCC, systemic therapy is indicated. For such cases, HH pathway inhibition with SMO antagonists, such as vismodegib or sonidegib, has been shown to be more effective than chemotherapy [64–66]. Although the proportion of BCC patients who are eligible for molecular therapy is small, the tremendous incidence of BCC cases each year makes the absolute number of patients who may be considered for vismodegib or sonidegib large. Unfortunately, systemic inhibition of the HH pathway can lead to adverse events, such as nausea, muscle cramps, loss of taste, weight loss, and alopecia [67]. Although relatively mild, these symptoms can cause patients to not adhere to treatment regimens, which may lead to BCC recurrence. Thus, the combination of radiotherapy with HH pathway inhibition may be used to achieve durable responses with cessation of systemic therapy for such patients [68].
In addition to recurrence due to lack of adherence, resistance to vismodegib and sonidegib has also been documented, typically via mutations in SMO, the target of both inhibitors [69, 70]. A frequent activating mutation in SMO is W535L, also known as SMOM2, which causes SMO to accumulate in the cilium even in the absence of HH ligands [71, 72]. In medulloblastoma, another HH-driven cancer where HH pathway inhibitors are used, there are examples of resistance that arise from amplification of targets downstream of SMO, such as GLI2 or cyclin D1 [73, 74]. Outside of alternative methods of HH pathway activation, rare examples of BCC resistance have been seen via loss of ciliation, loss of HH signaling, and subsequent activation of alternative signaling pathways, such as the Ras/MAPK pathway [7]. Overcoming resistance to SMO antagonists in BCC is an active area of research, with some efforts focused on targeting downstream elements of the HH pathway. HH pathway-independent treatment options, such as cancer immunotherapy, have also been proposed for resistant tumors. Given BCC’s high mutational burden and the correlation between mutational burden and the success of immunotherapy, clinical trials with anti-PD1 therapy have been initiated (NCT03132636, NCT03521830).
Melanoma Pathogenesis
There are diverse genetic changes and transcriptional programs that contribute to melanoma pathogenesis. Prominent activating mutations in key oncogenic driver genes, such as BRAF or NRAS, are present early in melanocytic nevi, but the cellular programs defining proliferative and invasive phenotypes are not defined by particular genetic changes, rather are governed by transcriptional master regulators [75–78]. It is the culmination of these complex changes that translates to biologic function, which is further refined by an individual’s immune system and microenvironment [79]. A detailed review of the genetic and phenotypic changes occurring in melanomagenesis is outside the scope of this review, but can be found elsewhere [75, 76]. Briefly, the initiating mutations result in uncontrolled proliferation of a melanocyte, followed by approximately 5–10 additional pathogenic alterations spread over several signaling pathways to result in melanoma [80]. It is also generally accepted that there are “intermediate” cell states between the “benign” and “malignant” categories of melanocytic neoplasms [79]. Although different combinations of mutations in a variety of genes underlie the diversity of melanocytic neoplasms, they all share the common denominator of activating the mitogen-activated protein kinase (MAPK) pathway. Beyond DNA mutations, deletions and amplifications, DNA methylation [81], microRNAs [82], transcription [83], and translation [84] also play important roles in determining the malignant potential of a particular melanocytic neoplasm [85]. Finally, the tumor’s microenvironment and interaction with an individual’s immune system also contribute to the clinical course of melanoma [86]. Importantly, regardless of the variability of the genetic landscape, there is downstream convergence on a malignant phenotype.
Primary Cilia in Melanocytic Neoplasms
The presence of primary cilia on the cell surface of lesional melanocytes within conventional melanocytic nevi was first reported in 2011 by Kim et al. [14]. This was in contrast to the near-complete loss of primary cilia on in situ, invasive, and metastatic melanoma. At that time, it was also investigated whether the observed loss of primary cilia might be due to ongoing cell cycle progression, as primary cilia reabsorption is a known physiologic response during the interphase portion of the cell cycle [2]. In their study, staining for Ki-67, a protein present during the cell cycle and absent during G0, was performed and found to vastly under-represent the percentage of non-ciliated cells. This was the initial evidence that active cell cycle progression was unlikely to account for the almost complete loss of primary cilia observed in melanoma.
An independent group, Snedecor et al., validated the original findings by evaluating a cohort of melanocytic nevi compared to in situ, invasive, and metastatic melanoma [15]. Interestingly, although still significantly higher than their melanoma cohort, the average percentage of ciliated melanocytes within the nevus cohort was 25%. This is notably lower than that of the Kim et al. study demonstrating an average of 94% ciliated melanocytes in nevi. A direct comparison of these results may be limited by variations in immunofluorescence staining methods used as well as possible variability in the types of melanocytic nevi included. However, Snedecor et al. additionally contributed to the hypothesis that cilia loss in melanoma was unlikely to be solely due to an elevated proliferation rate.
The underlying mechanism of primary cilia loss in melanoma remained a mystery until a recent study examining the role of “enhancer of zeste homolog 2” gene (EZH2) overexpression/amplification in melanoma [87]. Zingg et al. uncovered that many cilia-associated genes were transcriptionally repressed by EZH2, which functions as the histone methyltransferase unit of polycomb repressive complex 2 (PRC2) [88]. PRC2 is a complex that methylates histone H3 to promote transcriptional silencing. As a global regulator of gene transcription, amplification or gain-of-function of EZH2 has significant downstream effects that have been associated with the transition to a melanoma state [79]. This study also experimentally demonstrated activation of the beta-catenin pathway as a consequence of primary cilia knockdown. These data provide insights into the intricacies of potentially targeting the Wnt/beta-catenin signaling pathway in melanoma therapy. Given the correlation of beta-catenin pathway activation and melanoma immune evasion [89], primary cilia-related proteins may also provide novel candidate targets for patients resistant to immunotherapies.
The function of primary cilia in the context of melanocyte biology and melanomagenesis is still in early phases of investigation; however, it appears to have an active role in suppressing the oncogenic activity of the Wnt/beta-catenin pathway. Importantly, linking the loss of primary cilia to upstream global effectors of melanomagenesis brings us closer to appreciating the potential value of using primary cilia staining as a diagnostic tool in ambiguous melanocytic neoplasms.
Melanoma Molecular Diagnostic Tools
The complexity and diversity of biologic checkpoints involved in the development of melanoma reflects the challenges that both clinicians and pathologists can face in making an accurate diagnosis. In particular, the “intermediate” lesions that do not meet histopathologic criteria for melanoma but have atypical features raise the question of their true biologic potential. Many of the most well-established adjunct diagnostic tools available to dermatopathologists for the diagnosis of ambiguous melanocytic neoplasms involve either genetic/genomic analysis or single protein detection by immunohistochemistry [13, 14]. There is now a greater appreciation that a purely genetic analysis may miss critical post-transcriptional and post-translational changes. Techniques such as mass spectrometry [90] and micro-RNA profiling [82] are emerging to address this gap. Each of these tests provides additional information that the pathologist can then use to integrate with the histopathology to reach their best estimate of malignant potential. The decision to initiate additional molecular analysis beyond routine histopathologic assessment largely rests on the ease of performing the test, the availability of sufficient tissue, and expertise for interpretation.
Performing primary cilia staining is no more technically challenging than standard immunohistochemical staining for single proteins, which is currently the mainstay of additional workup for histopathologically challenging melanocytic neoplasms. Immunofluorescence staining for primary cilia requires a single unstained standard thickness tissue section from a paraffin-embedded formalin-fixed tissue block (detailed methods found in Kim et al. [14]), and can be easily incorporated into a routine diagnostic workup. Three primary antibodies are combined in a single cocktail for the identification of melanocytes (SOX-10), the ciliary axoneme (acetylated alpha-tubulin), and centrioles (gamma-tubulin). The corresponding three secondary antibodies are conjugated to fluorophores with distinct excitation and emission wavelengths, which allows for concurrent identification of all three components using an immunofluorescence microscope attached to a camera. An example of this immunofluorescence stain is shown in Fig. 4, which highlights primary cilia in a melanocytic nevus and cilia depletion in melanoma.
As this cell surface structure is clearly absent in melanoma when compared to conventional melanocytic nevi, it has the potential for being used in a similar manner as immunohistochemical stains for p16 or PRAME are currently being used [79, 91]. The most common acquired genetic change distinguishing precursor lesions, such as melanocytic nevi or melanoma in situ (MIS), from invasive melanomas is loss of the CDKN2A locus, which is considered an important driver of melanoma [92]. Loss of immunohistochemical staining for the p16 protein can act as a surrogate of the underlying genetic event; however, negative staining for p16 does not always correlate with an underlying mutation being present and conflicting data argues against its use [93]. Conversely, preserved p16 staining does not exclude the possibility of melanoma, and in fact approximately 25% of metastatic melanoma can retain this tumor suppressor gene (TCGA Research Network). With respect to PRAME immunohistochemical staining, there has been rapid adaptation of this stain for clinical use, but as with any single protein, the results must be interpreted with caution in the context of all clinical and histopathological findings. Overall, the cumulative literature results support the need for additional biomarkers, such as primary cilia staining, to help in cases when distinguishing benign from malignant by current immunohistochemical staining practices is insufficient.
Conclusions
Basic science research in the field of primary cilia biology continues to have implications for translational research and ultimately advances in patient care; therefore, clinicians will need to have a basic understanding of this cell surface organelle. The importance of this organelle is a relatively new discovery, but ongoing research is demonstrating how it relates to cellular function in a context-dependent way.
Acknowledgements
Funding
This work was generously supported by a Dermatology Foundation Career Development Award. This study was supported by the UCSF Physician Scientist Scholar Program and the NIH Grant K08 CA212279-01 to David R. Raleigh. No Rapid Service Fee was received by the journal for the publication of this article.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
Abrar Choudhury, Neil M. Neumann, David R. Raleigh, and Ursula E. Lang have nothing to disclose.
Compliance with Ethics Guidelines
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964, as revised in 2013.
Data Availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Footnotes
Enhanced Digital Features
To view enhanced digital features for this article go to 10.6084/m9.figshare.11522553.
References
- 1.Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 2.Seeley ES, Nachury MV. The perennial organelle: assembly and disassembly of the primary cilium. J Cell Sci. 2010;123(Pt 4):511–518. doi: 10.1242/jcs.061093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oh EC, Katsanis N. Context-dependent regulation of Wnt signaling through the primary cilium. J Am Soc Nephrol. 2013;24(1):10–18. doi: 10.1681/ASN.2012050526. [DOI] [PubMed] [Google Scholar]
- 4.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11(5):331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen BS, Malinda RR, Schmid FM, Pedersen SF, Christensen ST, Pedersen LB. PDGFRβ and oncogenic mutant PDGFRα D842V promote disassembly of primary cilia through a PLCγ- and AURKA-dependent mechanism. J Cell Sci. 2015;128(19):3543–3549. doi: 10.1242/jcs.173559. [DOI] [PubMed] [Google Scholar]
- 6.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317(5836):372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 7.Kuonen F, Huskey NE, Shankar G, et al. Loss of primary cilia drives switching from Hedgehog to Ras/MAPK pathway in resistant basal cell carcinoma. J Investig Dermatol. 2019;139(7):1439–1448. doi: 10.1016/j.jid.2018.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guy GP, Thomas CC, Thompson T, et al. Vital signs: melanoma incidence and mortality trends and projections—United States, 1982–2030. MMWR Morb Mortal Wkly Rep. 2015;64(21):591–596. [PMC free article] [PubMed] [Google Scholar]
- 9.Gerami P, Busam K, Cochran A, et al. Histomorphologic assessment and interobserver diagnostic reproducibility of atypical spitzoid melanocytic neoplasms with long-term follow-up. Am J Surg Pathol. 2014;38(7):934–940. doi: 10.1097/PAS.0000000000000198. [DOI] [PubMed] [Google Scholar]
- 10.Elmore JG, Barnhill RL, Elder DE, et al. Pathologists’ diagnosis of invasive melanoma and melanocytic proliferations: observer accuracy and reproducibility study. BMJ. 2017;28(357):j2813. doi: 10.1136/bmj.j2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiavash K, Bluth MH, Thompson AD. An update regarding the molecular genetics of melanocytic neoplasms and the current applications of molecular genetic technologies in their diagnosis and treatment. Clin Lab Med. 2018;38(2):385–399. doi: 10.1016/j.cll.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Lang UE, Yeh I, McCalmont TH. Molecular melanoma diagnosis update: gene fusion, genomic hybridization, and massively parallel short-read sequencing. Clin Lab Med. 2017;37(3):473–484. doi: 10.1016/j.cll.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Jackson PK. EZH2 inactivates primary cilia to activate Wnt and drive melanoma. Cancer Cell. 2018;34(1):3–5. doi: 10.1016/j.ccell.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Dabiri S, Seeley ES. Primary cilium depletion typifies cutaneous melanoma in situ and malignant melanoma. PLoS One. 2011;6(11):e27410. doi: 10.1371/journal.pone.0027410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snedecor ER, Sung CC, Moncayo A, et al. Loss of primary cilia in melanoma cells is likely independent of proliferation and cell cycle progression. J Investig Dermatol. 2015;135(5):1456–1458. doi: 10.1038/jid.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lang UE, Love NR, Cheung C, McCalmont TH, Kim J. Use of the ciliation index to distinguish invasive melanoma from associated conventional melanocytic nevi. Am J Dermatopathol. 2019;42:11–15. doi: 10.1097/DAD.0000000000001459. [DOI] [PubMed] [Google Scholar]
- 17.Love NR, Lang UE, Cheung C, Kim J. Depletion of primary cilium in acral melanoma. J Cutan Pathol. 2019;46(9):665–671. doi: 10.1111/cup.13484. [DOI] [PubMed] [Google Scholar]
- 18.Garcia G, Raleigh DR, Reiter JF. How the ciliary membrane is organized inside-out to communicate outside-in. Curr Biol. 2018;28(8):R421–R434. doi: 10.1016/j.cub.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonçalves J, Pelletier L. The ciliary transition zone: finding the pieces and assembling the gate. Mol Cells. 2017;40(4):243–253. doi: 10.14348/molcells.2017.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiter JF, Leroux MR. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol. 2017;18(9):533–547. doi: 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Leeuwenhoek A. Concerning little animals observed in rain-, well-, sea- and snow-water; as also in water wherein pepper had lain infused. Philos Trans R Soc Lond. 1677;12:821–831. [Google Scholar]
- 22.Dobell C. Antony van Leeuwenhoek and his “little animals”. New York: Harcourt, Brace and Co; 1932. [Google Scholar]
- 23.Kowalevsky A. Entwickelungsgeschichte des Amphioxus lanceolatus. Memoires Acad Imp Sci St-Petersbourg VII. 1867;11:1–17. [Google Scholar]
- 24.Langerhans P. Zur Anatomie des Amphioxus. Arch Mikrokopische Anat. 1876;12:290–348. doi: 10.1007/BF02933895. [DOI] [Google Scholar]
- 25.Zimmermann K. Beitrage zur Kenntniss einiger Drusen und Epithelien. Arch Mikrosk Anat. 1898;52:552–706. doi: 10.1007/BF02975837. [DOI] [Google Scholar]
- 26.Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3(2):207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- 27.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci. 1993;90(12):5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pazour GJ, Dickert BL, Vucica Y, et al. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151(3):709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bloodgood RA. From central to rudimentary to primary: the history of an underappreciated organelle whose time has come. The primary cilium. Methods Cell Biol. 2009;94:3–52. doi: 10.1016/S0091-679X(08)94001-2. [DOI] [PubMed] [Google Scholar]
- 30.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364(16):1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raleigh DR, Reiter JF. Misactivation of Hedgehog signaling causes inherited and sporadic cancers. J Clin Investig. 2019;129(2):465–475. doi: 10.1172/JCI120850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higgins M, Obaidi I, McMorrow T. Primary cilia and their role in cancer. Oncol Lett. 2019;17(3):3041–3047. doi: 10.3892/ol.2019.9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.May-Simera HL, Kelley MW. Cilia, Wnt signaling, and the cytoskeleton. Cilia. 2012;1(1):7. doi: 10.1186/2046-2530-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grisanti L, Revenkova E, Gordon RE, Iomini C. Primary cilia maintain corneal epithelial homeostasis by regulation of the Notch signaling pathway. Development. 2016;143(12):2160–2171. doi: 10.1242/dev.132704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asgari MM, Moffet HH, Ray GT, Quesenberry CP. Trends in basal cell carcinoma incidence and identification of high-risk subgroups, 1998–2012. JAMA Dermatol. 2015;151(9):976–981. doi: 10.1001/jamadermatol.2015.1188. [DOI] [PubMed] [Google Scholar]
- 36.Bonilla X, Parmentier L, King B, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet. 2016;48(4):398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- 37.Gorlin RJ, Goltz RW. Multiple nevoid basal-cell epithelioma, jaw cysts and bifid rib. A syndrome. N Engl J Med. 1960;5(262):908–912. doi: 10.1056/NEJM196005052621803. [DOI] [PubMed] [Google Scholar]
- 38.Hahn H, Wicking C, Zaphiropoulous PG, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85(6):841–851. doi: 10.1016/S0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 39.Johnson RL, Rothman AL, Xie J, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272(5268):1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 40.Herzberg JJ, Wiskemann A. The fifth phakomatosis. Basal cell nevus with hereditary malformation and medulloblastoma. Dermatologica. 1963;126:106–123. doi: 10.1159/000254913. [DOI] [PubMed] [Google Scholar]
- 41.Schweisguth O, Gerard-Marchant R, Lemerle J. Basal cell nevus syndrome. Association with congenital rhabdomyosarcoma. Arch Fr Pediatr. 1968;25(9):1083–1093. [PubMed] [Google Scholar]
- 42.McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/S0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 43.Chiang C, Litingtung Y, Lee E, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383(6599):407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 44.Echelard Y, Epstein DJ, St-Jacques B, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75(7):1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 45.Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996;384(6605):176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 46.Smyth I, Narang MA, Evans T, et al. Isolation and characterization of human patched 2 (PTCH2), a putative tumour suppressor gene in basal cell carcinoma and medulloblastoma on chromosome 1p32. Hum Mol Genet. 1999;8(2):291–297. doi: 10.1093/hmg/8.2.291. [DOI] [PubMed] [Google Scholar]
- 47.Stone DM, Hynes M, Armanini M, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384(6605):129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 48.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DYR, Reiter JF. Vertebrate smoothened functions at the primary cilium. Nature. 2005;437(7061):1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 49.Huang P, Zheng S, Wierbowski BM, et al. Structural basis of smoothened activation in Hedgehog signaling. Cell. 2018;175(1):295–297. doi: 10.1016/j.cell.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12(6):393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 51.Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci USA. 2009;106(51):21666–21671. doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kogerman P, Grimm T, Kogerman L, et al. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1999;1(5):312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 53.Stone DM, Murone M, Luoh S, et al. Characterization of the human suppressor of fused, a negative regulator of the zinc-finger transcription factor Gli. J Cell Sci. 1999;112(Pt 23):4437–4448. doi: 10.1242/jcs.112.23.4437. [DOI] [PubMed] [Google Scholar]
- 54.Svärd J, Heby-Henricson K, Henricson KH, et al. Genetic elimination of suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10(2):187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 55.St-Jacques B, Dassule HR, Karavanova I, et al. Sonic hedgehog signaling is essential for hair development. Curr Biol. 1998;8(19):1058–1068. doi: 10.1016/S0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- 56.Chiang C, Swan RZ, Grachtchouk M, et al. Essential role for Sonic hedgehog during hair follicle morphogenesis. Dev Biol. 1999;205(1):1–9. doi: 10.1006/dbio.1998.9103. [DOI] [PubMed] [Google Scholar]
- 57.Oro AE, Higgins K. Hair cycle regulation of Hedgehog signal reception. Dev Biol. 2003;255(2):238–248. doi: 10.1016/S0012-1606(02)00042-8. [DOI] [PubMed] [Google Scholar]
- 58.Sato N, Leopold PL, Crystal RG. Induction of the hair growth phase in postnatal mice by localized transient expression of Sonic hedgehog. J Clin Investig. 1999;104(7):855–864. doi: 10.1172/JCI7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang LC, Liu ZY, Gambardella L, et al. Regular articles: conditional disruption of hedgehog signaling pathway defines its critical role in hair development and regeneration. J Investig Dermatol. 2000;114(5):901–908. doi: 10.1046/j.1523-1747.2000.00951.x. [DOI] [PubMed] [Google Scholar]
- 60.Wong SY, Seol AD, So P-L, et al. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15(9):1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang N, Leung EL-H, Liu C, et al. INTU is essential for oncogenic Hh signaling through regulating primary cilia formation in basal cell carcinoma. Oncogene. 2017;36(35):4997–5005. doi: 10.1038/onc.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peterson SC, Eberl M, Vagnozzi AN, et al. Basal cell carcinoma preferentially arises from stem cells within hair follicle and mechanosensory niches. Cell Stem Cell. 2015;16(4):400–412. doi: 10.1016/j.stem.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koelblinger P, Lang R. New developments in the treatment of basal cell carcinoma: update on current and emerging treatment options with a focus on vismodegib. OncoTargets Ther. 2018;23(11):8327–8340. doi: 10.2147/OTT.S135650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacobsen AA, Aldahan AS, Hughes OB, Shah VV, Strasswimmer J. Hedgehog pathway inhibitor therapy for locally advanced and metastatic basal cell carcinoma: a systematic review and pooled analysis of interventional studies. JAMA Dermatol. 2016;152(7):816–824. doi: 10.1001/jamadermatol.2016.0780. [DOI] [PubMed] [Google Scholar]
- 65.Sekulic A, Migden MR, Oro AE, et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med. 2012;366(23):2171–2179. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang JY, Mackay-Wiggan JM, Aszterbaum M, et al. Inhibiting the Hedgehog pathway in patients with the basal-cell nevus syndrome. N Engl J Med. 2012;366(23):2180–2188. doi: 10.1056/NEJMoa1113538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Von Hoff DD, LoRusso PM, Rudin CM, et al. Inhibition of the Hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009;361(12):1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 68.Raleigh DR, Algazi A, Arron ST, Neuhaus IM, Yom SS. Induction Hedgehog pathway inhibition followed by combined-modality radiotherapy for basal cell carcinoma. Br J Dermatol. 2015;173(2):544–546. doi: 10.1111/bjd.13748. [DOI] [PubMed] [Google Scholar]
- 69.Atwood SX, Sarin KY, Whitson RJ, et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell. 2015;27(3):342–353. doi: 10.1016/j.ccell.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharpe HJ, Pau G, Dijkgraaf GJ, et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell. 2015;27(3):327–341. doi: 10.1016/j.ccell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lam CW, Xie J, To KF, et al. A frequent activated smoothened mutation in sporadic basal cell carcinomas. Oncogene. 1999;18(3):833–836. doi: 10.1038/sj.onc.1202360. [DOI] [PubMed] [Google Scholar]
- 72.Xie J, Murone M, Luoh SM, et al. Activating smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391(6662):90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 73.Buonamici S, Williams J, Morrissey M, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med. 2010;2(51):51ra70. doi: 10.1126/scitranslmed.3001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dijkgraaf GJP, Alicke B, Weinmann L, et al. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res. 2011;71(2):435–444. doi: 10.1158/0008-5472.CAN-10-2876. [DOI] [PubMed] [Google Scholar]
- 75.Widmer DS, Cheng PF, Eichhoff OM, et al. Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment Cell Melanoma Res. 2012;25(3):343–353. doi: 10.1111/j.1755-148X.2012.00986.x. [DOI] [PubMed] [Google Scholar]
- 76.Verfaillie A, Imrichova H, Atak ZK, et al. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat Commun. 2015;9(6):6683. doi: 10.1038/ncomms7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoek KS, Schlegel NC, Brafford P, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19(4):290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 78.Arozarena I, Wellbrock C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat Rev Cancer. 2019;19(7):377–391. doi: 10.1038/s41568-019-0154-4. [DOI] [PubMed] [Google Scholar]
- 79.Shain AH, Joseph NM, Yu R, et al. Genomic and transcriptomic analysis reveals incremental disruption of key signaling pathways during melanoma evolution. Cancer Cell. 2018;34(1):45–55.e4. doi: 10.1016/j.ccell.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shain AH, Bastian BC. From melanocytes to melanomas. Nat Rev Cancer. 2016;16(6):345–358. doi: 10.1038/nrc.2016.37. [DOI] [PubMed] [Google Scholar]
- 81.Conway K, Edmiston SN, Parker JS, et al. Identification of a robust methylation classifier for cutaneous melanoma diagnosis. J Investig Dermatol. 2019;139(6):1349–1361. doi: 10.1016/j.jid.2018.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Torres R, Lang UE, Hejna M, et al. MicroRNA ratios distinguish melanomas from nevi. J Investig Dermatol. 2019;140:164–173.e7. doi: 10.1016/j.jid.2019.06.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ferris LK, Farberg AS, Middlebrook B, et al. Identification of high-risk cutaneous melanoma tumors is improved when combining the online American Joint Committee on Cancer Individualized Melanoma Patient Outcome Prediction Tool with a 31-gene expression profile-based classification. J Am Acad Dermatol. 2017;76(5):818–825.e3. doi: 10.1016/j.jaad.2016.11.051. [DOI] [PubMed] [Google Scholar]
- 84.Sengupta D, Tackett AJ. Proteomic findings in melanoma. J Proteom Bioinform. 2016;9(4):e29. [DOI] [PMC free article] [PubMed]
- 85.Moran B, Silva R, Perry AS, Gallagher WM. Epigenetics of malignant melanoma. Semin Cancer Biol. 2018;51:80–88. doi: 10.1016/j.semcancer.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 86.Marzagalli M, Ebelt ND, Manuel ER. Unraveling the crosstalk between melanoma and immune cells in the tumor microenvironment. Semin Cancer Biol. 2019;59:236–50. [DOI] [PubMed]
- 87.Zingg D, Debbache J, Peña-Hernández R, et al. EZH2-mediated primary cilium deconstruction drives metastatic melanoma formation. Cancer Cell. 2018;34(1):69–84.e14. doi: 10.1016/j.ccell.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 88.Kim KH, Roberts CWM. Targeting EZH2 in cancer. Nat Med. 2016;22(2):128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 90.Lazova R, Seeley EH. Proteomic mass spectrometry imaging for skin cancer diagnosis. Dermatol Clin. 2017;35(4):513–519. doi: 10.1016/j.det.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 91.Shain AH, Yeh I, Kovalyshyn I, et al. The genetic evolution of melanoma from precursor lesions. N Engl J Med. 2015;373(20):1926–1936. doi: 10.1056/NEJMoa1502583. [DOI] [PubMed] [Google Scholar]
- 92.Zeng H, Jorapur A, Shain AH, et al. Bi-allelic loss of CDKN2A initiates melanoma invasion via BRN2 activation. Cancer Cell. 2018;34(1):56–68.e9. doi: 10.1016/j.ccell.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koh SS, Cassarino DS. Immunohistochemical expression of p16 in melanocytic lesions: an updated review and meta-analysis. Arch Pathol Lab Med. 2018;142(7):815–828. doi: 10.5858/arpa.2017-0435-RA. [DOI] [PubMed] [Google Scholar]
- 94.Muller OF. Animalcula infusoria; fluvia tilia et marina, que detexit, systematice descripsit et ad vivum delineari curavit. Molleri, Havniae; 1786.
- 95.Purkinje JE, Valentin CG. Entdeckung continuerlicher durch Wimperhaare erzeugter Flimmerbewegungen. Arch Anat Physiol Wiss Med. 1834;1:391–400. [Google Scholar]
- 96.DuJardin F. Histoire Naturelle des Zoophytes (Infusoires). Paris: Roret; 1841.
- 97.Ecker A. Flimmerbewegung im Gehörorgan von Petromyzon marinus. Arch Anat Physiol Wiss Med Müller’s Arch. 1844;1:520–521. [Google Scholar]
- 98.Kolliker A. Manual of human microscopical anatomy (translation by George Busk of Mikroskopische Anatomie) Philadelphia: Lippincott, Grambo and Company; 1854. [Google Scholar]
- 99.Flemming W. Studien uber die Entwicklungsgeschichte der Najaden. Akad Wiss Wien. 1875;71:81–147. [Google Scholar]
- 100.Van Beneden E. Contribution a l’histoire de la vesiculaire germinative et du premier noyau embryonnaire. Bull Acad R Belg. 1876;2(42):35–97. [Google Scholar]
- 101.Henneguy LF. Sur les rapports des cils vibratiles avec les centrosomes. Arch Anat Microsc. 1898;1:481–496. [Google Scholar]
- 102.von Lenhossek M. Untersuchungen uber Spermatogenese. Arch Mikrosk Anat. 1898;51:215–318. doi: 10.1007/BF02977498. [DOI] [Google Scholar]
- 103.von Lenhossek M. Ueber Flimmerzellen. Verh Anat Ges. 1898;12:106–128. [Google Scholar]
- 104.Alverdes K. Zentralgeisselapparat der Epithelzellen im Rete testis des Menschen. Z Mikro Anat Frschg. 1927;11:172–180. [Google Scholar]
- 105.Sjostrand FS. The ultrastructure of the inner segments of the retinal rods of the guinea pig eye as revealed by electron microscopy. J Cell Comp Physiol. 1953;42:45–70. doi: 10.1002/jcp.1030420104. [DOI] [PubMed] [Google Scholar]
- 106.deHarven E, Bernhard W. Etude au microscope de l’ultrastructure du centriole chez les vertebres. Z Zellforsch Mikrosk Anat. 1956;45:378–398. doi: 10.1007/BF01106086. [DOI] [PubMed] [Google Scholar]
- 107.Bernhard W, deHarven E. L’ultrastructure du centriole et d’autres elements de l’appareil achromatique. In: Bargmann W, Peters D, Wolpers C, editors. Proceedings of the 4th international congress electron microscopy. Berlin: Springer; 1960. pp. 217–227. [Google Scholar]
- 108.Barnes BG. Ciliated secretory cells in the pars distalis of the mouse hypophysis. J Ultrastruct Res. 1961;5:453–467. doi: 10.1016/S0022-5320(61)80019-1. [DOI] [PubMed] [Google Scholar]
- 109.Wilson RB, McWhorter CA. Isolated flagella in human skin. Election microscopic observations. Lab Invest. 1963;12:242–249. [PubMed] [Google Scholar]
- 110.Archer FL, Wheatley DN. Cilia in cell-cultured fibroblasts II. Incidence in mitotic and post-mitotic BHK 21-C13 fibroblasts. J Anat. 1971;109(Pt 2):277–292. [PMC free article] [PubMed] [Google Scholar]
- 111.Afzelius BA. A human syndrome caused by immotile cilia. Science. 1976;193(4250):317–319. doi: 10.1126/science.1084576. [DOI] [PubMed] [Google Scholar]
- 112.Lehman JM, Laag E, Michaud EJ, Yoder BK. An essential role for dermal primary cilia in hair follicle morphogenesis. J Investig Dermatol. 2009;129(2):438–448. doi: 10.1038/jid.2008.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ezratty E, Stokes N, Chai S, Shah A, Williams S, Fuchs E. A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell. 2011;145(7):1129–1141. doi: 10.1016/j.cell.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.