Abstract
Objectives
Due to the expanded bacterial genetic tolerance to antibiotics through different mechanisms, infectious diseases of MDR bacteria are difficult for treatment. Consequently, we synthesized drug conjugated nanoparticles to dissolve this problem. Moreover, the present study aims to display the cell death status treated with cefotaxime-CS-AgNPs and also, apoptosis pathways of human RPE-1 normal cells and human MCF-7 breast cancer cells.
Methods
Here, we demonstrate the possibility to synthesize AgNPs and conjugate them with cefotaxime to survey the probability of cefotaxime-CS-AgNPs as an antimicrobial agent against cefotaxime-resistant strains E. coli and MRSA.
Results
TEM showed the size of AgNPs, CS-AgNPs and cefotaxime-CS-AgNPs ranged from 7.42 to 18.3 nm, 8.05–23.89 nm and 8.48–25.3 nm, respectively, with a spherical shape. The cefotaxime-CS-AgNPs enhanced the high antimicrobial properties compared to AgNPs or pure antibiotic. The MIC of Cefotaxime-CS-AgNPs ranged from 3 µg/mL to 8 µg/mL against tested E. coli and MRSA bacteria. Consequently, the highest reduction in the MIC of cefotaxime-CS-AgNPs was noted against tested strains ranging from 22% to 96%. Comparing cefotaime-CS-AgNPs to AgNPs we showed that cefotaime-CS-AgNPs have no cytotoxic effect on normal cells at even 12 µg/mL for 24 hrs. The IC50 for the AgNPs and cefotaxime-CS-AgNPs was 12 µg/mL for human RPE-1 normal cells and human MCF-7 breast cancer cell lines. The pro-apoptotic genes p53, p21, and Bax of cancer cell lines significantly upregulated followed by downregulated by anti-apoptotic gene Bcl-2 after 48 hrs at 24 µg/mL, and this concentration represents the most effective dose.
Conclusion
Results enhanced the conjugating utility in old unresponsive cefotaxime to AgNPs to restore its efficiency against previous strains and demonstrated potential therapeutic applications of cefotaxime-CS-AgNPs. Moreover, this research gives remarkable insights for designing nanoscale delivery and curative systems that have a pronounced cytotoxic activity on cancer cells and are safe to normal cells.
Keywords: AgNPs and cefotaxime-Cs-AgNPs, MRSA, E. coli, anticancer, cytotoxicity, genotoxicity
Introduction
Cefotaxime is one antibiotic of Oxyimino-cephalosporins. Consequently, they were the “most important antimicrobial” for human medicine.1 Moreover, there are varieties of resistance mechanisms depending on the specific microorganism involved against these antimicrobials. Unique of these mechanisms is enzymatic medicine degradation via extended-spectrum β-lactamases (ESBLs) production in Gram-negative pathogens especially pathogenic Escherichia coli.2 Moreover, the appearance of pathogenic resistant Staphylococcus aureus (S. aureus) isolates, especially methicillin-resistant (MRSA), is a global challenge to their infections.3,4 Ordinarily, strains of MRSA cause different infectious diseases in all age groups and consider one of the important human pathogens. Therefore, infectious diseases of these resistant bacteria extended hospital stay, increased the cost of health care services, and lead to a significant increase in morbidity and mortality rates.5
Nowadays, evolution of Microbial resistance mechanisms associated with medical procedures has become a serious problem with high concern to public health.6 Therefore, the researchers prepared new nano-antimicrobial drugs with high efficiency and improved delivery potential.7
Silver has been used in some medicinal applications for centuries. Regulators approved silver as an antibacterial agent in the early 1990s.8 Accordingly, Hippocrates obtained that silver had helpful and antimicrobial benefits.9 Ordinarily, to extend the milk freshness, silver coins were placed in containers of milk.10 Consequently, cost less and new antimicrobial drugs were improved by numerous scientists due to the focus on MDR microbe’s proliferation and lessening the health care expense. The advanced era finds new ways of antimicrobial agents and nanotechnology offers as one possible solution. Therefore, the revival of silver-based antibiotics led to resolve such problems.11,12 Accordingly, biological methods of AgNPs synthesis got more consideration since there is no use of any toxic chemicals in the synthesis process and they cost less, eco-friendly.13 The AgNPs biosynthesis is a simple process and extensive amounts. Nanoparticles (NPs) can be set up in a short time and the plant extract is considered as an energizing branch and used as a reducing and stabilizing agent in the biosynthesis processes.
Subsequently, antibiotic conjugated nanoparticle could be compelling bactericidal materials. By various researchers, the improved action of AgNPs and anti-microbial agents together has been reported earlier.14–18 For example, the conjugation of AgNPs with Doxycycline brought about the higher bactericidal effect on E. coli than when the components are administered separately.19
Toxicity studies are required for metal nanoparticles expected application on biomedical application. Consequently, metal nanoparticles as nanomaterials are including those in diagnostics, medical imaging, and tumor therapeutics. Moreover, metal nanoparticles have been applied as drug delivery, and cancer therapy in the early detection, and diagnosis.20
Thus, the present study is an attempt to synthesize Cefotaxime-CS-AgNPs for numerous applications such as a novel nano-antimicrobial agent for treating various bacterial diseases. Additionally, we study the cytotoxicity effect of these nanoparticles on Both MCF-7 and RPE-1 cells. We displayed the conjugation of AgNPs with cefotaxime, as a β-lactam antibiotic yields a hybrid agent for the inhibition of MRSA strains and MDR strains E. coli, especially to cefotaxime. We synthesized AgNPs, CS-AgNPs and Cefotaxime-CS-AgNPs and performed characterization by XRD, FTIR, UV–Vis spectroscopy and TEM followed by pertinent antibacterial assays. Interestingly, when compared to their unconjugated free forms (cefotaxime or AgNPs) against tested resistant strains, Cefotaxime-CS-AgNPs were found to have very low minimum inhibitory concentration (MIC) which adds novelty and significance of the study.
Material and Methodology
Bacteria and Growth Conditions
The MRSA strains and MDR E. coli obtained from our previous study. The MDR E. coli were ESBL positive strains and displayed complete resistance to antibiotic cefotaxime. Also, The MRSA strains were methicillin-resistant.21 At the beginning, a fresh inoculum of each bacterial strain (MRSA strains and MDR E. coli) was grown in nutrient broth for 18 h at 37°C. To arrive at the common standard of the turbidity (i.e., 1.5 × 108 CFU/mL), bacterial cells were adjusted with the nutrient broth.
Synthesis of AgNPs Using Rosa damascenes Extract
For biosynthesis of AgNPs, Rosa damascenes extract (5 mL, 25%) was mixed with 45 mL of AgNO3 aqueous solution (1 mm) under constant stirring with incubation time for 2 hrs. Moreover, the conditions were displayed at different temperatures (35–80°C) and different pH (6–9). Synthesized AgNPs using Rosa damascenes extract may be easily displayed using UV–Vis spectroscopy and the change of AgNPs color was monitored. After complete incubation of mixture, the synthesized AgNPs were washed with distilled water and centrifuged for 15 min at 15,000 xg. Finally, the AgNPs sample was washed with 50% ethanol.22
Preparing Chitosan-AgNPs
For preparation of Chitosan solution, the 0.5 g Chitosan powder (low molecular weight) Chitosan was dissolved in a 1% acetic acid solution. Moreover, the mixture of AgNPs with chitosan was stirred and mixed overnight, the chitosan-AgNPs (CS-AgNPs) were recovered using centrifuge.23
Conjugation of Cefotaxime with CS-AgNPs
The Cefotaxime-CS-AgNPs were prepared by mixing cefotaxime (1mg/mL) with CS-AgNPs (1mg/mL) in solution of phosphate buffer, followed by incubation time for 24 h, and in final step, Cefotaxime-CS-AgNPs centrifuged at 15,000 xg for 10 min.24 Moreover, ten kDa membrane of cellulose was used for dialysis of Synthesized Cefotaxime-CS-AgNPs against deionized water over-night. Once dialyzed to remove any unreacted substances, the Cefotaxime-CS-AgNPs were freeze-dried. The sample was stored to further characterization.
In vitro Drug Release Kinetics
Cefotaxime was performed by using the membrane dialysis technique. For dialysis, three milliliters of Cefotaxime-CS-AgNPs (2 mg/mL, in PBS, pH = 7.4) and were put in soaked dialysis bag (MW =12 KDa). In 30 mL of in phosphate buffer saline (pH = 7.4), the membrane bag was preserved in a shaking water bath operating at 100 rpm over-night at 37°C. Moreover, antioxidant ascorbic acid (0.2% w/v) was added to the Cefotaxime-CS-AgNPs solution to prevent cefotaxime degradation.25 At determined times, 1 mL of sample was withdrawn and replaced with a fresh PBS medium. Consequently, absorption of the sample was analyzed by UV–vis spectrophotometer at 298 nm, and constructed calibration curves were plotted.
Determination of Conjugating Efficiency of Cefotaxime-CS-AgNPs
Cefotaxime-CS-AgNPs conjugating efficiency was calculated as Shaker and Shaaban.26 The double beam spectrophotometer was used for quantification of free drug in the supernatant after centrifugation of the Cefotaxime-CS-AgNPs at 15,000×g for 10 min. Moreover, the peak of cefotaxime was obtained at ʎ max 298.27 Also, calibration curve of cefotaxime was plotted. To calculate the conjugating efficiency percentage of Cefotaxime-CS-AgNPs, we need to find out the cefotaxime amount which attached to the CS-AgNPs according to the following equation:
Conjugating efficiency of Cefotaxime-CS-AgNPs (%) = (Total amount of cefotaxime–Free cefotaxime in the supernatant/Total amount of cefotaxime) × 100.
Characterization of the AgNPs and Conjugates Cefotaxime-CS-AgNPs
UV-Visible Spectroscopy Analysis
The AgNPs aliquots were observed on double beam spectrophotometry (Shimadzu UV-1650) in 300–800 nm operated at a resolution of 1 nm.28 Moreover, UV–Vis spectroscopy was used for monitoring CS-AgNPs and Cefotaxime-CS-AgNPs.
Transmission Electron Microscopy (TEM)
To determine the shape or size of Cefotaxime-CS-AgNP, CS-AgNP as well as AgNPs, Transmission Electron Microscopy (TEM) technique was used. Furthermore, imaging of E. coli strain treated with Cefotaxime-CS-AgNPs was observed and the TEM (model JEOL 100CX II) was used to display the shape of Cefotaxime-CS-AgNP, CS-AgNP and AgNP. The sample was dropped on carbon-coated copper grids. Then the film on the grid was dried and examined by TEM.29
X-Ray Diffraction (XRD) Analysis
The nature and size of the Cefotaxime-CS-AgNPs, CS-AgNPs or AgNPs have determined the usage of XRD. This became executed the use of Shimadzu XRD (Shimadzu XD-3A, Japan) and the analyzed AgNPs or Cefotaxime-CS-AgNPs turned into fine powder and the compositions were determined. Moreover, the size of Cefotaxime-CS-AgNPs, CS-AgNPs or AgNPs has calculated using Debye Scherer’s equation.
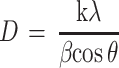 |
where β is the width of the maximum peak at half of the height at the diffraction angle Ө, D is the average crystalline size of the Cefotaxime-CS-AgNPs, CS-AgNPs or AgNPs, k is a geometric factor (0.9) and λ is the X-ray radiation source wavelength.30
Fourier Transform Infrared Spectroscopy (FTIR)
The powder of plant extract, AgNPs, CS-AgNPs or Cefotaxime-CS-AgNPs were collected using centrifuge at 15,000×g for 20 min. Consequently, the residue was then washed with de-ionized water to eliminate unattached biological components. Also, the samples were performed on a Shimadzu IR-470 Spectrometer (Shimadzu, Japan). The AgNPs, CS-AgNPs or Cefotaxime-CS-AgNPs peaks of FTIR were evaluated and expressed in wave numbers (cm−1). Cefotaxime was also analyzed.31
Antibacterial Activity of Cefotaxime-CS-AgNPs
Using the well agar diffusion method, the Cefotaime-CS-AgNP and synthesized AgNPs solutions were tested for their antibacterial efficacy against studied MRSA strains and MDR E.coli.27 Cefotaxime was used as a control. The inoculum of each MRSA strain or MDR E.coli cultures were initially inoculated in Müller-Hinton broth medium followed by incubation at 37°C for 18 hrs. Thus, 100 μL of each E. coli or MRSA culture was distributed onto the surface of Müller-Hinton agar medium. Thirty microliters of cefotaxime, synthesized AgNPs, and Cefotaxime-CS-AgNPs with different concentrations were pipetted into the 6-mm wells and the plates were incubated at 37°C for 24 hrs. Finally, the inhibition zone of previous nano-materials or cefotaxime were detected by recording the diameter of clear area around each well.
Determination of Cefotaxime-CS-AgNPs Minimum Inhibitory Concentration (MIC)
On a 96 well plate, the MIC evaluation of Cefotaxime-CS-AgNPs, synthesized AgNPs and cefotaxime was performed. Into the 6 wells in column 1, two hundred μL of Cefotaxime-CS-AgNPs, synthesized AgNPs or cefotaxime were piped. One hundred μL of suitable broth growth medium for each strain was piped into the wells in each row. Thereafter, 100 μL of the cefotaxime or previous nano-material solutions was taken from column 1 and was serially diluted along the row until column number 10. Thereafter, 5 μL of MRSA strains or MDR E.coli cultures were piped into each well containing the suitable broth medium except column number 10, which was considered as blank. Moreover, 50 μL of phenol red dye (2 mg/mL) solution has pipetted in each well to estimate the viability of bacteria. The plates of MRSA strains or MDR E.coli were incubated and red color of previous bacterial cell viability was detected after an incubation time of 24 h at 37°C. The minimum concentration of Cefotaxime-CS-AgNPs, AgNPs, or cefotaxime, has been considered as the MIC value at which no growth or no color was observed. The incubation of previous bacterial culture plates was obtained at 37°C for 24 h.32
Cell Lines and Culture
Cell Culture and Anticancer Drug Discovery Lab at National Research Center in Egypt nobily providing the Human RPE-1 normal cells and human breast cancer cells, MCF-7 that obtained from the center of the American Type Culture Collection (ATCC). Both MCF-7 and RPE-1 cells were grown, proliferate and preserved in Eagle’s minimal essential medium (EMEM); this media supplemented with fetal bovine serum (Gibco by life technologies, USA); in a concentration of 10% and antibiotics of penicillin-streptomycin at a concentration of 1% (Gibco by life technologies, USA), at 37.0°C, in air 95%/carbon dioxide (CO2) 5% and 100% comparable humidity.33 Then, after 24 hrs in the above-supplemented medium, cell lines were treated on day 0 with different treatments and then further incubated for another 24 and 48 hrs.
In vitro Cell Viability/Cytotoxicity Studies
Trypan Blue staining method and MTT assay were performed to investigate the in vitro cytotoxicity induced in the normal and cancer cells after treatment with different concentration from each targeted nanoparticles. The safety doses of these nanoparticles treatments on normal cell line (RPE-1) were also investigated.
Trypan Blue Dye Exclusion Assay
In the Trypan blue staining method; the healthy and viable cells have a clear cytoplasm, whereas the cells colored in blue are the dead one. In brief, 60 µL of 0.05% freshly prepared Trypan Blue solution was added to 60 µL of cell suspension within 5 mins, spread onto a microscopic slide and covered with a cover slip. After 24 hrs, cytotoxicity and cell viability were detected by count the viable cells (Trypan Blue excluding) in a hemocytometer.34
MTT Assay
In the present study the MTT assay was executed according to a modified method by Mosmann.35 Selected concentrations of AgNPs or Cefotaxime-CS-AgNPs (0, 3.0, 6.0, 12.0, and 24.0 µg/mL) were added to RPE-1 or MCF-7 cells after incubating for 24 hrs at 37°C. MTT (3 [4,5dimethylthiazol2yl] 2,5diphenyltetrazolium bromide) (5 mg/mL) at a volume of 10 µL added to all wells which contained RPE-1 or MCF-7 cells and further incubated for 24 h in the same previous conditions. The negative control represented by Eagle’s minimal essential medium without any treatments. The selected concentrations of both NPs were examined in triplicates.36 The percentage of the cell viability of normal and breast cancer cells, calculated using the equation:
 |
The IC50, the concentration inhibits 50% of cell growth, for RPE-1 or MCF-7 cells calculated from plotted dose–response curve.
Gene Expression Analysis for Apoptotic Related Genes
The extraction of total RNA from both cell lines performed by using TRIzole reagent (CA, USA). Instantly, about 2 μg of the isolated RNA from different groups were used in RT-PCR to synthesize the cDNA by Maxime RT, reverse transcription kit (Maxime RT PreMix – intron Biotechnology, Korea). The amplification reaction was proceeded by PXE 0.5 Thermo thermal cycler. The PCR products run on an agarose gel at a concentration of 1.5% in TBE buffer then visualized on a UV Transilluminator. The bands were scanned to quantify the signal intensities using Gel-Pro software (version 3.1 for Windows 3). The ratio between the levels of Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene and the target gene’s product after amplification was calculated.37,38 Gene expression was analyzed using the following primers pairs,39 listed in Table 1.
Table 1.
Summary of the Targeted Primers Used in the Study
| Genes | Forward Primer | Reverse Primer |
| P-53 | 5ʹ-TAACAGTTCCTGCATGGGCGGC-3’ | 5ʹAGGACAGGCACAAACACG CACC3’ |
| P21 | 5ʹ-ATG TGT GTG GAG AGC GTC AAC C-3’ | 5ʹ-GCA TCC CAG CCT CCG TTA TC-3’ |
| Bax | 5ʹ-CCT TTT CTA CTT TGC CAG CAA AC-3’ | 5ʹ-GAG GCC GTC CCA ACC AC-3’ |
| BCl2 | 5ʹ-AGGTGACACTATAGAATA-3’ | 5ʹ-GGGATATCACTCAGCATG-3’ |
| GAPDH | 5ʹ-GTGCCGGTTCAGGTACTCAGTCA-3’ | 5ʹ-TTGTGGCCTTCTTTGAGTTCGGTG-3’ |
Notes: Adefolaju GA, Theron KE, Hosie MJ. In vitro effects of protease inhibitors on BAX, BCL-2 and apoptosis in two human breast cell lines. S Afr J Sci. 2015; 111(11/12), Art. #2014-0417, 9 pages. http://dx.doi.org/10.17159/sajs.2015/20140417.
Statistical Analysis
The SPSS.11 program was used for statistical analysis. By One Way – ANOVA, the significance of the differences among treatment groups was determined then followed by Duncan’s multiple tests.40,41 The values of analysis expressed as Mean ± SE. All notifications of significance based on probability of P <0.05.
Results and Discussion
Biosynthesis of AgNPs and Conjugation of Cefotaxime with AgNPs
For AgNPs synthesis, with constant stirring, 5 mL of the prepared aqueous Rosa damascenes petals aqueous extract was mixed with 45 mL of 1 mM AgNO3 solution.42 Moreover, silver ions reduction into AgNPs during exposure to Rosa damascenes extracts was observed as change of color to a brown at optimum pH 8 and optimum temperature 80°C. Accordingly, these data are agreed with Gopinath et al.43 Also, cefotaxime was conjugated to CS-AgNPs to form Cefotaxime-CS-AgNPs.
Characterization of the AgNPs and Conjugates Cefotaxime-CS-AgNPs
UV-Visible Spectroscopy Analysis
The cefotaxime, AgNPs, CS-AgNPs and Cefotaxime-CS-AgNPs formations were dispersed and the absorbance of them was measured by using UV–visible spectrophotometer from 300 to 800 nm. The UV–Vis spectra of Cefotaxime-CS-AgNPs (Figure 1I) showed peaks at 298 nm and 414 nm. Accordingly, the peak at 445 nm for pure AgNPs and CS-AgNPs and peaks at 298 nm for pure cefotaxime. Furthermore, cefotaxime has a carboxylic acid group which can form amide bond with the amino group of chitosan to form Cefotaxime-Cs-AgNPs.44
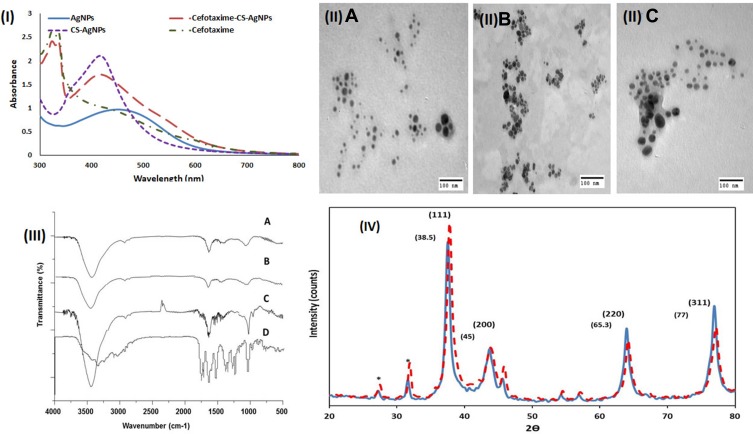
Figure 1.
(I) UV–visible spectrum of AgNPs (ـــــــــ), CS-AgNPs (—–), cefotaxime (ـــــ ـ ـــــ) and Cefotaxime-CS-AgNPs (ــــــ ــــــ); (II) TEM image AgNPs (A), cefotaxime-CS-AgNPs (B) and CS-AgNPs (C); (III) FT-IR analysis of AgNPs using Rosa damascenes petal extract (A); CS-AgNPs (B); Cefotaxime-CS-AgNPs (C) and cefotaxime (D); (IV) X-ray diffraction of AgNPs (ــــــــــ) Cefotaxime-CS-AgNPs (ــ ــ ــ).
Transmission Electron Microscopy (TEM)
The Cefotaxime-CS-AgNPs, CS-AgNPs, in addition to the AgNPs, were characterized using TEM evaluation (Figure 1 IIA–C). The different size and form of AgNPs, CS-AgNPs and Cefotaxime-CS-AgNPs were synthesized at the optimum conditions. Furthermore, the AgNPs, CS-AgNPs and Cefotaxime-CS-AgNPs were spherical shape as shown in Figure 1 IIA&IIB&IIC. The size of nanoparticles were in the range 7.42–18.3 nm, 8.05–23.89 and 8.48–25.3 nm for AgNPs, CS-AgNPs and Cefotaxime-CS-AgNPs, respectively. Accordingly, these data were correlated to the results obtained by Agnihotri et al.45
X-Ray Diffraction (XRD)
The XRD pattern of the Cefotaxime-CS-AgNPs, as well as synthesized AgNPs showed four intense peaks 37.51º, 44.12º, 64.00º and 76.91º (Figure 1IV) corresponds to (111), (200), (220) and (311), the Bragg’s reflection planes of face-centered cubic crystal structure silver, respectively.46
Fourier Transforms Infrared Spectroscopy (FT-IR)
FT-IR spectroscopy analysis was used to determine the functional groups of extract, cefotaxime, AgNPs, CS-AgNPs and Cefotaxime-CS-AgNPs and its interaction between each other are shown in (Figure 1III). FTIR spectra of AgNPs synthesized using plant extracts revealed major peaks at 3431.64, 1635.55, 1456.85, 1078.30, as well as at 594.95 cm−1 that corresponded to stretching peak O-H, N-H stretch, C-N stretch bond, C-O stretch and -C = C-H, respectively (Figure 1III-A).
FTIR spectra of CS-AgNPs appeared major absorption peaks at 3439.98, 1635.65, 1456.92, 1052.33 and at 575.95 cm−1 that revealed to stretching peak O-H, stretch N-H, C-N stretch bond, C–O stretch and -C = C-H, respectively (Figure 1III-B). Moreover, in case of AgNPs synthesized using Rosa damascenes extracts, broad, strong and characteristic peak for of this nanomaterial was observed at 3439.98 cm−1 representing 0-H stretching or H-bonding for alcohol or phenols indicating that OH bonding is enhanced during NPs formation. Furthermore, the band at 1635.65 cm−1 and 1052.33 cm−1 in the spectrum corresponds to amine group peak for chitosan. These peaks were a result of NH3+ group and glycosidic bond stretching vibrations, respectively.47–49 Therefore, the appearance of chitosan peaks displays that the AgNPs were successfully coated with a polymer of chitosan. The -C = C-H observed at 575.95 cm−1. Thus, the last functional group indicated that groups involved in the reduction of AgNPs.
FTIR spectroscopy of cefotaxime functional groups was also confirmed (Figure 1III-D). The absorption peaks that appeared in the FTIR analysis of cefotaxime were 3441.46 cm−1 and 1649.63 cm−1 which could be corresponded to the N-H groups and amide groups stretching vibrations, respectively. Moreover, the smaller peaks at 1382.45, 1047.11 and 980.048 cm−1 could be revealed to the C–N, C–O, and =C–H stretching vibrations, respectively. Also, the C–O and C–C groups’ vibration modes appeared at the regions from 600 cm−1 to 500 cm−1.50
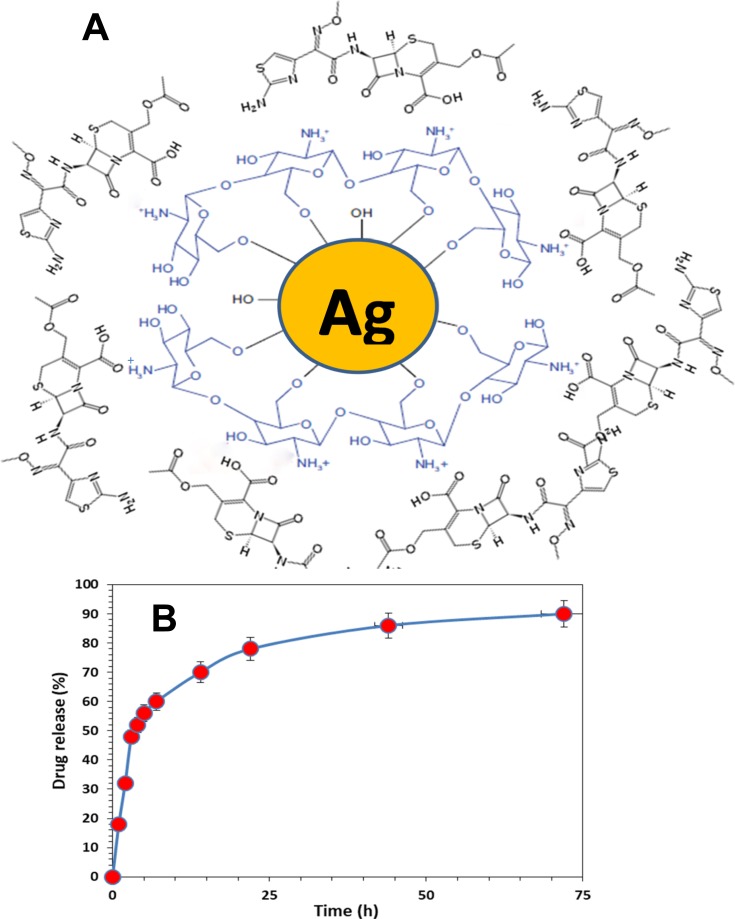
Upon mixing cefotaxime with CS-AgNPs, the peaks for amide C=O of cefotaxime shifted from 1649.63 cm−1 to 1662.79 cm−1. This result suggests that the conjugation of cefotaxime with CS-AgNPs could take place through the amide C=O (Figure 1III-C). Also, Sharp absorption peak at 2330 cm−1 appeared, which shows that the NH+ groups are crosslinked with chitosan molecules.51 Figure 2A shows the interaction between the cefotaxime, chitosan, and AgNPs in the Cefotaxime-CS-AgNP. The carboxylic acid C=O group of cefotaxime can form amide bond with NH2 group of chitosan to form cefotaxime-CS-AgNPs.44
Figure 2.
(A) Schematic representation of the interaction between cefotaxime, chitosan, and silver nanoparticles in the Cefotaxime-CS-AgNPs. (B) Drug release percent of cefotaxime, from the Cefotaxime-CS-AgNPs in PBS (pH = 7.4) as a release medium at 37°C.
In vitro Drug Release
Figure 2B shows the cefotaxime release from Cefotaxime-CS-AgNPs. The drug release results showed that the Cefotaxime-CS-AgNPs exhibited release profile described as an initial fast release rate, which continued to almost first 7 h followed by a steady release phase with a slow release rate that extended up to 72 h or the complete release of all of the loaded drug. Consequently, the release of cefotaxime from Cefotaxime-CS-AgNPs reached 78% within the first 24 h and reached almost 90% after 72 h. This behavior can be attributed to the particle size effect. Higher amount of drug associated with the same volume of small size nanoparticles thus higher probability of drug molecules repulsion, leading to faster diffusion through the dialysis membrane. Consequently, silver nanoparticles’ size provides a mean of controlling cefotaxime release rates.26
Determination of Conjugating Efficiency of Cefotaxime-CS-AgNPs
Conjugating efficiency is estimating an important parameter for the characterization of Cefotaxime-CS-AgNPs. The efficiency of Cefotaxime-CS-AgNPs became determined to be 76.95%. Accordingly, conjugating efficacy should be high so that cefotaxime is not lost during preparation of cefotaxime conjugated CS-AgNPs and the less amount is needed for its therapeutic application. Consequently, these data are correlated to the results obtained by Shaker and Shaaban.26
Antimicrobial Activity
At the beginning, the MDR E.coli and MRSA strains were resisted at least to 9 antibiotics as shown in Table 2. The multidrug-resistant E. coli bacteria are resisting to cefotaxime and MRSA bacteria are resisting methicillin. Moreover, these bacteria were identified using 16S rRNA analysis in the previous study with accession number as shown in Table 3.
Table 2.
Resistance Patterns of Bacterial Isolates
| Bacterial Isolates | Antibiotic Resistance Pattern | No. of Antibiotics |
| *STA7 | AMP, AMC, CEC, FOX, CXM, MEM, OXA, PEN, ERY | 9 |
| *STA5 | AMP, AMC, CEC, FOX, CXM, MEM, OXA, PEN, LEX, CRO,CFR | 11 |
| *STA8 | AK, AMC AZM, CEC, FOX, CXM, CIP, LVX, OXA, PEN, CLR, CLI, ERY | 13 |
| aESC7 | AMP, AMC, CEC, FEP, CLR, CXM, CAZ, CRO, CTX, CIP, LVX, DO, SXT, LEX, FUX | 15 |
| aESC3 | AMP, AMC, CEC, FOX, FEP, CXM, CAZ, CRO, CTX, CIP, LVX, DO, SXT, LEX, CLR, AZM, CLI, FUX, CFR | 19 |
| aESC5 | AMP, AMC, CEC, FOX, FEP, CXM, CAZ, CRO, CTX, CIP, LVX, DO, SXT, LEX, CLR, AZM, CLI, CFR, AK | 19 |
Notes: *MRSA: Methicillin-resistant Staphylococcus aureus. aMDR resistant E. coli especially to cefotaxime.
Abbreviations: AK, Amikacin; AMC, Amoxy/Clavulanic acid; CEC, Cefaclor; FEP, Cefepime; CTX, Cefotaxime; FOX, Cefoxitin; CAZ, Ceftazidime; CRO, Ceftriaxone; CXM, Cefuroxime; DO, Doxycycline/HCL; CIP, Ciprofloxacin; LVX, Levofloxacin; CIR, Clarithromycin; CLI, Clindamycin; AMP, Ampicillin; LEX, Cefalexin; AZM, Azithromycin; CFR, Cefadroxil; FEP, Cefepim; MEM, Meropenem; SXT, Sulpha/Trimethoprim; FUX, Fluxocillin.
Table 3.
Antibacterial Activity of Cefotaxime and Cefotaxime-CS-AgNPs Against Multidrug-Resistant Bacteria
| Bacterial Strains | Accession Number | AgNPs Inhibitory Zones (mm) | Cefotaxime Inhibitory Zones (mm) | Conjugates Inhibitory Zones (mm) |
|---|---|---|---|---|
| * STA5 | LC189111.1 | 11 | 8 | 36 |
| * STA7 | LC189113.1 | 12 | 8 | 36 |
| * STA8 | LC189114.1 | 13 | 9 | 37 |
| aESC3 | LC189094.1 | 10 | 0 | 23 |
| aESC5 | LC189096.1 | 12 | 0 | 25 |
| aESC7 | LC189098.1 | 11 | 0 | 24 |
Notes: *Methicillin-resistant Staphylococcus aureus (MRSA).aMDR resistant E. coli especially to cefotaxime.
Furthermore, Cefotaxime-CS-AgNPs appeared high antimicrobial activity against tested strains compare to free cefotaxime. The inhibitory diameter of Cefotaxime-CS-AgNPs ranged from 23 mm to 37 mm against MDR E. coli and MRSA strains (Table 3). Moreover, the MIC of Cefotaxime-CS-AgNPs ranged from 3 µg/mL to 8 µg/mL against tested bacteria (Table 4). MIC for Cefotaxime-CS-AgNPs was reduced significantly compared to free cefotaxime. Consequently, the Cefotaxime-CS-AgNPs MIC value decreased in range 22–96% against E. coli and MRSA strains. Moreover, the Cefotaxime-CS-AgNPs antibacterial efficiency was higher than synthesized AgNPs alone and free cefotaxime against all E. coli and MRSA strains. Conversely, the CS-AgNPs were screened, using the bacteria, and did not display an inhibitory zone, indicating that the antibacterial activity was a result of the slow release of loaded cefotaxime from the Cefotaxime-CS-AgNPs.
Table 4.
Percentage Change in MIC Value Cefotaxime-CS-AgNPs
| Bacterial Strains | AgNPs (µg/mL) | Cefotaxime (µg/mL) | AgNPs (µg/mL)+Cefotaxime (µg/mL) | Conjugates (µg/mL) | % Change in MIC |
|---|---|---|---|---|---|
| * STA5 | 55 | 30 | 40 | 4 | −26 |
| * STA7 | 49 | 30 | 36 | 3 | −27 |
| * STA8 | 62 | 30 | 42 | 8 | −22 |
| aESC3 | 57 | 100 | 79 | 4 | −96 |
| aESC5 | 65 | 100 | 81 | 8 | −92 |
| aESC7 | 58 | 100 | 78 | 4 | −96 |
Notes: Percentage change in MIC was calculated by using formula: b − a/a × 100, where a is cefotaxime alone and b is nanoparticles conjugates (Cefotaxime-CS-AgNPs). *Methicillin-resistant Staphylococcus aureus (MRSA). aMDR resistant E. coli especially to cefotaxime.
Mechanism of Action
Figure 3 shows TEM of E. coli treated with 4 μg/mL Cefotaxime-CS-AgNPs. Conjugated nanoparticles accumulated both in the cell walls of bacteria and inside the cells. Thus, the antimicrobial efficiencies of Cefotaxime-CS-AgNPs depend on an important matter that chitosan has antimicrobial efficacy due to various factors, such as the physicochemical characteristics of chitosan, polymerization, molecular weight, and it is inversely affected by pH. Further study into the molecular mechanisms for both the cefotaxime and the CS-AgNPs to produce Cefotaxime-CS-AgNPs may reveal a high antimicrobial effect due to the increased cefotaxime molecules presence per unit volume of the system so cefotaxime retained untouched to β-lactamases. Also, the Cefotaxime-CS-AgNPs stability in the presence of β-lactamases (penicillinases) will be important for applying this nanoantibiotic to treat β-lactamase-mediated resistant bacteria in serious infections.52 Ordinarily, cefotaxime inhibited mucopeptide formation in the bacterial cell wall53 so, the cell membrane disruption due to Cefotaxime-CS-AgNPs. When the previous drug enters bacteria, they interrupt all membrane regulatory functions such as protein synthesis and interact with phosphorus in RNA or DNA. The high effectiveness of Cefotaxime-CS-AgNPs due to bounding action between CS-AgNPs and cefotaxime as they contain many active groups like amide C=O, which react with the amine group of CS-AgNPs.44,54
Figure 3.
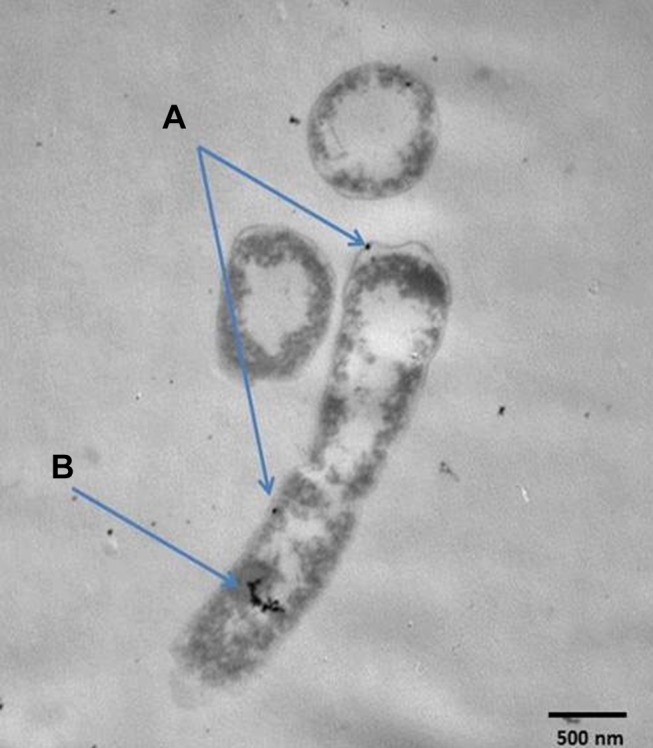
TEM analysis of E. coli after exposure to Cefotaxime-CS-AgNPs, and blue arrows explain its accumulation in cell wall (A) and inside the cell (B).
In vitro Cytotoxicity Assessment and Determination of IC50
The effect of synthesized AgNPs and Cefotaxime-CS-AgNPs on RPE-1 or MCF-7 cell survival was conducted in the concentration range of 0–24 μg/mL to determine the cytotoxic effects and doses corresponding to 50% of proliferated cells were reduced when treated with these NPs. RPE-1 or MCF-7 cells were cultured and treated at selected doses for either 24 or 48 h then cell assessed for viability, count and morphology by Trypan blue staining and MTT assay. The results displayed in Table 5.
Table 5.
The Effect of AgNPs and Cefotaxime-CS-AgNPs Treatment on the Proliferative Percentage of MCF-7 and RPE-1 Cells at Different Concentrations and Times Intervals
| Treatments Groups μg/mL | Proliferative Percentage of Treated Cells Lines | |||
|---|---|---|---|---|
| RPE-1 Cells | MCF-7 Cells | |||
| After 24 hrs | After 48 hrs | After 24 hrs | After 48 hrs | |
| Untreated Cells | 98.2 ± 0.98 a | 94.8 ± 0.72a | 95.30 ± 0.75a | 97.54 ± 1.51a |
| AgNPs 3.0 | 78.50 ± 1.44 d | 72.8 ± 0.10c | 76.10 ± 0.95cd | 71.80 ± 0.66 c |
| AgNPs 6.0 | 70.80 ± 1.54 b | 62.8 ± 0.10 d | 65.60 ± 0.72e | 60.10 ± 1.67 d |
| AgNPs 12.0 | 64.60 ± 0.57 e | 50.40 ± 0.65 ef | 59.50 ± 0.37e | 53.10 ± 0.68 d |
| AgNPs 24.0 | 50.90 ± 1.09 c | 44.90 ± 0.79 f | 47.60 ± 0.54f | 39.30 ± 0.90 e |
| Cefotaxime-CS-AgNPs 3.0 | 97.60 ± 0.67 a | 93.44 ± 0.54 a | 94.60 ± 0.43a | 96.50 ± 0.57 a |
| Cefotaxime-CS-AgNPs 6.0 | 93.20 ± 0.72 a | 86.75 ± 0.52 b | 89.80 ± 0.75b | 86.5 ± 0.46 b |
| Cefotaxime-CS-AgNPs 12.0 | 87.61 ± 0.37 ab | 79.74 ± 0.56 bc | 81.60 ± 0.53c | 79.90 ± 0.77 bc |
| Cefotaxime-CS-AgNPs 24.0 | 79.20 ± 0.94 b | 74.52 ± 0.31c | 80.17 ± 0.95c | 73.10 ± 0.88 c |
Notes: Means with different superscripts (a, b, c, d, e, and f) between groups in the same column are significantly different at P< 0.05. Cell numbers were counted and data are expressed as the percentage of untreated control.
Effects of AgNPs and Cefotaxime-CS-AgNPs on RPE-1 Normal Cells Line
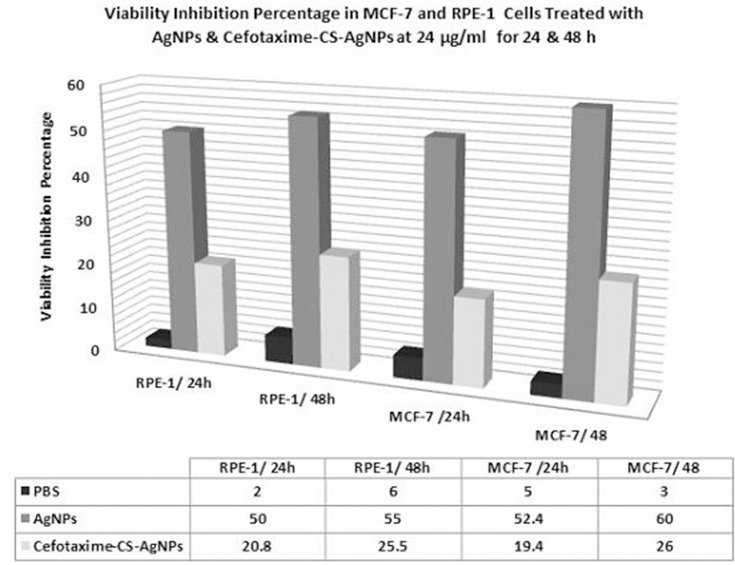
Treatment of RPE-1 normal cells with AgNPs caused decrease proliferation rates. The results confirmed that the proliferation of RPE-1 cells was inhibited in a concentration and time exposed-dependent manner after treated with AgNPs at different concentrations (3, 6, 12 and 24 µg/mL) (Table 5). However, at 12 and 24 µg/mL treatment for 48 h, showed significant antiproliferative activities on RPE-1 normal cells (50.40 ± 0.65 and 44.90 ± 0.79, respectively) compared to treatment at 3 and 6 µg/mL (72.8 ± 0.10 and 62.8 ± 0.10, respectively). These indicated that AgNPs at doses from 3 to 24 µg/mL were significant in inhibiting cellular proliferation rate when compared with control cells. On the other hand, the treatment of RPE-1 normal cells with Cefotaxime-CS-AgNPs was safe up to a final concentration of 24 µg/mL which showed viability inhibition effects by 20.8 and 25.5% when treated with RPE-1 normal cells for 24 hrs and 48 hrs, respectively (Figure 4).
Figure 4.
Histogram represents the effect of CS-AgNPs and Cefotaxime-CS-AgNPs treatments at 24 µg/mL for 24 and 48 hrs on viability inhibition percentage in MCF-7 and RPE-1 cells.
Effects of AgNPs and Cefotaxime-CS-AgNPs on MCF-7 Breast Cancer Cell Line
The results in Table 5 indicated that MCF-7 treated with AgNPs exhibited dose-dependent inhibition against the proliferation rate. After 48 h of AgNPs treatment, the IC50 value was 12µg/mL for MCF-7 cell line (Table 5) and the untreated cells were used as a negative control. When the MCF-7 cell line treated for 48 h, with AgNPs at 24.0 µg/mL a sever cytotoxicity induced in MCF-7 cell and the growth inhibitory effect reaches 60% against MCF-7 compared to the untreated cells with nanoparticles (Figure 4). In this concern, Kishore et al55 indicated that the in vitro anticancer effect of biosynthesized AgNPs performed on MCF-7 cell line using MTT assay showed 99.82% cell death at 10 µg/mL of AgNPs with IC50 value of 1.723 µg/mL.55 However, the conjugated NPs (Cefotaime-CS-AgNPs) induced in a significant inhibition in the proliferation percentage for both cell lines at doses 3.0 and 6.0 µg/mL when treated for period time 24 h. Comparing nano-drug, Cefotaime-CS-AgNPs to AgNPs we showed that Cefotaime-CS-AgNPs having no cytotoxic effect on normal cells at even 12.0 µg/mL for 24 hrs. When Cefotaime-CS-AgNPs treated at 24.0 µg/mL for 48 hrs induced moderate cytotoxic effects in MCF7 reaching 25.5%. Interestingly, the cytotoxic properties of AgNPs and Cefotaxime-CS-AgNPs are essentially dependent on cell type. In this domain, higher cytotoxicity was recorded against cancer cells when comparing to normal cells56 and this confirms the present result.
On the other side, the AgNPs treatment exhibited inhibition, based on the dose and time exposure, against the proliferation of MCF-7 cell line in this research. After 48 h of treatment, the IC50 value was 12.0 µg/mL of AgNPs for MCF-7 cells (Table 5). However, the growth inhibitory effect was no prominent against MCF-7 cells when treated with Cefotaxime-CS-AgNPs at the same concentration (12 µg/mL) reaching 15 and 20% after treatment with Cefotaxime-CS-AgNPs for a period of 24 and 48 h, respectively (Figures 4 and 5). In the same manner, Rejeeth et al57 reported that the green-based synthesis of silver nanoscale molecule induced in vitro proliferation inhibition on the breast cancer cell. Also, Kuppusamy et al58 investigated that the biosynthesized AgNPs using the aqueous extract of Commelina nudiflora L increased cytotoxicity and significantly reduced cell viability of colon cancer cells. Bio-functional silver nanoparticles synthesized within various plant extracts of clove and guava evidenced the satisfying anti-cancer impact versus four different human cancer cell lines; colorectal adenocarcinoma, kidney, chronic myelogenous, leukemia, bone marrow, and cervix.59 Moreover, Kishore et al55 reported that apoptotic morphological changes were caused by AgNPs using DAPI staining method.57
Effects of Target AgNPs and Cefotaxime-CS-AgNPs on the mRNA Expression of Apoptosis Related Genes in MCF-7 Cell Line
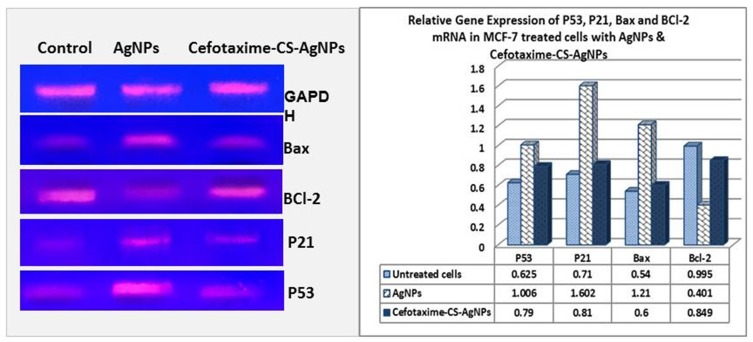
To understand and recognize the cell death pathway induced by both selected nanoparticles, we examined the expression levels of apoptosis marker genes. The present study utilized sqRT-PCR to determine the mRNA expression levels of apoptosis marker genes such as Bax, p53, p21, and Bcl-2, in cancerous cells, MCF-7, exposed to both AgNPs and Cefotaxime-CS-AgNPs at 24.0 µg/mL for 48 h (Figure 5). Cefotaxime-CS-AgNPs induced slight increase in cell apoptosis after treatment for 48 hrs at a dose of 24.0 µg/mL, whereas significant cell apoptosis was observed when MCF-7 cells treated with AgNPs (Figure 5). Further, the expression of the p53 gene of MCF-7 cancer cells treated with AgNPs was shown to be upregulated by 1.6 fold and the mRNA expression p21 gene level was remarkably upregulated by approximately 2.3 fold. Moreover, a notable increase in mRNA expression of Bax resulted in MCF-7 cancer cells after treatment with AgNPs followed by significant down-regulation in BCl-2 mRNA expression by a percentage of 65%; compared to untreated cells; these results leading to Bax/Bcl-2 ratio to be significantly increased (Figure 5).
Figure 5.
Photograph and Histogram showing the effect of AgNPs and Cefotaxime-Cs-AgNPs treatment on the relative transcript level of P53, P21, Bax and BCl-2 genes in MCF-7 Cells after 48 hrs. Results were obtained from three independent experiments and expressed as Mean ± SEM.
The upregulation of p53 by NPs has been pointed to trigger p21 protein accumulation, resulting in a sub-G1 phase cell cycle for apoptosis induction in MCF-7 cancerous cells.39,60–62 Conventional cancer therapy often fails to cause cell death in p53-nude cancer cells. Furthermore, it was indicated that nanoparticles of size ranging from 5 to 35 nm primarily induced cell death by targeting the function and structure of mitochondria.63
These findings indicate that nanoparticles manufactured using green nanotechnology and its conjugates can represent an ideal and effective strategy for combating epidemic diseases and cancer. The AgNPs treatment was more toxic than Cefotaxime-CS-AgNPs for both cell lines. They battle the cancer cell lines and have a low effect on normal cells and these findings are in agreement with the previous research.63–65
Herein, biosynthesis of highly stable AgNPs the usage of Rosa damascenes extract and coated with chitosan. It was conjugated with cefotaxime and evaluated as novel drug delivery. AgNPs and its conjugates had been characterized by the usage of UV–Vis spectroscopy, TEM, XRD, and FTIR. The later confirm the conjugation of CS-AGNPs and cefotaxime. Cefotaxime-CS-AgNPs showed high efficacy against tested E. coli and MRSA strains compared to pure cefotaxime drug with reduction of MIC dose. Thus, Cs-AgNPs carriers represent an efficient, viable, safe, and cheaper novel-targeted delivery system of cefotaxime. Also, we evaluated the cytotoxic effects of 0 to 24.0 µg/mL for AgNPs and Cefotaxime-CS-AgNPs on normal and tumor cells particularly breast cancer cells. The effect of the AgNPs and Cefotaxime-Cs-AgNPs on cell death and apoptosis was investigated and the role of anti-proliferative and the cytotoxic effect of these nanoparticles was determined. This research gives remarkable insights for designing nanoscale delivery and curative systems that have a pronounced cytotoxic activity on cancer cells and are safe to normal cells. Further, such carriers hold potential to increase the efficacy against MDR bacteria and reduce the toxicity of delivery of bioactive against normal cells and might be effective for other deadly diseases such as cancer.
Acknowledgment
This project was funded by A. Jameel Scientific Chair of Prophetic Medical Applications, King Abdulaziz University, Jeddah. The authors acknowledge and thanks to DSR technical and financial support.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
- 1.World Health Organization WHO. Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance Due to Non-Human Use. Geneva: World Health Organization; 2017. [Google Scholar]
- 2.Giske CG, Sundsfjord AS, Kahlmeter G, et al. Redefining extended-spectrum β-lactamases: balancing science and clinical need. J Antimicrob Chemother. 2009;63:1–4. doi: 10.1093/jac/dkn444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VGJ. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–666. doi: 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szczepanik A, Kozioł-montewka M, Al-doori Z, Morrison D, Kaczor D. Spread of a single multi resistant methicillin-resistant Staphylococcus aureus clone carrying a variant of staphylococcal cassette chromosome mec type III isolated in a university hospital. Eur J Clin Microbiol Infect Dis. 2007;26:29–35. doi: 10.1007/s10096-006-0237-5 [DOI] [PubMed] [Google Scholar]
- 5.Sancak B. Staphylococcus aureus and antibiotic resistance Mikrobiyol. Bul. 2011;45:565–576. [PubMed] [Google Scholar]
- 6.Huh AJ, Kwon YJ. “Nanoantibiotics”: a new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J Controlled Release. 2011;156:128–145. doi: 10.1016/j.jconrel.2011.07.002 [DOI] [PubMed] [Google Scholar]
- 7.Roshmi T, Soumya KR, Jyothis M, Radhakrishnan EK. Effect of biofabricated gold nanoparticle-based antibiotic conjugates on minimum inhibitory concentration of bacterial isolates of clinical origin. Gold Bull. 2015;48(1–2):63–71. doi: 10.1007/s13404-015-0162-4 [DOI] [Google Scholar]
- 8.Reddy DHK, Seshaiah K, Reddy AVR, Lee SM. Optimization of Cd (II), Cu (II) and Ni (II) biosorption by chemically modified Moringa oleifera leaves powder. Carbohydr Polym. 2012;88:1077–1086. doi: 10.1016/j.carbpol.2012.01.073 [DOI] [Google Scholar]
- 9.Nowack B, Krug HF, Height M. 120 years of nanosilver history: implications for policy makers. Environ Sci Technol. 2011;44:1177–1183. doi: 10.1021/es103316q [DOI] [PubMed] [Google Scholar]
- 10.Rao ML, Bhumi G, Savithramma N. Green synthesis of silver nanoparticles by Allamanda cathartica L. leaf extract and evaluation for antimicrobial activity. Nanotechnol. Int J Pharm Sci. 2013;6:2260–2268. [Google Scholar]
- 11.Konop M, Damps T, Misicka A. Certain aspects of silver and silver nanoparticles in wound care: a minireview. J Nanomater. 2016;2016:1–10. doi: 10.1155/2016/7614753 [DOI] [Google Scholar]
- 12.Rigo C, Ferroni L, Tocco I, et al. Active silver nanoparticles for wound healing. Int J Mol Sci. 2013;14(3):4817–4840. doi: 10.3390/ijms14034817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammed AE. Green synthesis, antimicrobial and cytotoxic effects of silver nanoparticles mediated by Eucalyptus camaldulensis leaf extract. Asian Pac J Trop Biomed. 2015;5(5):382–386. doi: 10.1016/S2221-1691(15)30373-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dar MA, Ingle A, Rai M. Enhanced antimicrobial activity of silver nanoparticles synthesized by Cryphonectria sp. evaluated singly and in combination with antibiotics. Nanomedicine. 2013;9(1):105–110. doi: 10.1016/j.nano.2012.04.007 [DOI] [PubMed] [Google Scholar]
- 15.Fayaz AM, Balaji K, Girilal M, Yadav R, Kalaichelvan PT, Venketesan R. Biogenic synthesis of silver nanoparticles and their synergistic effect with antibiotics: a study against gram-positive and gram-negative bacteria. Nanomedicine. 2010;6(1):103–109. doi: 10.1016/j.nano.2009.04.006 [DOI] [PubMed] [Google Scholar]
- 16.Geoprincy G, Saravanan P, Gandhi NN, Renganathan S. A novel approach for studying the combined antimicrobial effects of silver nanoparticles and antibiotics through agar over layer method and disk diffusion method. Dig J Nanomater Bios. 2011;6:1557–1565. [Google Scholar]
- 17.Shahverdi AR, Fakhimi A, Shahverdi HR, Minaian S. Synthesis and effect of silver nanoparticles on the antibacterial activity of different antibiotics against Staphylococcus aureus and Escherichia coli. Nanomedicine. 2007;3(2):168–171. doi: 10.1016/j.nano.2007.02.001 [DOI] [PubMed] [Google Scholar]
- 18.Devi LS, Joshi SR. Antimicrobial and synergistic effects of silver nanoparticles synthesized using soil fungi of high altitudes of Eastern Himalaya. Mycobiology. 2012;40(1):27–34. doi: 10.5941/MYCO.2012.40.1.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva HFO, Lima KMG, Cardoso MB, et al. Doxycycline conjugated with polyvinylpyrrolidone-encapsulated silver nanoparticles: a polymer’s malevolent touch against Escherichia coli. RSC Adv. 2015;5(82):66886–66893. doi: 10.1039/C5RA10880B [DOI] [Google Scholar]
- 20.Henriksen-lacey M, Carregal-romero S, Liz-marzán LM. Current challenges toward in vitro cellular validation of inorganic nanoparticles. Bioconjug Chem. 2017;28(1):212–221. doi: 10.1021/acs.bioconjchem.6b00514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gad El-Rab SMF, Halawani EM. Effect of antibiotic conjugated nanoparticles on multidrug resistant bacteria. J Microbiol Biotechnol. 2018. [Google Scholar]
- 22.Pugazhendhi S, Sathya P, Palanisamy PK, Gopalakrishnan R. Synthesis of silver nanoparticles through green approach using Dioscorea alata and their characterization on antibacterial activities and optical limiting behavior. J Photochem Photobiol B. 2016;159:155–160. doi: 10.1016/j.jphotobiol.2016.03.043 [DOI] [PubMed] [Google Scholar]
- 23.Inbaraj BS, Tsai TY, B HC. Synthesis, characterization and anti-bacterial activity of superparamagnetic nanoparticles modified with glycol chitosan. Sci Technol Adv Mater. 2012;13:015002. doi: 10.1088/1468-6996/13/1/015002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harshiny MMM, Arthanareeswaran G, Kumaran S, Rajasree S. Enhancement of antibacterial properties of silver nanoparticles–ceftriaxone conjugate through Mukia maderaspatana leaf extract mediated synthesis. Ecotoxicol Environ Saf. 2015;121:135–141. doi: 10.1016/j.ecoenv.2015.04.041 [DOI] [PubMed] [Google Scholar]
- 25.Rao BS, Murthy KV. Preparation and in vitro evaluation of chitosan matrices cross-linked by formaldehyde vapors Drug development and industrial pharmacy. Drug develop Ind Pharm. 2000;26(10):1085–1090. doi: 10.1081/ddc-100100272 [DOI] [PubMed] [Google Scholar]
- 26.Shakera M, Shaaban MI. Formulation of carbapenems loaded gold nanoparticles to combat multi-antibiotic bacterial resistance: in vitro antibacterial study. Int J Pharm. 2017;525:71–84. doi: 10.1016/j.ijpharm.2017.04.019 [DOI] [PubMed] [Google Scholar]
- 27.Kanmani PLS. Synthesis and characterization of pullulan-mediated silver nanoparticles and its antimicrobial activities. Carbohydr Polym. 2013;97(2):421–428. doi: 10.1016/j.carbpol.2013.04.048 [DOI] [PubMed] [Google Scholar]
- 28.Ahmed S, Manzoor K, Ikram S. Synthesis of silver nanoparticles using leaf extract of Crotolaria retusa as antimicrobial green catalyst. J Bionanosci. 2016;10:282–287. doi: 10.1166/jbns.2016.1376 [DOI] [Google Scholar]
- 29.Verma SK, Gond SK, Mishra A, et al. Biofabrication of antibacterial and antioxidant silver nanoparticles (Agnps) by an endophytic fungus Pestalotia Sp. isolated from Madhuca Longifolia. J Nanomater Mol Nanotechnol. 2016;5:1–7. doi: 10.4172/2324-8777.1000189 [DOI] [Google Scholar]
- 30.Dubey SP, Lahtinen M, Sillanpaa M. Tansy fruit mediated greener synthesis of silver and gold nanoparticles. Process Biochem. 2010;45:1065–1071. doi: 10.1016/j.procbio.2010.03.024 [DOI] [Google Scholar]
- 31.Anandalakshmi K, Venugobal J, Ramasamy V. Characterization of silver nanoparticles by green synthesis method using Pedalium murex leaf extract and their antibacterial activity. Appl Nanosci. 2016;6:399–408. doi: 10.1007/s13204-015-0449-z [DOI] [Google Scholar]
- 32.Pani ALJ, Yun S. Autoclave mediated one-pot-one-minute synthesis of AgNPs and Au-Ag nanocomposite from Melia azedarach bark extract with antimicrobial activity against food pathogens. Chem Cent J. 2016;10:1–11. doi: 10.1186/s13065-016-0157-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas W, Nga T, Toshihiko K, Xia W, Petra AT, Kristina UW. Cancer chemo preventive properties of orally bioavailable flavonoids – methylated versus unmethylated flavones. Biochem Pharmacol. 2007;73:1288–1296. doi: 10.1016/j.bcp.2006.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szende B, Tyihak E, Trezl L. Role of arginine and its methylated derivatives in cancer biology and treatment. Cancer Cell Int. 2001;17:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4 [DOI] [PubMed] [Google Scholar]
- 36.Badakhshan MP, Sreenivasan S, Jegathambigai RN, Surash R. Anti-leukemia activity of methanolic extracts of Lantana camara. Pharmacogn. Res. 2009;1:274–279. [Google Scholar]
- 37.Brun ME, Gasca S, Girard C, Bouton K, De Massy B, De Sario A. Characterization and expression analysis during embryo development of the mouse ortholog of MLL3. Gene. 2006;371:25–33. [DOI] [PubMed] [Google Scholar]
- 38.Hassan AM, Abdel-aziem SH, Abdel-wahhab MA. Modulation of DNA damage and alteration of gene expression during aflatoxicosis via dietary supplementation of Spirulina (Arthrospira) and whey protein concentrate. Ecotoxicol Environ Saf. 2012;79:294–300. doi: 10.1016/j.ecoenv.2012.01.017 [DOI] [PubMed] [Google Scholar]
- 39.Moghaddam AB, Moniri M, Azizi S, et al. Eco-friendly formulated zinc oxide nanoparticles: induction of cell cycle arrest and apoptosis in the MCF-7 cancer cell line. Genes. 2017;8,(281):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walter RA, Duncan DB, Bayes A. Rule for the symmetric multiple comparison problems. J Am Stat Assoc. 1969;64:1484–1503. [Google Scholar]
- 41.Snedecor GW, Cochran WG. Statistical Methods. 7th ed. Ames, Iowa: Iowa State University Press,1980; 334–364. [Google Scholar]
- 42.Kumar R, Ghoshal G, Jain A, Goyal M. Rapid green synthesis of silver nanoparticles (AgNPs) using (Prunus persica) plants extract: exploring its antimicrobial and catalytic activities. J Nanomed Nanotechnol. 2017;8:1–8. [Google Scholar]
- 43.Gopinath V, Priyadarshini S, Priyadharssini N, Pandian K, Velusamy P. Biogenic synthesis of antibacterial silver chloride nanoparticles using leaf extracts of Cissus quadrangularis Linn. Mater Lett. 2013;91:224–227. doi: 10.1016/j.matlet.2012.09.102 [DOI] [Google Scholar]
- 44.Perveen S, Safdar N, Chaudhry G, Yasmin A. Antibacterial evaluation of silver nanoparticles synthesized from lychee peel: individual versus antibiotic conjugated effects. World J Microbiol Biotechnol. 2018;34, 118:1–12. [DOI] [PubMed] [Google Scholar]
- 45.Agnihotri S, Mukherji S. Size-controlled silver nanoparticles synthesized over the range 5–100 nm using the same protocol and their antibacterial efficacy. RSC Adv. 2014;4:3974–3983. doi: 10.1039/C3RA44507K [DOI] [Google Scholar]
- 46.Jyoti K, Baunthiyal M, Singh A. Characterization of silver nanoparticles synthesized using Urtica dioica Linn. leaves and their synergistic effects with antibiotics. J Radiat Res. 2016;9:217–227. [Google Scholar]
- 47.Khatami M, Pourseyedi S, Khatami M, Hamidi H, Zaeifi M, Soltani L. Synthesis of silver nanoparticles using seed exudates of Sinapis arvensis as a novel bioresource, and evaluation of their antifungal activity. Bioresour Bioprocess. 2015;2,(19):1–7. doi: 10.1186/s40643-015-0043-y [DOI] [Google Scholar]
- 48.Silva SML, Braga CRC, Fook MVL, et al. Application of infrared spectroscopy to analysis of chitosan/clay nanocomposite In: Theophile T, editor. Infrared Spectroscopy - Materials Science, Engineering and Technology. Rijeka: InTech; 2012. [Google Scholar]
- 49.Qu JB, Shao HH, Jing GL, Huang F. PEG-chitosan-coated iron oxide nanoparticles with high saturated magnetization as carriers of 10-hydroxycamptothecin: preparation, characterization and cytotoxicity studies. Colloids Surf B Biointerfaces. 2013;102:37–44. doi: 10.1016/j.colsurfb.2012.08.004 [DOI] [PubMed] [Google Scholar]
- 50.Zakaria AS, Afifi SA, Elkhodairy KA. Newly developed topical cefotaxime sodium hydrogels: antibacterial activity and in vivo evaluation. BioMed Res Int. 2016;2016:1–15. doi: 10.1155/2016/6525163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jamil B, Habib H, Abbasi SA, Ihsan A, Nasir H, Imran M. Development of cefotaxime impregnated chitosan as nano-antibiotics: de Novo strategy to combat biofilm forming multi-drug resistant pathogens. Front Microbiol. 2016;7(Article):330. doi: 10.3389/fmicb.2016.00330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goy RC, Britto D, Assis OBG. A review of the antimicrobial activity of chitosan. Polimeros Cie Tec. 2009;3:1–7. [Google Scholar]
- 53.Kalita SKR, Sharma KK, Kataki AC, Deka M, Kotoky J. Amoxicillin functionalized gold nanoparticles revertsMRSA resistance. Mater Sci Eng C Mater Biol Appl. 2016;61:720–727. doi: 10.1016/j.msec.2015.12.078 [DOI] [PubMed] [Google Scholar]
- 54.Krishna G, Kumar SS, Pranitha V, Alha M, Charaya S. Biogenic synthesis of silver nanoparticles and their synergistic effect with antibiotics: a study against Salmonella sp. Int J Pharm Pharm Sci. 2015;7:84–88. [Google Scholar]
- 55.Kishore M, Abdulqader AT, Ahmad HS, Hanumantharao Y. Anticancer and antibacterial potential of green silver nanoparticles synthesized from Maytenus senegalensis (L.) leaf extract and their characterization. Drug Invention Today. 2018;10:554–561. [Google Scholar]
- 56.Zhang XF, Liu ZG, Shen W, Gurunathan S. Silver nanoparticles: synthesis, characterization, properties, applications, and therapeutic approaches. Int J Mol Sci. 2016;17,(1534):1–34. doi: 10.3390/ijms17091534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rejeeth C, Nataraj B, Vivek R, Sakthivel M. Biosynthesis of silver nanoscale particles using spirulina platensis induces growth-inhibitory effect on human breast cancer cell line MCF 7. Med Aromat Plants. 2014;3:1–7. doi: 10.4172/2167-0412.1000163 [DOI] [Google Scholar]
- 58.Kuppusamy P, Ichwan SJ, Al-zikri PN, et al. In vitro anticancer activity of au, Ag nanoparticles synthesized using Commelina nudiflora L. aqueous extract against HCT-116 colon cancer cells. Biol Trace Elem Res. 2016;173:297–305. doi: 10.1007/s12011-016-0666-7 [DOI] [PubMed] [Google Scholar]
- 59.El-naggar NE, Hussein MH, El-sawah AA. Bio-fabrication of silver nanoparticles by phycocyanin, characterization, in vitro anticancer activity against breast cancer cell line and in vivo cytotoxicity. Sci Rep. 2017;7(10844):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia M, Knezevic D, Vassilev LT. p21 does not protect cancer cells from apoptosis induced by non-genotoxic p53 activation. Oncogene. 2011;30(3):346–355. doi: 10.1038/onc.2010.413 [DOI] [PubMed] [Google Scholar]
- 61.Hacker M. Adverse drug reactions. In: Hacker M, Messer WS II, Bachmann KA, editors. Pharmacology: Principles and Practice. New York, NY, USA: Elsevier; 2009;327–352. [Google Scholar]
- 62.Alkhalaf M, El-mowafy AM. Overexpression of wild-type p53 gene renders MCF-7 breast cancer cells more sensitive to the anti-proliferative effect of progesterone. J Endocrinol. 2003;179(1):55–62. doi: 10.1677/joe.0.1790055 [DOI] [PubMed] [Google Scholar]
- 63.Abdel-fattah W, Ali GW. On the anti-cancer activities of silver nanoparticles. J Appl Biotechnol Bioeng. 2018;5:43‒46. [Google Scholar]
- 64.Khorrami S, Zarrabi A, Khaleghi M, Danaei M, Mozafari MR. Selective cytotoxicity of green synthesized silver nanoparticles against the MCF-7 tumor cell line and their enhanced antioxidant and antimicrobial properties. Int J Nanomedicine. 2018;13:8013–8024. doi: 10.2147/IJN.S189295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He Y, Du Z, Ma S, et al. Effects of green-synthesized silver nanoparticles on lung cancer cells in vitro and grown as xenograft tumors in vivo. Int J Nanomedicine. 2016;11:1879–1887. doi: 10.2147/IJN.S103695 [DOI] [PMC free article] [PubMed] [Google Scholar]