Abstract
Introduction
Lung adenocarcinoma (LUAD), which is associated with high morbidity and mortality, is prone to cisplatin resistance, resulting in poor patient prognosis. Long non-coding RNAs (lncRNAs) have complex biological functions in a variety of tumors. Elucidating the underlying molecular mechanisms between lncRNA and cisplatin resistance in LUAD is expected to enable identification of new targets for drug development.
Methods
Cell proliferation was measured by CCK-8 assay and cell apoptosis was detected using flow cytometry analysis. Luciferase reporter assay was conducted to determine the interaction between lncRNA and MicroRNA. Gene expression was evaluated by Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction and Western blot analysis.
Results
Long non-coding RNA activated by TGF-β (lncRNA-ATB) was shown to be significantly up-regulated in A549 cells resistant to cisplatin/cis-dichlorodiammineplatinum (II) (cis-DDP) (A549/CDDP cells), compared with corresponding levels in parental A549 cells. Overexpression of lncRNA-ATB significantly elevated cisplatin resistance in LUAD cell lines (A549 and H1975 cells), and this was associated with activation of apoptosis-related genes. Conversely, silencing of lncRNA-ATB decreased cisplatin resistance in LUAD cells. Mechanistically, lncRNA-ATB increased expression of β-catenin by directly binding to MicroRNA-200a (miR-200a), thereby promoting cell survival and cisplatin resistance. Transfection with a miR-200a mimic or treatment with the β-catenin downstream pathway inhibitor IWR-1 could reverse the phenotypes induced by lncRNA-ATB overexpression.
Conclusion
In summary, this study revealed that lncRNA-ATB is dramatically up-regulated in cisplatin-resistant LUAD cell lines, and that lncRNA-ATB facilitates cell survival by targeting the miR-200a/β-catenin pathway in these cells.
Keywords: lncRNA-ATB, miR-200a, β-catenin, lung adenocarcinoma, cisplatin resistance
Introduction
Lung cancer has the highest morbidity and mortality of all cancers, and is the most common malignant tumor globally, with nearly one million patients diagnosed each year and hundreds of thousands of patients dying annually.1,2 Lung adenocarcinoma (LUAD) accounts for approximately 40% of lung cancer cases.3 The primary treatment for lung cancer is surgery accompanied by radiotherapy, chemotherapy or neoadjuvant therapy.4,5 However, since most patients are diagnosed when they are at an advanced stage of lung cancer, often with multiple metastases, the 5-year survival rate of patients with lung cancer remains low.6,7
The first-line chemotherapy for lung cancer is predominantly platinum-based drugs, including cisplatin.8,9 However, the occurrence of drug resistance reduces the effectiveness of chemotherapy, leading to a significant decrease in quality of life and reduced compliance among patients.10,11 There is emerging evidence that non-coding RNAs, such as miR-5100,12 lncRNA-HOTAIR,13 and Hsa_circ_0001946,14 are implicated in drug resistance in lung cancer. Herein, elucidating the mechanism of cisplatin resistance will be of benefit to patients with LUAD.
Long non-coding RNA activated by TGF-β (lncRNA-ATB) is a recently discovered long non-coding RNA (lncRNA) that is greatly elevated in various tumor tissues, and acts as a cancer-promoting factor,15,16 LncRNA-ATB promotes tumor cell proliferation and metastasis by regulating the p38 signaling pathway and epithelial-to-mesenchymal transition (EMT).17,18 Previous work in LUAD cells suggested that lncRNA-ATB could also promote cell migration and invasion.19,20 In this study, lncRNA-ATB was significantly up-regulated in A549 cells resistant to cisplatin/cis-dichlorodiammineplatinum (II) (cis-DDP) (A549/CDDP cells), compared with corresponding levels in parental A549 cells. Notably, high expression of lncRNA-ATB was associated with cisplatin resistance of LUAD cells. These findings provide insights into the molecular mechanisms of cisplatin resistance in LUAD.
Materials and Methods
Cell Culture
Human lung adenocarcinoma cell lines A549 and H1975, and A549/CDDP (resistant to cisplatin/cis-dichlorodiammineplatinum (II) (cis-DDP)) cells were obtained from Cell Bank of Type Culture Collection of Chinese Academy of Sciences. Non-resistant LUAD cell lines were cultured in RPMI 1640 medium (HyClone, Logan, Utah, USA) containing 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin (Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA). Drug-resistant LUAD cell lines were cultured in the above-mentioned media supplemented with 0.05 µg/mL cisplatin. Cells were maintained at 37°C in a 5% CO2 incubator. The culture medium was changed every two days.
Lentivirus Infection
The lentiviruses lncRNA-ATB-OE and LV-si-lncRNA-ATB, their matched control viruses, and polybrene were purchased from GenePharma (Shanghai, China). A549, A549/CDDP, or H1975 cells were seeded in 6-well plates at a density of 5×105 cells per well. Experimental groups in this study comprised lncRNA-ATB overexpression group (LV-lncRNA-ATB-OE), empty lentivirus vector group (LV-NC), and lncRNA-ATB silencing group (LV-si-lncRNA-ATB). Culture medium, polybrene (8 μg/mL), and virus solution (MOI=100) were successively added to the cells (total volume 1 mL) following the manufacturer’s instructions (GenePharma). After incubation for 48 h, total RNA was extracted, and the infection efficiency was evaluated by RT-qPCR.
Cell Transfection
Cells were seeded in 6-well plates at a density of 5×105 cells per well. Next, 2 μg miR-200a mimics or miR-200a antagomir (Shanghai Sangon Biotech Co., Ltd, Shanghai, China) and miR-nonsense sequence control (miR-NC) (Shanghai Sangon Biotech Co., Ltd) were mixed with 2 μL LipofectamineTM 2000 (Beijing Solarbio Science & Technology Co., Ltd, Beijing, China) according to the manufacturer’s guidelines, and were incubated at room temperature for 20 min before being adding to the cells.
CCK-8 Cell Proliferation Assay
Cells were seeded in 96-well plates (six well per group) at a density of 1×104 cells per well. After transfection, infection or treatment with IWR-1 (catalog no. bs810444; Absin Bioscience Inc., Shanghai, China), 10% of CCK-8 reagent (Beijing Zoman Biotechnology Co., Ltd, Beijing, China) was added at pre-set time points and plates were incubated for 2 hrs at 37°C. The absorbance of each well at 450 nm was measured using a microplate reader (iMARK, Bio-Rad, Hercules, California, USA).
Flow Cytometry Analysis of Apoptosis
Cells were seeded in 6-well plates at a density of 5×105 cells per well. After treatment with or without cisplatin for 24 h, cells were collected by trypsinization and washed twice with PBS. Cells were then fixed with ice-cooled 75% ethanol for 30 min at 4°C, incubated with propidium iodide (PI; Beijing Solarbio Science & Technology Co., Ltd, China) and RNase A (Beijing Solarbio Science & Technology Co., Ltd) in the dark for 30 min at 4°C, and apoptosis was measured by flow cytometry. Data were analyzed by FlowJo 7.6.1 software (Becton, Dickinson & Company, State of New Jersey, USA).
Luciferase Reporter Assay
Luciferase assays were conducted using the Dual-Luciferase Assay kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Briefly, the predicted binding sequence of lncRNA-ATB or miR-200a or 3′UTR of β-catenin mRNA, and the mutated sequence of lncRNA-ATB or β-catenin were separately cloned into pmirGLO dual-luciferase vectors (GenePharma). Using Lipofectamine 2000, 293T cells were co-transfected with wild-type pmirGLO-lncRNA-ATB-Wt/miR-200a mimics, the mutated lncRNA-ATB-Mut/miR-200a mimics, WT-β-catenin-3′UTR-pmirGLO/miR-200a mimics, Mut-β-catenin-3′UTR-pmirGLO/miR-200a mimics, or the negative control (Renilla luciferase vectors). After 48 h, luciferase activity was detected using the dual-luciferase reporter kit (Promega; Madison, Wisconsin, USA). Relative firefly luciferase activity was quantified by normalizing to Renilla luciferase activity.
Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from cells with TRIzol Reagent (Life Technologies, Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. Subsequently, cDNA was synthesized using TransScript® miRNA First-Strand cDNA Synthesis SuperMix (TransGen Biotechnology, Beijing, China) or PrimeScriptTM RT Master Mix (Perfect Real Time) (code no. RR036A; Takara Bio Inc., Japan) according to the manufacturer’s instructions. One Step TB Green® PrimeScriptTM RT-PCR Kit (Perfect Real Time) (code no. RR066A; Takara) was used for RT-qPCR. For microRNA, U6 was used as an internal reference. For the protein-coding gene, β-actin was used as an internal reference. All primers were synthesized by Shanghai Sangon Biotech Co., Ltd, China and are listed in Table 1. The PCR amplification procedure comprised a denaturation step at 95°C for 2 min, followed by 35 cycles of denaturation at 95°C for 30 sec, annealing at 58°C for 30 sec and extension at 72°C for 1 min. Relative expression was calculated by the 2−ΔΔCt method.
Table 1.
Primers for Real-Time PCR
| Gene | Forward Sequence | Reverse Sequence |
|---|---|---|
| U6 | 5ʹ-CTCGCTTCGGCAGCACA-3’ | 5ʹ-AACGCTTCACGAATTTGCGT-3’ |
| β-actin | 5ʹ-CCTCGCCTTTGCCGATCC-3’ | 5ʹ-GGATCTTCATGAGGTAGTCAGTC-3’ |
| Vimentin | 5ʹ-GCCCTAGACGAACTGGGTC-3’ | 5ʹ-GGCTGCAACTGCCTAATGAG-3’ |
| Cyclin D1 | 5ʹ-TGGAGCCCGTGAAAAAGAGC-3’ | 5ʹ- TCTCCTTCATCTTAGAGGCCAC-3’ |
| β-catenin | 5ʹ-CCTATGCAGGGGTGGTCAAC-3’ | 5ʹ-CGACCTGGAAAACGCCATCA-3’ |
| Bax | 5ʹ-CCCGAGAGGTCTTTTTCCGAG-3’ | 5ʹ-CCAGCCCATGATGGTTCTGAT-3’ |
| Bcl-2 | 5ʹ-GGTGGGGTCATGTGTGTGG-3’ | 5ʹ-CGGTTCAGGTACTCAGTCATCC-3’ |
| Bcl-xl | 5ʹ-GACTGAATCGGAGATGGAGACC-3’ | 5ʹ-GCAGTTCAAACTCGTCGCCT-3’ |
Western Blot Analysis
The following primary antibodies were purchased from Cell Signaling Technology, Inc (Boston, USA) and were each used at a 1:1000 dilution: rabbit anti-human Bcl-xl (catalog no. 2764), rabbit anti-human Bcl-2 (catalog no. 15071), rabbit anti-human Bax (catalog no. 2774), rabbit anti-human caspase-3 (catalog no. 9662), rabbit anti-human caspase-9 (catalog no. 9504), rabbit anti-human cleaved caspase-3 (catalog no. 9664), rabbit anti-human cleaved caspase-9 (catalog no. 9509), rabbit anti-human cyclin D1 (catalog no. 2978), and rabbit anti-human vimentin (catalog no. 5741). Rabbit anti-human β-catenin primary antibody (1:2000 dilution; catalog no. ab32572), rabbit anti-human β-actin primary antibody (1:5000; catalog no. ab179467), and goat anti-rabbit IgG H&L (HRP) secondary antibody (1:5000; catalog no. ab6721) were purchased from Abcam (Cambridge, UK). Briefly, cell lysates were generated using RIPA lysis buffer (Beijing Solarbio Science & Technology Co., Ltd) containing 1% phenylmethylsulfonyl fluoride (PMSF; Beijing Solarbio Science & Technology Co., Ltd). The protein concentration of each lysate was analyzed using the bicinchoninic acid (BCA) method, and 30 μg of each lysate was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Merck, Darmstadt, Germany). Membranes were blocked with 5% skim milk powder for 2 h at room temperature, followed by incubation with the corresponding primary antibodies at 4°C overnight. After washing three times with TBST, membranes were then incubated with HRP-labeled IgG secondary antibodies (1:5000 dilution) for 2 h at room temperature. Proteins were visualized using a Millipore Western blot kit (Merck), and quantified by Quantity One 4.6.7 software.
Statistical Analysis
SPSS 21.0 software was used for statistical analysis. Data were expressed as mean ± standard deviation (SD). At least three independent replicates were performed for each experiment. Multiple comparisons among groups were conducted using one-way ANOVA with an LSD test. The independent-samples t-test was used to analyze the statistical difference between two groups. P<0.05 is regarded as statistically significant.
Results
Up-Regulation of lncRNA-ATB Promotes Cisplatin Resistance in LUAD Cells
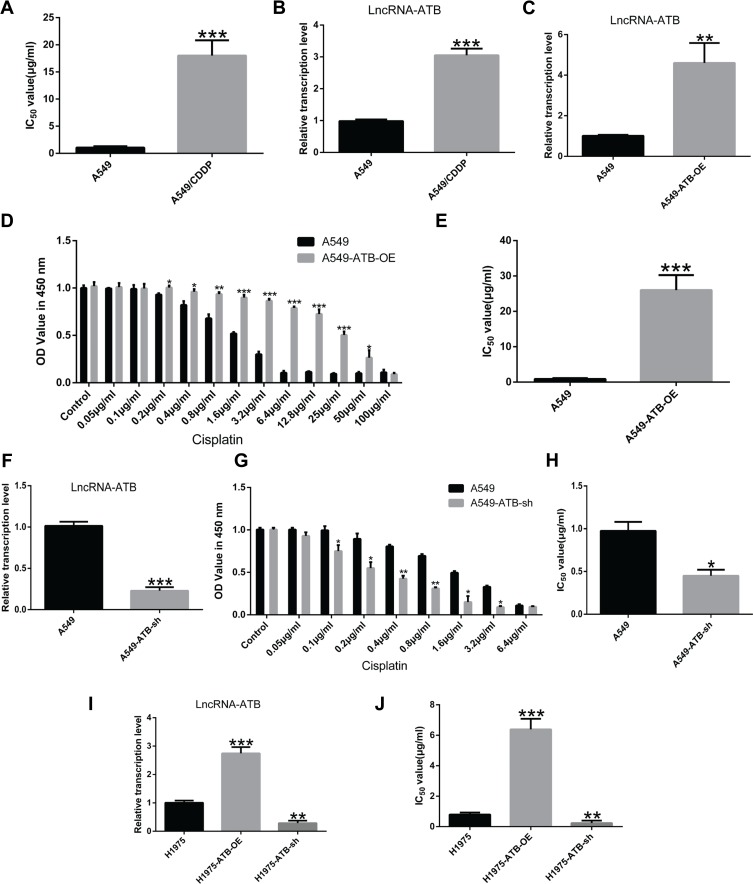
The role of lncRNA-ATB in cisplatin resistance of LUAD cells was evaluated by determining the half-maximal inhibitory concentration (IC50) of cisplatin, and quantifying expression of lncRNA-ATB. The IC50 an important pharmacodynamic index of drug effectiveness.21 Both the IC50 of cisplatin (Figure 1A) and expression of lncRNA-ATB (Figure 1B) were significantly elevated in A549/CDDP cells when compared with parental A549 cells. This indicated that the A549/CDDP cells were less sensitive to cisplatin than the parental A549 cells were. Generation of lentivirus constructs and successful overexpression of lncRNA-ATB in A549 cells (A549-ATB-OE) (Figure 1C) and H1975 cells (H1975-ATB-OE) (Figure 1I) was confirmed by dramatically elevated levels of lncRNA-ATB expression compared with the matched control cells. Meanwhile, the sensitivity of A549-ATB-OE cells to cisplatin was strikingly reduced compared with control cells (Figure 1D). In addition, the IC50 of LUAD cells overexpressing lncRNA-ATB was significantly increased compared with that measured in matched control cells (Figure 1E and J). Conversely, silencing lncRNA-ATB (Figure 1F and I) significantly sensitized LUAD cells to cisplatin (Figure 1G, H and J).
Figure 1.
Up-regulation of lncRNA-ATB promotes cisplatin resistance in LUAD cells. (A) The half-maximal inhibitory concentration (IC50) of cisplatin in A549/CDDP cells was significantly greater than in A549 cells. (B) LncRNA-ATB was strikingly elevated in A549/CDDP cells compared with parental A549 cells. (C) Successful construction of A549-ATB-OE cells was validated by RT-qPCR. (D) The effects of different concentrations of cisplatin on proliferation of A549-ATB-OE and A549 cells were detected by CCK-8 assay. (E) IC50 of cisplatin in A549-ATB-OE cells was remarkably higher than that in A549 cells. (F) Successful construction of A549-ATB-sh cells was validated by RT-qPCR. (G) The effects of various concentrations of cisplatin on proliferation of A549-ATB-sh and A549 cells were detected by CCK-8 assay. (H) IC50 of cisplatin in A549-ATB-sh cells was significantly lower than that in A549 cells. (I) Successful construction of H1975-ATB-OE and H1975-ATB-sh cells was validated by RT-qPCR. (J) IC50 of cisplatin in H1975-ATB-OE and H1975-ATB-sh cells. Data are expressed as mean ± SD from three independent experiments. *P<0.05; **P<0.01; ***P<0.001.
LncRNA-ATB Inhibits Cisplatin-Induced Apoptosis in LUAD Cells
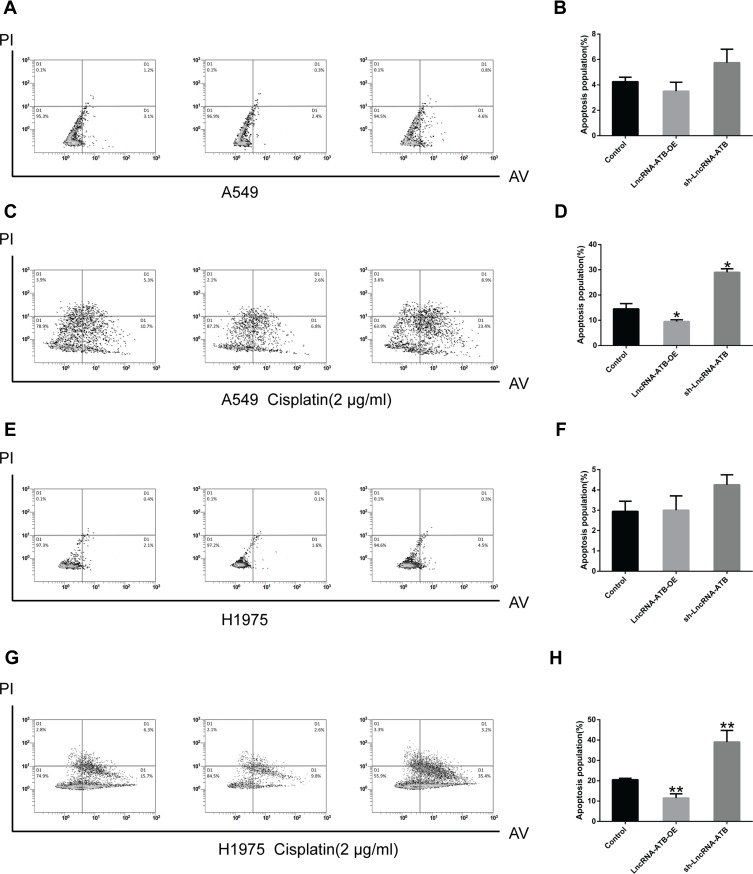
To understand the mechanism underlying the role of lncRNA-ATB in cisplatin resistance, apoptosis of LUAD cells overexpressing lncRNA-ATB was assessed by flow cytometry analysis. The apoptotic rate of A549-ATB-OE cells tended to be decreased compared with the control cells, whereas the apoptotic rate of A549 cells silencing lncRNA-ATB (A549-ATB-sh cells) had a tendency to be increased (Figure 2A and B). However, after treatment with cisplatin, the apoptotic rate of A549-ATB-OE cells was significantly lower than that of the control cells, while the rate of apoptosis was remarkably increased in A549-ATB-sh cells (Figure 2C and D). H1975 cells overexpressing or silencing lncRNA-ATB, with and without cisplatin treatment, yielded results consistent with those of A549-ATB-OE cells and A549-ATB-sh cells (Figure 2E–H).
Figure 2.
LncRNA-ATB inhibits cisplatin-induced apoptosis. (A) Total apoptotic rates (including early and late apoptotic cells) of A549-ATB-OE, A549-ATB-sh, and A549 cells detected by flow cytometry, and (B) statistically analyzed. (C) Total apoptotic rates of A549-ATB-OE, A549-ATB-sh, and A549 cells treated with cisplatin (2 μg/mL) for 24 h, then detected by flow cytometry, and (D) quantified. (E) Apoptosis of H1975 cells overexpressing or silencing lncRNA-ATB, and (F) subsequent statistical analysis. (G) Effect of cisplatin (2 μg/mL) treatment on apoptosis of H1975 cells overexpressing or silencing lncRNA-ATB compared with control cells, and (H) subsequent quantification. Data are expressed as mean ± SD (n=3). *P<0.05; **P<0.01.
LncRNA-ATB Suppresses Activation of Apoptosis-Related Genes
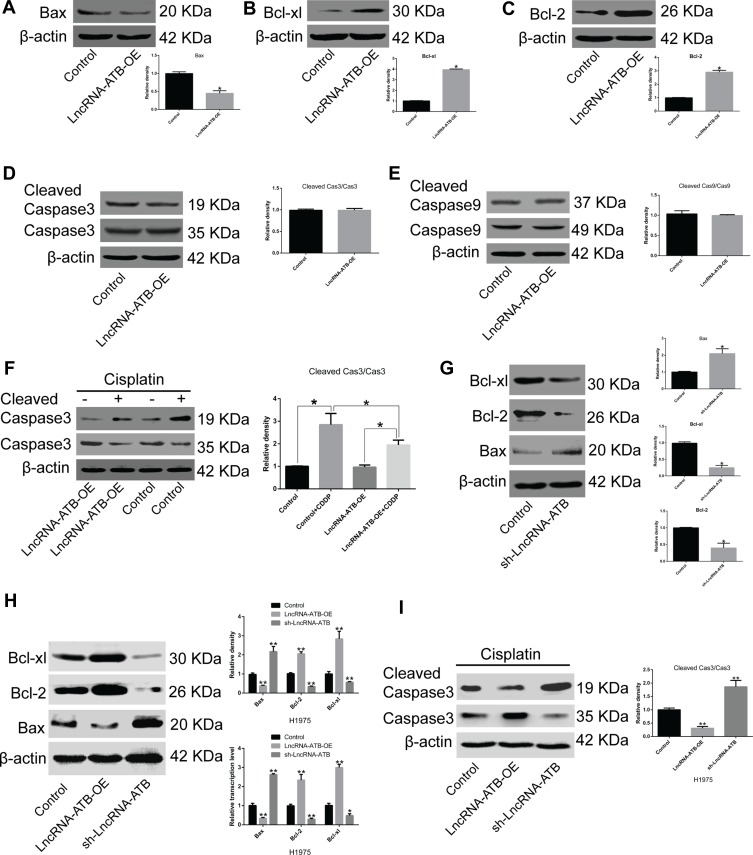
To elucidate the molecular mechanism of lncRNA-ATB in cisplatin resistance, the expression of genes related to apoptosis and cell survival was measured. Bax expression was strikingly attenuated and expression of Bcl-xl and Bcl-2 was significantly up-regulated in A549-ATB-OE (Figure 3A–C) and H1975-ATB-OE (Figure 3H) cells compared with expression levels in matched control groups. Furthermore, there were no significant differences in expression and activation of caspase-3 and caspase-9 in A549-ATB-OE cells (Figure 3D and E). However, when A549-ATB-OE (Figure 3F) and H1975-ATB-OE (Figure 3I) cells were treated with cisplatin, Western blot analysis showed that levels of cleaved caspase-3, the activated form of caspase-3, were significantly reduced compared with levels in cisplatin-treated control cells. In contrast, LUAD cells silencing lncRNA-ATB had significantly down-regulated expression of Bcl-xl and Bcl-2, and significantly up-regulated Bax expression, compared with matched control cells (Figure 3G and H). Collectively, these findings suggested that enforced lncRNA-ATB expression suppressed activation of apoptosis-related genes.
Figure 3.
LncRNA-ATB suppresses activation of apoptosis-related proteins. mRNA and protein expression levels of (A) Bax, (B) Bcl-xl, and (C) Bcl-2 in A549-ATB-OE cells were measured by RT-qPCR and Western blotting. No difference was observed in the activation of (D) caspase-3 and (E) caspase-9 between A549-ATB-OE cells and control cells. (F) Active caspase-3 levels were lower in A549-ATB-OE cells treated with cisplatin than in untreated cells. (G) Protein expression levels of Bax, Bcl-xl and Bcl-2 in A549-ATB-sh cells and control cells. (H) Protein and mRNA expression levels of Bax, Bcl-xl and Bcl-2 in H1975 cells overexpressing or silencing lncRNA-ATB. (I) Ratio of cleaved caspase-3/caspase-3 in H1975 cells overexpressing or silencing lncRNA-ATB following treatment with cisplatin (2 μg/mL). Data are expressed as mean ± SD (n=3). *P<0.05; **P<0.01.
LncRNA-ATB Targets the miR-200a/β-Catenin Pathway
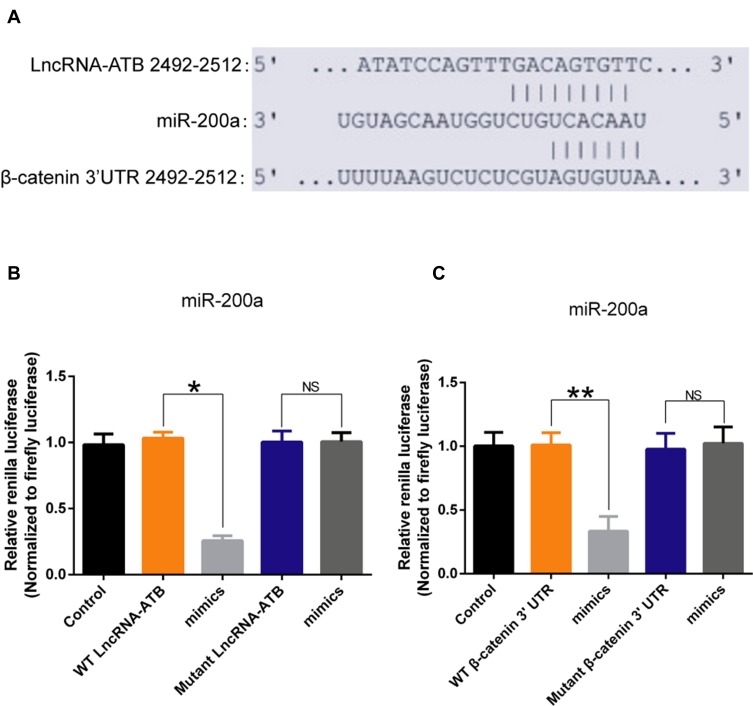
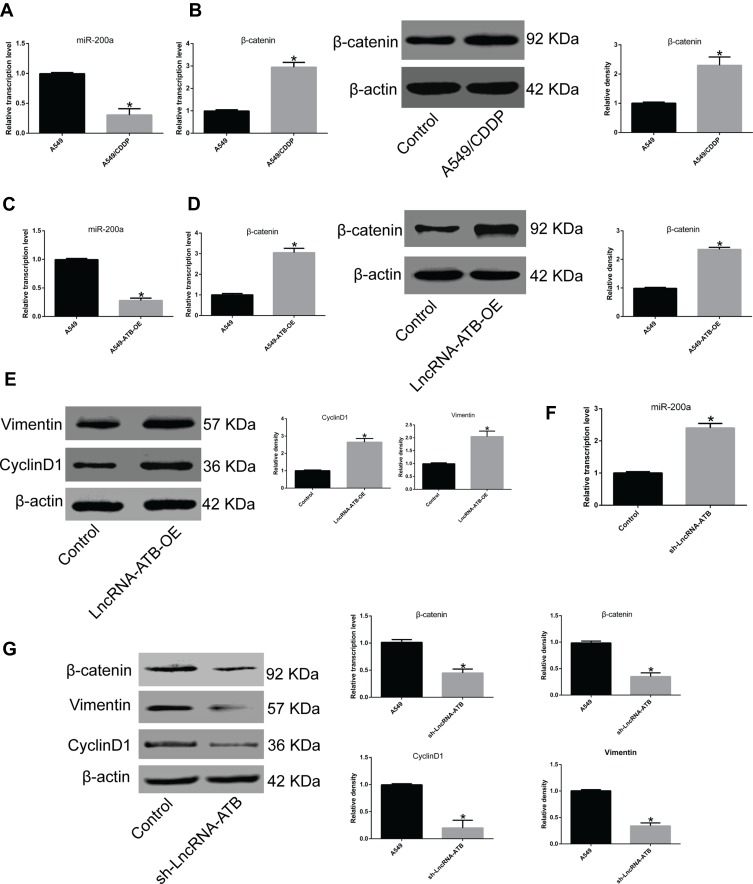
To understand how lncRNA-ATB exerts its biological function, the lncRNA and Disease Database (http://www.cuilab.cn/lncrnadisease) and TargetScan (http://www.targetscan.org/vert_72/) were used to search for miRNAs with complementary bases to lncRNA-ATB. This analysis predicted the binding sites between lncRNA-ATB and miR-200a, and miR-200a and β-catenin (Figure 4A). Luciferase reporter assays revealed that lncRNA-ATB could directly bind to miR-200a (Figure 4B), and miR-200a could directly bind to β-catenin (Figure 4C). The level of miR-200a in A549/CDDP cells was strikingly reduced in comparison to that in A549 cells (Figure 5A), and β-catenin expression was significantly up-regulated (Figure 5B). Moreover, the miR-200a level was confirmed to be significantly down-regulated in A549-ATB-OE cells compared with the level in control cells (Figure 5C), while expression of β-catenin (Figure 5D) and its downstream molecules cyclin D1 and vimentin (Figure 5E) were remarkably increased. In contrast, miR-200a was greatly increased in A549-ATB-sh cells (Figure 5F), whereas expression of β-catenin, cyclin D1 and vimentin was significantly down-regulated in these cells compared with expression in control cells (Figure 5G).
Figure 4.
The interaction between lncRNA-ATB and miR-200a, and miR-200a and its target molecule β-catenin. (A) Predicted binding sites between lncRNA-ATB and miR-200a, and miR-200a and β-catenin. (B) Relative Renilla luciferase activity of constructs with wild-type lncRNA-ATB or its mutant in 293T cells. (C) Relative Renilla luciferase activity of constructs with wild-type 3′UTR of β-catenin or its mutant in 293T cells. Data are expressed as mean ± SD (n=3). *P<0.05; **P<0.01; NS, not significant.
Figure 5.
LncRNA-ATB regulates the miR-200a/β-catenin pathway. (A) miR-200a was significantly down-regulated in A549/CDDP cells compared to A549 cells. (B) β-Catenin expression was greatly up-regulated in A549/CDDP cells compared with A549 cells at both transcriptional and translational levels. (C) miR-200a was strikingly reduced in A549-ATB-OE cells in comparison with control cells. (D) β-Catenin was highly expressed in A549-ATB-OE cells compared with control cells. (E) Vimentin and cyclin D1 expression was decreased in A549-ATB-OE cells compared with control cells. (F) miR-200a was greatly increased in A549-ATB-sh cells compared with control cells. (G) mRNA and protein expression levels of β-catenin, vimentin and cyclin D1 in A549-ATB-sh cells and A549 cells were quantified by RT-qPCR and Western blotting assays. Data are expressed as mean ± SD (n=3). *P<0.05.
Overexpression of miR-200a Down-Regulates β-Catenin Expression
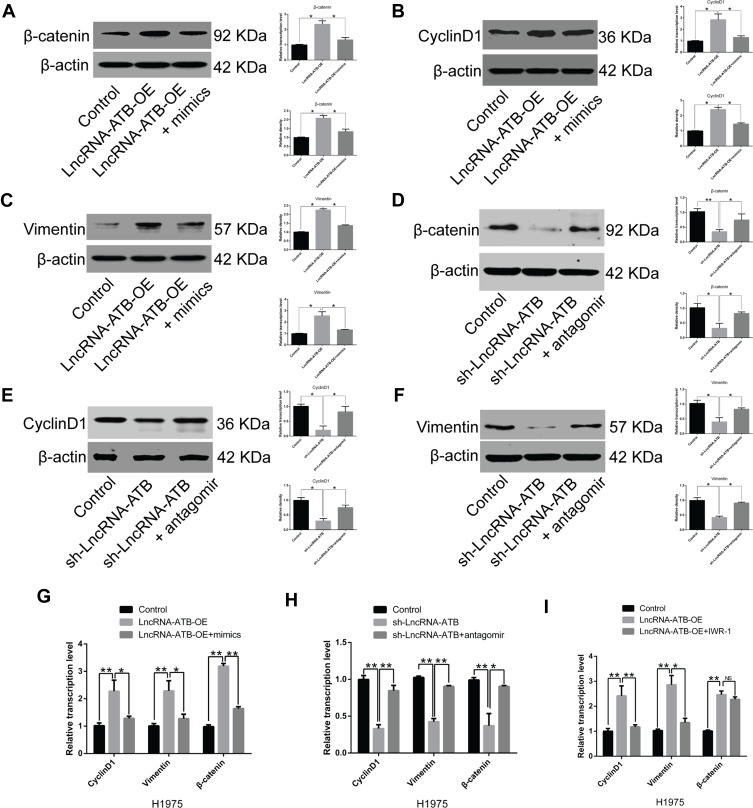
LncRNA-ATB overexpression was found to decrease miR-200a expression, but up-regulate expression of β-catenin, cyclin D1 and vimentin in A549-ATB-OE and H1975-ATB-OE cells compared with matched control cells. After transfection of A549-ATB-OE and H1975-ATB-OE cells with miR-200a mimics (Figure 6A–C and Figure 6G, respectively), the expression levels of β-catenin, cyclin D1 and vimentin were partially reversed as compared with the matched control cells. However, expression of β-catenin, cyclin D1 and vimentin was significantly lower in A549-ATB-sh (Figure 6D–F) and H1975-ATB-sh (Figure 6H) cells than in matched control cells, whereas further transfection with a miR-200a antagomir could elevate expression of these molecules. In addition, H1975-ATB-OE cells were treated with 500 nM IWR-1, a tankyrase inhibitor that blocks the Wnt/β-catenin signaling pathway, for 48 h according to a previous study.22 This IWR-1 treatment markedly attenuated mRNA levels of cyclin D1 and vimentin, whereas there was no significant difference in β-catenin mRNA levels compared with levels in control cells (Figure 6I). Collectively, lncRNA-ATB exerts its biological function by regulating the miR-200a/β-catenin axis, thereby modulating expression of cyclin D1 and vimentin in LUAD cells.
Figure 6.
Effects of miR-200a overexpression on expression of β-catenin, vimentin and cyclin D1. mRNA and protein expression levels of (A) β-catenin, (B) cyclin D1 and (C) vimentin in A549-ATB-OE cells were significantly higher than in control cells, while further treatment with miR-200a mimics (24 h) could partially reverse their expression. Expression of (D) β-catenin, (E) cyclin D1 and (F) vimentin was significantly lower in A549-ATB-sh cells than in control cells, whereas treatment with an miR-200a antagomir could partially reverse their expression. (G) mRNA expression levels of β-catenin, cyclin D1 and vimentin in H1975-ATB-OE cells treated with or without miR-200a mimics. (H) mRNA expression levels of β-catenin, cyclin D1 and vimentin in H1975-ATB-sh cells with or without miR-200a antagomir treatment. (I) mRNA expression levels of β-catenin, cyclin D1 and vimentin in H1975-ATB-OE cells with or without IWR-1 (500 nM) treatment for 48 h. Data are expressed as mean ± SD (n=3). *P<0.05; **P<0.01; NS, not significant.
Overexpression of miR-200a Reduces Cisplatin Resistance of LUAD Cells
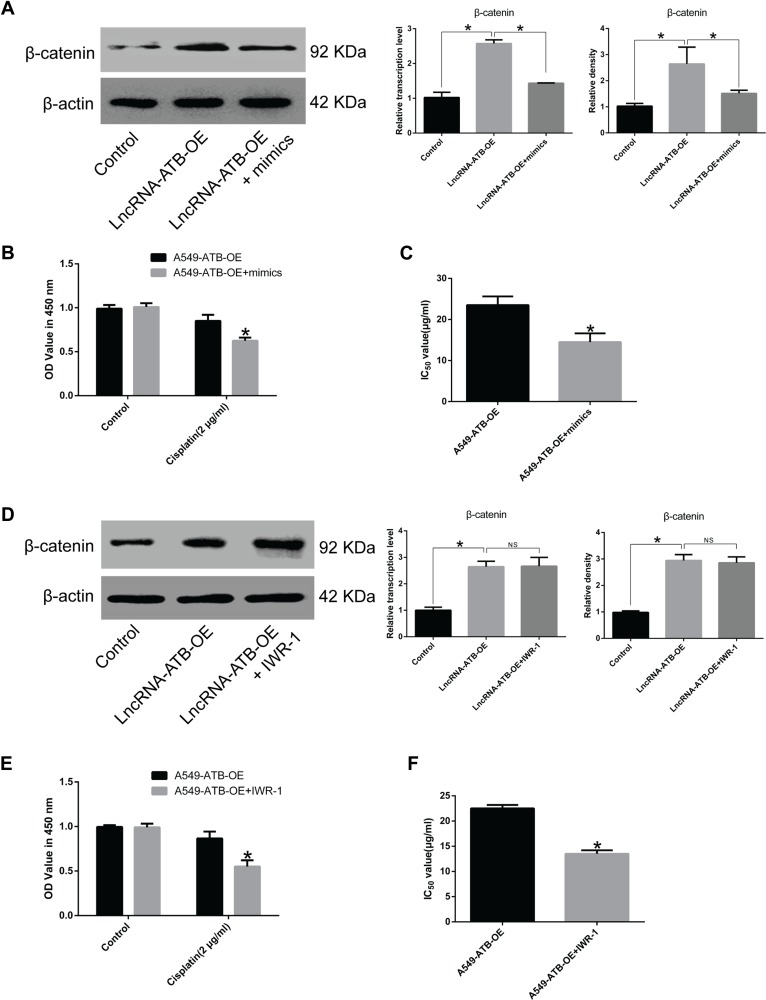
Overexpression of miR-200a in A549-ATB-OE cells inhibited β-catenin expression (Figure 7A). A CCK-8 assay confirmed that miR-200a overexpression increased the sensitivity of A549-ATB-OE cells to cisplatin (Figure 7B), and the IC50 of cisplatin in A549-ATB-OE cells transfected with miR-200a mimics was significantly lower than that measured in A549-ATB-OE cells (Figure 7C). Furthermore, although treatment of A549-ATB-OE cells with 500 nM IWR-1 for 48 h did not significantly alter β-catenin expression (Figure 7D), the sensitivity of the cells to cisplatin was elevated (Figure 7E), and the IC50 of cisplatin was strikingly reduced compared with untreated cells (Figure 7F). Collectively, these findings indicated that lncRNA-ATB plays a role in cisplatin resistance of LUAD cells, and lncRNA-ATB exerts its function through the miR-200a/β-catenin axis.
Figure 7.
Effects of miR-200a overexpression on cisplatin resistance of LUAD cells. (A) β-Catenin expression in A549-ATB-OE cells transfected with or without miR-200a mimics was detected using RT-qPCR and Western blot assays. (B) Proliferation of A549-ATB-OE cells transfected with or without miR-200a mimics following cisplatin treatment (2 μg/mL) for 48 h was validated by CCK-8 assay. (C) IC50 of cisplatin in A549-ATB-OE cells transfected with or without miR-200a mimics. (D) β-Catenin expression in A549-ATB-OE cells treated with or without Wnt/β-catenin signaling pathway inhibitor IWR-1 (500 nM) for 48 h. (E) Proliferation of A549-ATB-OE cells treated with or without IWR-1 (500 nM) for 48 h following cisplatin treatment (2 μg/mL) for 48 h was validated by CCK-8 assay. (F) IC50 of cisplatin in A549-ATB-OE cells treated with or without IWR-1 (500 nM) for 48 h. Data are expressed as mean ± SD (n=3). *P<0.05; NS, not significant.
Discussion
LUAD is the most common lung cancer subtype.23 Surgery combined with adjuvant therapy is currently the first-line treatment for lung cancer. Platinum-based (cisplatin) chemotherapy is more common in patients with advanced lung cancer;24 however, cisplatin resistance in lung cancer cells leads to treatment failure.25
LncRNA-ATB is a recently discovered lncRNA related to tumor progression, whereby lncRNA-ATB promotes the proliferation, migration and invasion of LUAD, gastric cancer and prostate cancer cells.26 However, the role of lncRNA-ATB in drug resistance of LUAD has not been elucidated. In the present study, lncRNA-ATB was significantly up-regulated in A549/CDDP (cisplatin-resistant) cells. Overexpression of lncRNA-ATB promoted cisplatin resistance in LUAD cells, while lncRNA-ATB silencing significantly sensitized A549 cells to cisplatin, suggesting that lncRNA-ATB plays a role in cisplatin resistance of LUAD. Cisplatin treatment contributes to apoptosis of tumor cells.27 Further experiments in the present study revealed that the apoptotic rate of A549 and H1975 cells overexpressing lncRNA-ATB was significantly down-regulated following cisplatin treatment, while the apoptotic rate of lncRNA-ATB-silenced A549 cells was greatly increased. Mechanistically, overexpression or silencing of lncRNA-ATB significantly alters expression levels of the apoptosis-related genes Bcl-cl, Bax and Bcl-2, and inhibits activation of caspase-3 and caspase-9, which are both key molecules in apoptosis.28,29
miR-200a targets multiple genes to regulate the biological functions of tumor cells, and β-catenin is one of these regulated genes.30 A key molecule in the Wnt signaling pathway, β-Catenin has a significant impact on tumor resistance.31,32 A previous study implicated the lncRNA-ATB/miR-200a/β-catenin regulatory axis in hepatitis C virus (HCV)-related liver fibrosis.33 In addition, work in glioma tissues and cell lines revealed that lncRNA-ATB promoted glioma malignancy by negatively regulating miR-200a.34 Findings from the present study indicate that enforced expression of lncRNA-ATB down-regulates miR-200a, thereby increasing expression of β-catenin, which may contribute to the survival of tumor cells under cisplatin treatment. To further confirm the association between lncRNA-ATB and miR-200a or β-catenin, experiments using miR-200a mimics and the Wnt/β-catenin signaling pathway inhibitor IWR-1 were conducted. β-Catenin expression was strikingly reduced in A549 and H1975 cells overexpressing lncRNA-ATB and treated with miR-200a mimics. In addition, IWR-1 treatment significantly increased the sensitivity of A549/CDDP cells to cisplatin, indicating that lncRNA-ATB regulates the miR-200a/β-catenin axis, and this is likely to be responsible for cisplatin resistance in LUAD. However, further work is needed to confirm the interactions between lncRNA-ATB/miR-200a/β-catenin axis and cisplatin resistance in LUAD.
In summary, this study revealed that lncRNA-ATB was highly expressed in A549/CDDP (cisplatin-resistant) cells. Overexpression of lncRNA-ATB decreased the chemosensitivity of A549 and H1975 cells, whereas silencing of lncRNA-ATB increased it. The function of lncRNA-ATB in LUAD cells is partially exerted via sponging of miR-200a, resulting in inhibition of β-catenin expression. This study provides the novel insight that dysregulation of the lncRNA-ATB/miR-200a/β-catenin axis may be one of the key mechanisms of cisplatin resistance in LUAD.
Acknowledgments
We wish to thank Dr. Ye Feng for his technical guidance and critical reading of the manuscript.
Funding Statement
This study was supported by the National Natural Science Foundation of China (Grant No.81702414), National Natural Science Foundation of China (Grant No.81802323), Natural Science Foundation of Fujian Province of China (Grant No.2017Y0084), Natural Science Foundation of Fujian Province (Grant No. 2016J01636), Xiamen Science and Technology Planning Project (Grant No. 3502Z20194003) and National Major Scientific and Technological Special Project for “Significant New Drugs Development” (2020ZX09201005).
Author Contributions
Study concept and design: Lilin Chen. Performed the experiments: Weiwei Tang and Xiuyi Yu and Ru Zeng. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
All authors declare no conflicts of interest in this work.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 3.Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018;9(2):117. doi: 10.1038/s41419-017-0063-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemjabbar-Alaoui H, Hassan OU, Yang Y-W, Buchanan P. Lung cancer: biology and treatment options. Biochim Biophys Acta. 2015;1856(2):189–210. doi: 10.1016/j.bbcan.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edelman MJ. Highlights in the treatment of lung cancer: what’s new? Not much. JAMA oncol. 2017;3(3):301–302. doi: 10.1001/jamaoncol.2016.3984 [DOI] [PubMed] [Google Scholar]
- 6.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. [DOI] [PubMed] [Google Scholar]
- 7.Bilfinger T, Keresztes R, Albano D, Nemesure B. Five-year survival among Stage IIIA lung cancer patients receiving two different treatment modalities. Med Sci Monitor. 2016;22:2589–2594. doi: 10.12659/MSM.898675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stenzel-Bembenek A, Sagan D, Guz M, Stepulak A. [Single nucleotide polymorphisms in lung cancer patients and cisplatin treatment]. Postepy Hig Med Dosw (Online). 2014;68:1361–1373. Polish. doi: 10.5604/17322693.1129820 [DOI] [PubMed] [Google Scholar]
- 9.Hughes BG, Ahern E, Lehman M, et al. Concurrent chemoradiation with cisplatin and vinorelbine followed by consolidation with oral vinorelbine in locally advanced non-small cell lung cancer (NSCLC): the Phase II CONCAVE study. Asia Pac J Clin Oncol. 2017;13(3):137–144. doi: 10.1111/ajco.2017.13.issue-3 [DOI] [PubMed] [Google Scholar]
- 10.Cortes-Funes H. New treatment approaches for lung cancer and impact on survival. Semin Oncol. 2002;29(3 Suppl 8):26–29. doi: 10.1053/sonc.2002.33530 [DOI] [PubMed] [Google Scholar]
- 11.Qin X, Yu S, Zhou L, et al. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. Int J Nanomedicine. 2017;12:3721–3733. doi: 10.2147/IJN.S131516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Lin Z, Wang Y, et al. MiR-5100 increases the cisplatin resistance of the lung cancer stem cells by inhibiting the Rab6. Mol Carcinog. 2018;57(3):419–428. doi: 10.1002/mc.v57.3 [DOI] [PubMed] [Google Scholar]
- 13.Guo F, Cao Z, Guo H, Li S. The action mechanism of lncRNA-HOTAIR on the drug resistance of non-small cell lung cancer by regulating Wnt signaling pathway. Exp Ther Med. 2018;15(6):4885–4889. doi: 10.3892/etm.2018.6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang MS, Liu JY, Xia XB, et al. Hsa_circ_0001946 inhibits lung cancer progression and mediates cisplatin sensitivity in non-small cell lung cancer via the nucleotide excision repair signaling pathway. Front Oncol. 2019;9:508. doi: 10.3389/fonc.2019.00508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Li Z, Zheng W, et al. LncRNA-ATB: an indispensable cancer-related long noncoding RNA. Cell Prolif. 2017;50:6. doi: 10.1111/cpr.12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue B, Qiu S, Zhao S, et al. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol. 2016;31(3):595–603. doi: 10.1111/jgh.2016.31.issue-3 [DOI] [PubMed] [Google Scholar]
- 17.Wei L, Wu T, He P, Zhang JL, Wu W. LncRNA ATB promotes the proliferation and metastasis of lung cancer via activation of the p38 signaling pathway. Oncol Lett. 2018;16(3):3907–3912. doi: 10.3892/ol.2018.9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar MM, Goyal R. LncRNA as a therapeutic target for angiogenesis. Curr Top Med Chem. 2017;17(15):1750–1757. doi: 10.2174/1568026617666161116144744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao Y, Luo X, Ding X, Cui S, Guo C. LncRNA ATB promotes proliferation and metastasis in A549 cells by down-regulation of microRNA-494. J Cell Biochem. 2018;119(8):6935–6942. doi: 10.1002/jcb.v119.8 [DOI] [PubMed] [Google Scholar]
- 20.Ke L, Xu SB, Wang J, Jiang XL, Xu MQ. High expression of long non-coding RNA ATB indicates a poor prognosis and regulates cell proliferation and metastasis in non-small cell lung cancer. Clin trans oncol. 2017;19(5):599–605. doi: 10.1007/s12094-016-1572-3 [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Holden-Wiltse J, Wang J, Liang H. A strategy to model nonmonotonic dose-response curve and estimate IC50. PLoS One. 2013;8(8):e69301. doi: 10.1371/journal.pone.0069301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busch AM, Johnson KC, Stan RV, et al. Evidence for tankyrases as antineoplastic targets in lung cancer. BMC Cancer. 2013;13:211. doi: 10.1186/1471-2407-13-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inamura K. Clinicopathological characteristics and mutations driving development of early lung adenocarcinoma: tumor initiation and progression. Int J Mol Sci. 2018;19:4. doi: 10.3390/ijms19041259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuwen D, Mi S, Ma Y, et al. Andrographolide enhances cisplatin-mediated anticancer effects in lung cancer cells through blockade of autophagy. Anticancer Drugs. 2017;28(9):967–976. doi: 10.1097/CAD.0000000000000537 [DOI] [PubMed] [Google Scholar]
- 25.Dai CH, Chen P, Li J, et al. Co-inhibition of pol theta and HR genes efficiently synergize with cisplatin to suppress cisplatin-resistant lung cancer cells survival. Oncotarget. 2016;7(40):65157–65170. doi: 10.18632/oncotarget.11214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao H, Zhang F, Zou Y, Li J, Liu Y, Huang W. The function and mechanism of long non-coding RNA-ATB in Cancers. Front Physiol. 2018;9:321. doi: 10.3389/fphys.2018.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pabla N, Dong Z. Cisplatin nephrotoxicity: mechanisms and renoprotective strategies. Kidney Int. 2008;73(9):994–1007. doi: 10.1038/sj.ki.5002786 [DOI] [PubMed] [Google Scholar]
- 28.Fan TJ, Han LH, Cong RS, Liang J. Caspase family proteases and apoptosis. Acta Biochim Biophys Sin (Shanghai). 2005;37(11):719–727. doi: 10.1111/j.1745-7270.2005.00108.x [DOI] [PubMed] [Google Scholar]
- 29.Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9(3):459–470. doi: 10.1016/S1097-2765(02)00482-3 [DOI] [PubMed] [Google Scholar]
- 30.Hu B, Qiu-Lan H, Lei RE, Shi C, Jiang HX, Qin SY. Interleukin-9 promotes pancreatic cancer cells proliferation and migration via the miR-200a/beta-catenin axis. Biomed Res Int. 2017;2017:2831056. doi: 10.1155/2017/2831056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emons G, Spitzner M, Reineke S, et al. Chemoradiotherapy resistance in colorectal cancer cells is mediated by Wnt/beta-catenin signaling. Mol Cancer Res. 2017;15(11):1481–1490. doi: 10.1158/1541-7786.MCR-17-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Liu HC, Wang C, et al. Overexpression of beta-catenin induces cisplatin resistance in oral squamous cell carcinoma. Biomed Res Int. 2016;2016:5378567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu N, Zhao SX, Kong LB, et al. LncRNA-ATB/microRNA-200a/beta-catenin regulatory axis involved in the progression of HCV-related hepatic fibrosis. Gene. 2017;618:1–7. doi: 10.1016/j.gene.2017.03.008 [DOI] [PubMed] [Google Scholar]
- 34.Ma CC, Xiong Z, Zhu GN, et al. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J Exp Clin Cancer Res. 2016;35(1):90. doi: 10.1186/s13046-016-0367-2 [DOI] [PMC free article] [PubMed] [Google Scholar]