Abstract
Small interfering RNAs (siRNAs) are potential therapeutic substances due to their gene silencing capability as exemplified by the recent approval by the US Food and Drug Administration (FDA) of the first siRNA therapeutic agent (patisiran). However, the delivery of naked siRNAs is challenging because of their short plasma half-lives and poor cell penetrability. In this study, we used vesicles made from bolaamphiphiles (bolas), GLH-19 and GLH-20, to investigate their ability to protect siRNA from degradation by nucleases while delivering it to target cells, including cells in the brain. Based on computational and experimental studies, we found that GLH-19 vesicles have better delivery characteristics than do GLH-20 vesicles in terms of stability, binding affinity, protection against nucleases, and transfection efficiency, while GLH-20 vesicles contribute to efficient release of the delivered siRNAs, which become available for silencing. Our studies with vesicles made from a mixture of the two bolas (GLH-19 and GLH-20) show that they were able to deliver siRNAs into cultured cancer cells, into a flank tumor and into the brain. The vesicles penetrate cell membranes and the blood-brain barrier (BBB) by endocytosis and transcytosis, respectively, mainly through the caveolae-dependent pathway. These results suggest that GLH-19 strengthens vesicle stability, provides protection against nucleases, and enhances transfection efficiency, while GLH-20 makes the siRNA available for gene silencing.
Keywords: Bolaamphiphiles, Vesicles, Delivery Systems, siRNA, Targeting, Blood Brain Barrier (BBB), Transfection, Gene Silencing, Molecular Dynamics, RNA Nanotechnology
Graphical Abstract
Introduction
RNA interference (RNAi), a process of sequence-specific post-transcriptional gene silencing, has been intensively studied due to its therapeutic potential,1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 as confirmed by the US Food and Drug Administration (FDA) recently approved small interfering RNA (siRNA) therapeutic agent (patisiran).14 RNAi can be triggered by siRNAs, which are capable of silencing specific genes.15 Therefore, siRNA can be used to treat cancer and other diseases by injecting the synthesized sequences of siRNA that silence specific target genes. However, in order to be functional, naked siRNAs have to overcome critical biological barriers during the delivery process. First, naked unmodified siRNA molecules are significantly degraded in blood circulation and in tissues by RNases before they reach the target cells. In addition, the typical size of an siRNA molecule, which contains 21–23 nt in each strand, can be easily filtered and excreted by the kidney.16,17 Even if the siRNA reaches the target cell after surviving the nucleases and the elimination processes, its negative charge prevents its adherence to the negatively charged cell membrane and, thus, its penetration into the cell is perturbed.18 To overcome these biological barriers, siRNA requires delivery agents that (1) have positively charged atoms to neutralize the negatively charged backbone of the siRNA, (2) have an ability for efficient cell internalization, (3) efficiently bind to siRNA, (4) are capable of protecting the siRNA against nuclease degradation in blood, (5) avoid rapid filtration by kidney and liver accumulation, and (6) efficiently release siRNA into the cytoplasm after penetrating into the target cell.
Examples of delivery carriers are liposomes,19 polymers,3 magnetic nanoparticles,20 protein particles, dendrimers, inorganic materials, and viral particles.21, 22, 23, 24 In addition, instead of using plain siRNAs, RNA nanostructures have been studied and have shown potential for controlled and efficient delivery of RNAi inducers.25
In our previous studies, we synthesized bolaamphiphiles (bolas) with positively charged head groups that have relatively low toxicity and high stability in vivo and in vitro.26,27 We have shown that such bolas form vesicles that can encapsulate and deliver a variety of compounds, such as small molecules, peptides, and proteins, across the blood-brain barrier (BBB) into the brain.28, 29, 30, 31 We also examined the possibility of delivering siRNAs and RNA nanoparticles functionalized with RNAi inducers by complexing them with bola micelles. In these studies, we used four different bolas: GLH-19, GLH-20, GLH-58, and GLH-60.32, 33, 34, 35 In general, these bolas consist of hydrophilic head groups that are connected to a hydrophobic chain. The GLH-19 head groups are attached by the acetyl group of acetylcholine (ACh), while those of GLH-20 are attached via the nitrogen atom of the choline moiety (see molecular structures of GLH-19 and GLH-20 in Figure S1). GLH-20 has a unique feature, as its ACh head group can be hydrolyzed by choline esterase (ChE).29,30 In addition, vesicles made from GLH-20 penetrate the BBB and release drugs that are associated with the vesicles via vesicle destabilization due to hydrolysis of the GLH-20 head groups that occurs when vesicles are exposed to tissues that highly express ChE, such as the brain.29 GLH-58 and GLH-60 have two and four ACh head groups, respectively, which are connected to the two ends of a relatively short hydrophobic aliphatic chain via the nitrogen atom of the choline moiety.33
In previous studies, we formed micelles from the bolas complexed with siRNAs and studied the effect of the geometry and total charge of the bola head groups and the length of the hydrophobic chain on the binding affinity of the micelle to the siRNA, the ability of the micelles to protect siRNA against RNase, the ability of the micelles to transfect siRNA efficiently, and the ability to release the siRNA from the bola/siRNA complex for gene silencing.32,33 In addition to these findings, which are related to the interactions between siRNA and the bolas, we also found that, due to their long hydrophobic chains, GLH-19 and GLH-20 can form stable vesicles.26,27 In comparison, both GLH-58 and GLH-60 do not form stable vesicles because of their short hydrophobic chains and the large amount of positive charge per molecule.33
Inspired by previous work, we now focused on cationic vesicles made from bolas for their efficiency in delivering siRNA through biological barriers, particularly into the brain and tumors. Using computational and experimental methods, we examined the properties of vesicles made from various formulations containing the bolas GLH-19 and GLH-20 together with stabilizing components with respect to their stability, protection of associated siRNA from nucleases, transfection, and silencing capability, as well as in vivo siRNA distribution following intravenous injection of GLH-19/GLH-20/siRNA vesicles into mice. Our in silico and in vitro studies showed that cholesterol (CHOL) and cholesteryl hemisuccinate (CHEMS) are required to stabilize vesicle formation. In addition, we found that in comparison to GLH-20 vesicles, GLH-19 vesicles show better stability, provide better protection for siRNA against the action of RNases, and enable better transfection into cells. However, both vesicles showed independently similar silencing efficiencies. However, when the two bolas, GLH-19 and GLH-20, were mixed together to form vesicles, GLH-19 increased the stability, the binding affinity, the protection of the siRNA from the action of nucleases, and the transfection efficiency, while GLH-20 allowed the GLH-19/GLH-20/siRNA vesicles to deliver the associated siRNAs into the mouse brain across the BBB and possibly make them available for gene silencing.
Results
MD Simulations of Bola Vesicles
The Stability of Bola Membranes
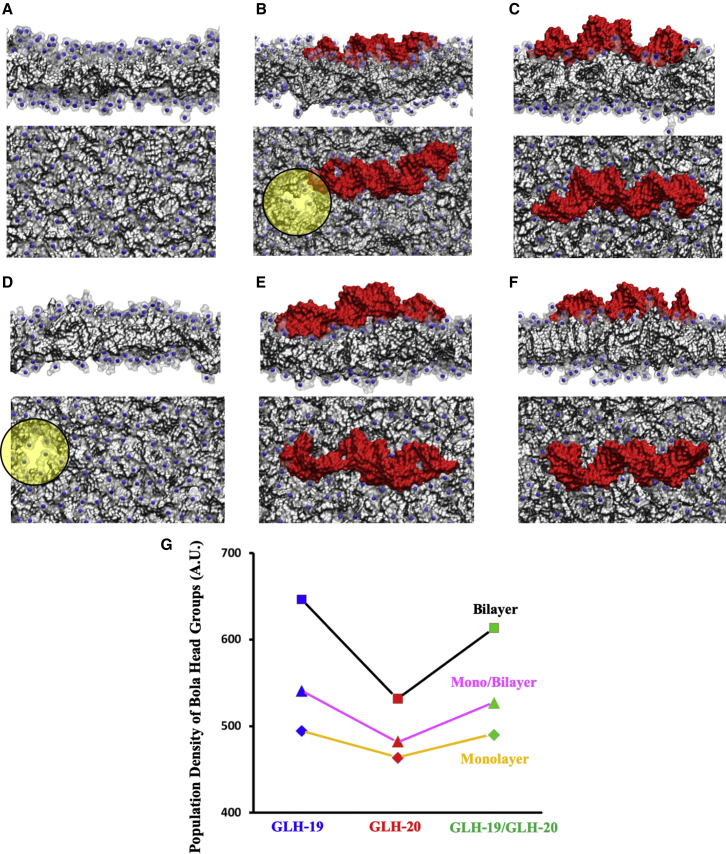
The stability of bola membranes was examined by molecular dynamics (MD) simulations with and without the presence of CHOL and CHEMS. CHOL is electrostatically neutral and is able to aggregate with CHEMS and bolas via hydrophobic interactions. CHEMS is also hydrophobic but has negatively charged oxygen due to deprotonation (Figure S1). In the absence of CHOL and CHEMS, the GLH-19 bola monolayer maintained a stable membrane conformation for 100 ns (Figure 1A), while the bilayer was significantly bent (Figure S3). These results indicate that the high density of the GLH-19 head groups on the bilayer surface triggered destabilization of the balance between the hydrophilic surface and the hydrophobic membrane body, resulting in severe bending. The surface densities of the bola head groups are summarized in Table S2. Although the GLH-19 monolayer maintained the membrane conformation without CHOL and CHEMS when an siRNA was associated with its surface, the electrostatic interactions between the siRNA and the bola head groups destabilized the membrane surface and caused a water channel near one end of the siRNA (Figure 1B). Our dynamic light scattering (DLS) measurements and cryogenic transmission electron microscopy (cryo-TEM) images also showed the collapse of the pure GLH-19 vesicles due to their association with siRNA (data not shown). When CHOL and CHEMS were introduced, monolayer, bilayer, and the mixture of monolayer/bilayer conformations of GLH-19 remained stable regardless of the association with siRNA (see Figures 1C and S4).
Figure 1.
Molecular Dynamics Shows Differences in Membrane Stability and siRNA Interaction that is Dependent Upon Composition
(A–F) The side and top view of membrane snapshots of (A) GLH-19 monolayer without CHOL and CHEMS, (B) GLH-19/siRNA without CHOL and CHEMS, (C) GLH-19/siRNA with CHOL and CHEMS, (D) GLH-20 monolayer without CHOL and CHEMS, (E) GLH-20/siRNA with CHOL and CHEMS, and (F) GLH-19/GLH-20/siRNA with CHOL and CHEMS. Gray regions, transparent surfaces, and blue spheres represent hydrophobic areas, head groups, and positively charged nitrogen (N) atoms, respectively. Water holes are marked with yellow circles. siRNA is represented by a red surface. (G) Total population density of bola head groups near siRNA phosphate groups.
In silico studies showed that GLH-20 membranes without CHOL and CHEMS were quickly destabilized and collapsed. Without CHOL and CHEMS, a water channel was created in the GLH-20 monolayer at the early stage of the MD simulation (Figure 1D) while the bilayer membrane was significantly distorted in a 30-ns MD simulation (see Figure S3). However, when CHOL and CHEMS were included, the monolayer, bilayer, and monolayer/bilayer mixtures all maintained stable membrane conformations regardless of the association with siRNA (see Figures 1E and S5). The enhancement of the stability of GLH-20 vesicles due to the presence of CHOL and CHEMS was also experimentally monitored (Figure 2A).
Figure 2.
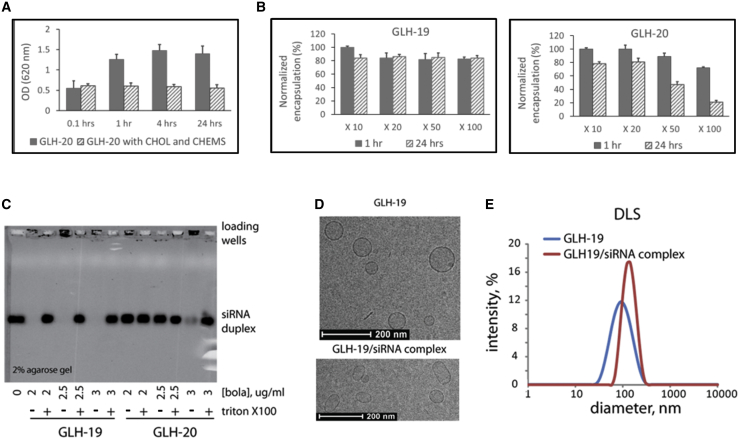
Characterization of the Bola Vesicles in Terms of Stability, Affinity to siRNA, Morphology, and Size Distribution
(A) Stability of the vesicles in solution as measured by the development of turbidity due to vesicle aggregation. Vesicles were made from 10 mg/mL GLH-20 without CHOL and CHEMS (filled bars) and from 10 mg/mL GLH-20 with CHOL and CHEMS at a molar ratio of 2:1:1 (striated bars). Turbidity was measured by absorbance at 620 nm. (B) Stability of vesicles after dilution. Vesicles were prepared from 10 mg/mL GLH-19 (left panel) and GLH-20 (right panel) in PBS/10 buffer containing 0.1 mg/mL CF. Percent CF encapsulation was measured immediately (filled bars) and 24 h (striated bars) after dilution. A series of dilutions (10- to 100-fold) was done in PBS/10. (C) Agarose gel electrophoresis of GLH-19/siRNA and GLH-20/siRNA vesicles. siRNA (400 nM final concentration) was mixed with the bola at final amounts, which are indicated in the figure (in μg). For each amount of bola, 0.1% Triton X-100 was added in order to disintegrate the vesicles and release the encapsulated siRNA. (D) Cryo-TEM images of GLH-19 and GLH-19/siRNA vesicles. (E) Size distribution measured by dynamic light scattering (DLS) indicates average diameters for GLH-19 and GLH-19/siRNA to be ∼85 and ∼125 nm, respectively. These sizes are in a good agreement with the observation made by the cryo-TEM. All tested bola vesicles in (B)–(E) contain CHOL and CHEMS. Note that all depicted error bars (A and B) represent the mean of 3 replicates + or - the standard deviation (SD).
When GLH-19 and GLH-20 were mixed together (designated as GLH-19/GLH-20) to form a monolayer, bilayer, and monolayer/bilayer, all of these systems maintained stable membrane conformations in the presence of CHOL and CHEMS, regardless of the association with siRNA (Figures 1F and S6).
Thus, our MD simulations indicate that without CHOL and CHEMS, the strong electrostatic repulsion between the positively charged head groups destabilized the membrane and caused a water channel to penetrate the membrane or induce severe membrane bending. However, when CHOL and CHEMS were introduced for vesicle formation, the stability of the membrane increased due to the enhanced hydrophobic interactions and reduced electrostatic repulsion caused by the negatively charged CHEMS that interacted with the positively charged bola head groups. The electrosttic surface potentials of the bola membranes in Figure S7 show the effect of the electrostatic relaxation due to CHOL and CHEMS. This was shown also to be the case experimentally (see The Stability of Bola Membranes above and Figure 2A).
Cl− Ion Distribution on the Surface of Bola Membranes
The surface properties of each bola membrane were characterized by the mobility of the bola head groups and the average occupation time of Cl− ions in the vicinity of the membrane surface. The distributions of the bola head groups on each membrane surface are summarized in Table S2 and Figure S8. Since the length of the GLH-19 head group is longer than that of GLH-20, the positively charged nitrogen (N) atoms of GLH-19 extend farther from the membrane surface than do those of GLH-20. In addition, GLH-19 head groups have 5- and 1.5-fold higher linear and angular mobility, respectively, than do those of GLH-20. Interestingly, the characteristics of the bola head group mobility are independent of the membrane conformation as well as of the components of the membranes. For instance, when GLH-19 and GLH-20 were mixed, the dynamics of the GLH-19 head groups did not interfere with those of GLH-20, regardless of the membrane conformations, that is, monolayer, bilayer, or monolayer/bilayer (see Table S1 and Figure S8).
The average occupancy time of Cl− ions in the vicinity of the membrane surface was measured by the residence time of Cl− ions that remained in the vicinity (12 Å) of the positively charged nitrogen atoms during 5-ns intervals from 50 to 100 ns (Figure S2). Thus, the maximum residence time of Cl− ions that was monitored was 5 ns. The average occupancy time of Cl− ions was normalized by the total number of nitrogen atoms in the bola and Cl− ions in each system. The average Cl− ion density was the highest on the GLH-20 surface, medium on the GLH-19/GLH-20 surface, and the lowest on the GLH-19 surface (Figure S9). In addition, the bilayer surface had the highest, that of the monolayer/bilayer mixture had medium, and the monolayer had the lowest Cl− ions density. Furthermore, the higher surface density of the bola head groups on the bilayer surface among the test groups provided a more stable electrostatic attraction for the Cl− ions. These results indicate that the lower mobility of GLH-20 head groups induced a denser Cl− ion cloud on its surface than that of GLH-19. Therefore, our in silico studies explain the weaker siRNA association with GLH-20 than with GLH-19 in gels (see Affinity of the Bolas to siRNA below).
MD Simulations of Bola/siRNA Complexes
The association of the siRNA with different conformations of bola membranes was characterized by the population density of each bola head group near the siRNA phosphate groups (see Figure S2C). The number and population density of bola head groups in the vicinity of each phosphate atom (Pi) were monitored for the last 30 ns of each MD trajectory (70–100 ns range) and are plotted in Figure S10. Both GLH-19 and GLH-20 head groups were highly populated in the area of the minor grooves. In the major groove regions, small amounts of the GLH-19 head group populations were still observed due to the relatively long length and high mobility of the GLH-19 head groups. However, the major groove of the siRNA near the GLH-20 membrane was more exposed to water than that of GLH-19 because the relatively short GLH-20 head groups on the membrane surface were not able to reach the major groove. Therefore, independent of the orientation of the siRNA, GLH-19 showed better coverage of siRNA than did GLH-20.
The total population density of the bola head groups around the siRNA phosphate groups (∑Pi, where i indicates all phosphate groups in siRNA) is displayed in Figure 1G. For each tested group, the GLH-19 membranes showed the highest head group population densities around the siRNA while those of the GLH-20 membranes had the lowest. When both bolas were mixed together, due to the GLH-19, the population density of the GLH-19/GLH-20 head groups around the siRNA phosphate groups was increased compared to that of the GLH-20 membranes. In addition, bilayer membranes, which have the highest surface density of bola head groups, showed the highest population of head group density around the siRNA phosphate groups while those of the monolayer had the smallest. Therefore, these results indicate that GLH-19 head groups in bilayer membrane conformations can provide better siRNA coverage than do other test groups. This in silico study explains why GLH-19 and GLH-19/GLH-20 vesicles showed better protection of vesicle-associated DNA against DNase than did GLH-20 vesicles (see Protection against Nucleases below).
In Vitro Characterization of the Bola Vesicles
Vesicle Stability in Solution
The stability of vesicles that were made from GLH-20 with and without CHOL and CHEMS in solution was compared. As shown in Figure 2A, vesicles made from a formulation that contained the bola without CHOL and CHEMS aggregated rapidly, as shown by the increased absorbance of the vesicle suspension at 620 nm over time. The inclusion of CHOL and CHEMS in the vesicle formulation resulted in a stable vesicle suspension that did not aggregate and maintained its stability for at least 24 h after vesicle preparation. Our in silico studies also showed that CHOL and CHEMS are required for vesicle stability (see The Stability of Bola Membranes above). As a measure of vesicle stability, we also looked at the amount of carboxyfluorescein (CF) encapsulation at various times after vesicle preparation with CHOL and CHEMS. We found that the vesicles maintained their encapsulated CF for a long period (up to 6 months after preparation, which was our last measurement) when the vesicles were kept at a high concentration of 10 mg of bolas per mL (data not shown). However, we noticed that the GLH-20 vesicles lost their stability when they were diluted. Since our MD simulations indicated that GLH-19 vesicles may be more stable than GLH-20 vesicles, we compared the stability of GLH-19 and GLH-20 vesicles after diluting them to various degrees. As shown in Figure 2B, when both bolas were diluted 100-fold for 24 h, the GLH-19 vesicles maintained their encapsulated CF while the GLH-20 vesicles lost almost all encapsulated CF.
Based on these results, all bola vesicles that were associated with siRNA described below contain CHOL and CHEMS.
Affinity of the Bolas to siRNA
To study the binding affinity of the bolas to siRNA, we studied the association of 400 nM siRNA with three different concentrations of bolas (2.0, 2.5, and 3.0 μg/mL). The results, which are presented in Figure 2C, show that the affinity of GLH-19 to siRNA is significantly greater than that of GLH-20. For all three concentrations, siRNA was associated with GLH-19 vesicles and did not migrate on the electrophoretic gel as free siRNA. The siRNA was dissociated from the GLH-19 vesicles only after adding the detergent Triton X-100 that disrupted the vesicles. In comparison, it was found that siRNA was associated with GLH-20 only at the highest concentration that was used in this experiment (3.0 μg/mL). We estimated the approximate percentage of loading efficiency as follows: the molecular mass of bolas is around 1,200 Da, which means that 2 μg/mL corresponds to 1,666 nM. As shown in Figure 2C, 400 nM siRNA is fully associated with 1,666 nM GLH-19, meaning that the loading efficiency for GLH-19 is at least 24%, whereas 400 nM siRNA is associated (partially) with 3 μg/mL GLH-20 (equal to 2,500 nM GLH-20), namely, less than 16% the loading efficiency for GLH-20. Our MD simulations explained the differences in the binding affinities between GLH-19 and GLH-20. GLH-20 vesicles induce higher density and more stable Cl− ions on their surface than on the GLH-19 surface, so that siRNA cannot easily associate with the GLH-20 surface at low concentrations (see Cl− Ion Distribution on the Surface of Bola Membranes above).
Cryo-TEM and DLS Measurements
Cryo-TEM showed that both GLH-19 and GLH-20 formed spherical vesicles with average diameters of 110 nm (see, for example, the GLH-19 vesicles in Figure 2D). When siRNAs were associated with the GLH-19 vesicles, the overall shape of the vesicles was slightly deformed to an oval shape (Figure 2D), and the zeta potential (Figure S11) of the vesicles (∼37 mV) decreased when siRNA was added (∼32 mV). DLS measurements also showed consistent results with the cryo-TEM (Figure 2E). Without siRNAs, the diameter of GLH-19 vesicles was measured to be 85 nm while it was increased to 125 nm when siRNAs were associated with the GLH-19 vesicles. These results indicate that the association with siRNA may alter the surface tension of the bola vesicles and induce slight deformations.
Protection against Nucleases
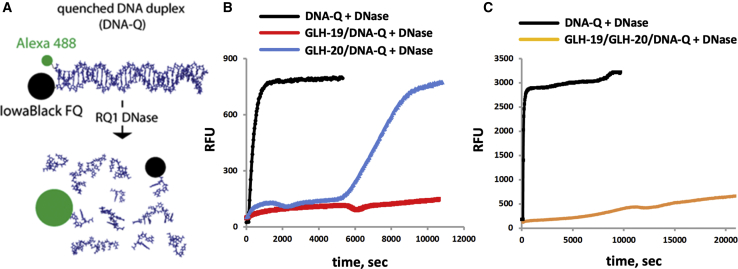
The ability of the vesicles to protect associated nucleic acids from the hydrolytic action of nucleases was tested by exposing DNA-containing vesicles to DNase. The 5′ end of the sense strand of the DNA was tagged with an Iowa Black FQ (fluorescence quencher), and the 3′ end of the antisense strand of the DNA was tagged with Alexa Fluor 488 (Al488) (Figure 3A). The hydrolysis of the DNA by the DNase was detected by monitoring the emission of the fluorescence, which occurs when the Al488 and the quencher separate due to the hydrolysis of the DNA. The hydrolysis of naked DNA was relatively fast, as shown by the black line in Figures 3B and 3C, and it was digested in about 15 min. In comparison, the hydrolysis of the DNA associated with GLH-19 vesicles was very slow; even at 180 min after the addition of the DNase, only about 10% of the DNA underwent hydrolysis (Figure 3B). Also, the hydrolysis of the DNA that was associated with GLH-20 vesicles was slow, but it was significantly faster than the hydrolysis of the DNA that was associated with GLH-19 vesicles, and it was digested after about 160 min (Figure 3B). We also tested whether vesicles made of a mixture of GLH-19 and GLH-20 will still protect the associated nucleic acids from hydrolysis while GLH-20 plays its role in BBB penetration and efficient siRNA release. As shown in Figure 3C, vesicles made from a mixture of GLH-19 and GLH-20 protected the associated DNA from hydrolysis by DNase almost as well as did the GLH-19 vesicles. Our computational studies were consistent with these experimental results, and they explain why GLH-19 provides better protection than GLH-20. During the MD simulations it was found that GLH-19 head groups cover a larger area of the siRNA than the area covered by GLH-20, and therefore the siRNA can be protected more efficiently when it is associated with GLH-19 vesicles.
Figure 3.
Protection of Encapsulated Polynucleotides against Nucleases
(A) The digestion of DNA by DNase can be detected by the fluorescence that is triggered when the polynucleotide is degraded by the nuclease and separation between the Al488 and the Iowa Black FQ occurs. (B) Relative stabilities of DNA duplexes associated with either GLH-19 (red line) or GLH-20 (blue line) and (C) GLH-19/GLH-20 (orange line) in the presence of DNase. Quenched DNA duplex (100 nM final) labeled with Al488 and Iowa Black FQ was mixed with the bolas (10 μg final), and DNase was added after 2 min of incubation at 37°C. As a control, naked DNA duplex was completely digested by DNase (black line). Excitation was set at 460 nm and emission was measured at 520 nm. All tested bola vesicles contain CHOL and CHEMS.
Transfection and Gene Silencing
Transfection Experiments
Our previous data showed that internalization of the vesicles occurs via endocytosis since uptake of vesicles in a variety of cell types has been observed at 37°C but not at 4°C.30,36 Therefore, in order to determine the mechanism of endocytosis, we performed some additional experiments with specific inhibitors for different pathways of endocytosis, similar to experiments with polyethylenimine, another delivery system for nucleic acids.37 Our results clearly demonstrate that the internalization of vesicles made of GLH-19/GLH-20 proceeds mainly via the caveolae-dependent pathway and not via the clathrin-dependent pathway, as Filipin III, a specific inhibitor of the caveolae-dependent pathway, significantly inhibited vesicle internalization whereas chlorpromazine, a specific inhibitor of the clathrin-dependent pathway, had no effect on vesicle internalization (Figure S12).
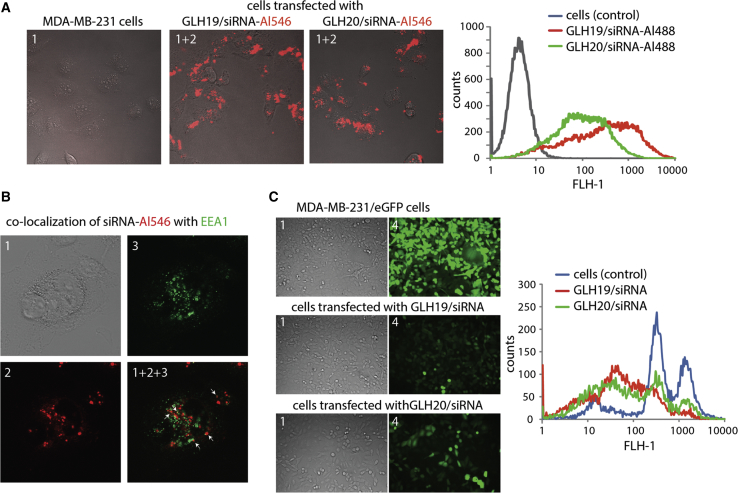
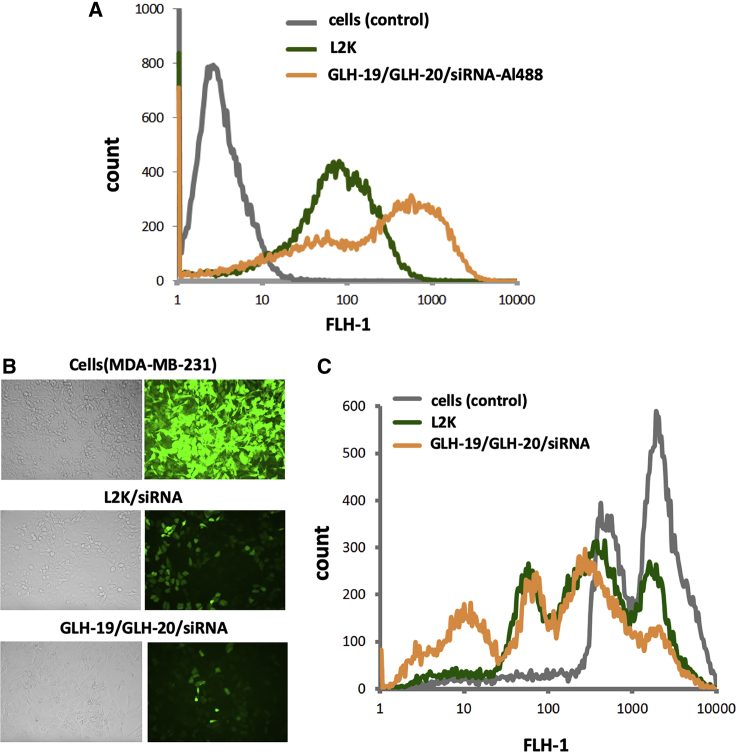
The transfections of bola vesicles (made from GLH-19, GLH-20, or GLH-19/GLH-20, each associated with siRNA) were examined. The 3′ end of the siRNA antisense strands was fluorescently labeled with Al488 or Alexa Fluor 546 (Al546). The sequence of the sense strands was designed to silence the enhanced green fluorescent protein (EGFP) gene. Transfection was tested using human breast cancer cells, MDA-MB-231. The transfection efficiencies were visualized by confocal fluorescence microscopy and further analyzed by fluorescence-activated cell sorting (FACS). The results of the transfection studies showed that the uptake of GLH-19/siRNA vesicles was higher than that of the GLH-20/siRNA vesicles (Figure 4A), possibly due to the higher binding affinity of GLH-19 to the siRNA, leading to less dissociation of the siRNA (see Figure 2C). The transfection proceeds via the endosomal system as indicated by the co-localization of the siRNA with EEA1. EEA1 localizes exclusively to early endosomes and has an important role in endosomal trafficking. EEA1 binds directly to the phospholipid phosphatidylinositol 3-phosphate through its C-terminal FYVE domain and forms a homodimer. EEA1 acts as a tethering molecule that couples vesicle docking with soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) such as N-ethylmaleimide-sensitive fusion protein, bringing the endosomes physically closer and ultimately resulting in the fusion and delivery of endosomal cargo (Figure 4B), which is now available to act on its specific gene as indicated in the present study by the ability of the delivered siRNA to silence specific genes.
Figure 4.
Transfection and Silencing Efficiencies for GLH-19/siRNA and GLH-20/siRNA Complexes (siRNAs = 100 nM)
(A) Transfection efficiencies of human breast cancer cells (MDA-MB-231) with GLH-19/siRNA and GLH-20/siRNA were visualized by confocal fluorescence microscopy (left) and statistically analyzed with flow cytometry experiments (right). To show the generality, two different duplexes labeled with Al546 and Al488 were used for imaging and flow cytometry experiments, respectively. (B) Study of the co-localization of fluorescently labeled siRNA with commonly used marker EEA1 for early endosomal compartments. (C) EGFP knockdown assays for human breast cancer cells (MDA-MB-231/EGFP) that stably express EGFP. Three days after the transfection of cells with siRNAs, EGFP expression was observed by fluorescence microscopy (left) and statistically analyzed with flow cytometry experiments (right). Note that the initial fluorescent cells had two populations represented by two maximums. All tested bola vesicles contain CHOL and CHEMS. Image numbers in (A)–(C) correspond to differential interference contrast (DIC) images (1), Al546 emission (2), EAA1 antibody staining (3), and GFP signal (4). Images 1+2 and 1+2+3 are superimpositions of two or three different images.
Specific Gene Silencing Experiments
Silencing was examined by the ability of the siRNA to suppress the expression of the EGFP gene, which is constitutively expressed in human breast cancer cells (MDA-MB-231/EGFP). The efficiency of the EGFP silencing for GLH-19/siRNA and GLH-20/siRNA was measured by fluorescence microscopy and, further, by FACS (Figure 4C). GLH-19/siRNA and GLH-20/siRNA vesicles demonstrated similar silencing performance. Considering the better uptake of GLH-19/siRNA, we conclude that the similar silencing efficiency possibly stems from the lower binding affinity of GLH-20 with siRNA, which was shown in both the experimental results (see Figure 2C) and the MD simulation studies (see MD Simulations of Bola/siRNA Complexes above).
We also tested vesicles made from a mixture of GLH-19 and GLH-20 for their transfection and silencing efficiencies. As shown in Figure 5A, the transfection efficiency of vesicles made from a mixture of GLH-19 and GLH-20 was good and, in fact, it was better than the commercial delivery agent Lipofectamine 2000 (L2K). Also, compared to L2K, GLH-19/GLH-20/siRNA vesicles demonstrated better silencing efficiency (Figures 5B and 5C). It seems that in vesicles made from a mixture of GLH-19 and GLH-20, each bola contributes to transfection and silencing based on its own characteristics to improve the resulting silencing efficiency. Thus, GLH-19 contributes to stability and binding affinity, while GLH-20 contributes to efficient release of the siRNAs, which becomes available for silencing.
Figure 5.
Transfection and Silencing Efficiencies for GLH-19/GLH-20/siRNA-Al488 Vesicles (siRNA = 100 nM)
(A) Transfection (uptake) and (B and C) silencing efficiencies of L2K and GLH-19/GLH-20/siRNA vesicles shown by fluorescence microscopy in B and flow cytometry in (C). All tested bola vesicles contain CHOL and CHEMS.
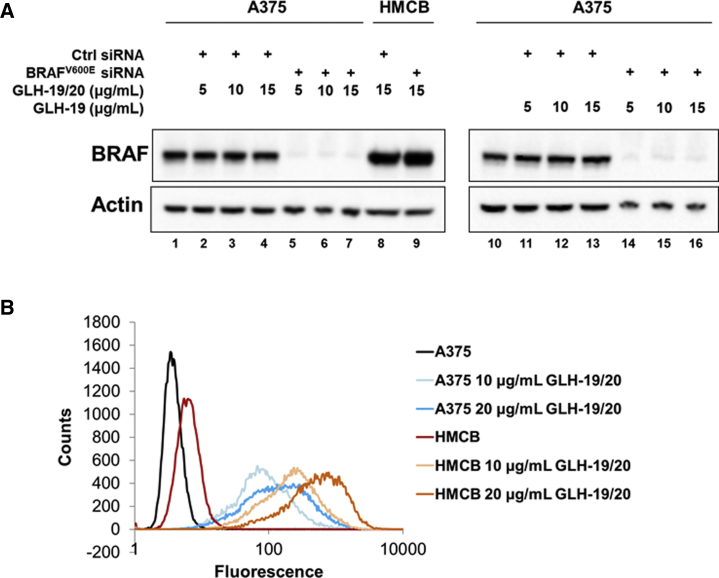
To show the generality of the reported platform and the high potential of bola vesicles for siRNA delivery, we explored the ability of bola vesicles to deliver therapeutic siRNAs to human melanoma cells. Approximately 50% of malignant melanomas carry mutations in BRAF kinase (BRAFV600E), leading to constitutive activation of MEK (mitogen-activated protein kinase kinase)-ERK (extracellular signal-regulated kinase) downstream signaling, which drives melanomagenesis.38 We transfected A375 melanoma cells carrying the homozygous BRAFV600E mutation with a BRAFV600E-specific siRNA using GLH-19/GLH-20 bola vesicles. HMCB melanoma cells, which carry wild-type BRAF, were used as a non-target control. As shown in Figure 6A, efficient knockdown was observed for the melanoma-associated BRAFV600E allele (92% ± 3%, SEM; n = 4) but not for wild-type BRAF in HMCB cells, even though both cells showed similar siRNA-Al488 uptake with GLH-19/GLH-20 vesicles (Figure 6B). Thus, bola agents can efficiently deliver an siRNA that specifically targets a relevant oncogenic mutation in tumor cells.
Figure 6.
Silencing Oncogenic BRAF with siRNAs Delivered with Bola Vesicle Formulations
(A) Specific gene silencing of mutated BRAF in A375 cells (BRAFV600E) but not in HMCB cells (BRAFWT). (B) Relative uptakes of siRNA-Al488 with different GLH-19/20 concentrations in human A375 and HMCB melanoma cells. All siRNAs were used for transfection at a final concentration of 100 nM.
In Vivo and Ex Vivo Experiments
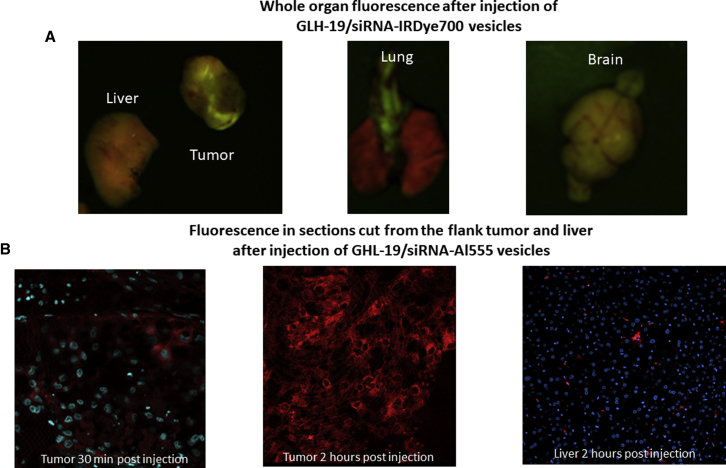
In the initial in vivo experiments, we tested GLH-19/siRNA-IRDye 700 and GLH-19/siRNA-Al555 vesicles injected into the tail vein of mice with a flank tumor in two different experiments. At 2 h after the injection, we collected tumor, liver, lung, and brain and performed fluorescence imaging on the whole organ (Figure 7A). Fluorescence imaging of the collected organs showed significant accumulation of siRNA-IRDye 700 in the implanted flank tumor and in the brain, but not in the liver or the lung after the tail vein injection of the GLH-19/siRNA-IRDye 700 vesicles. In another experiment, we injected GLH-19/siRNA-Al555 and cryosectioned the organs to obtain thin sections (40 μm thick) from the tumor and the liver at 30 min and 2 h after the injection. Fluorescence microscopy of these sections showed progressive infiltration of the siRNA-Al555 into the tumor parenchyma. At 30 min after the injection, the Al555 fluorescence was seen in blood vessels that supply blood to the tumor and some also in the tumor parenchyma in proximity to the blood vessel. Significant accumulation of the dye in all of the cells within the tumor was also seen 2 h after systemic tail vein injection (Figure 7B). In comparison, the liver showed little fluorescence that seemed to be accumulated mainly near or in phagocytic Kupffer cells (Figure 7B).
Figure 7.
Fluorescence Imaging of Tumor, Liver, Lung, and Brain of Mice after Intravenous Injection of GLH-19 Vesicles Containing siRNA Coupled to a Fluorescent Probe
(A) Fluorescence imaging of whole organs 2 h after tail vein injection of GLH-19/siRNA-IRDye 700 vesicles, showing significant accumulation of fluorescence in the flank implanted tumor and in the brain. Note that the fluorescence is seen in green over a brown background, but not in lung and liver, and that only brown background is seen in the liver and the lung, except for green fluorescence in blood vessels of the lung. (B) Fluorescence imaging of sections that were cut from a flank implanted tumor and the liver at 30 min and 2 h after tail vein injection of GLH-19/siRNA-Al555, showing progressive accumulation of siRNA-Al555 in the tumor, but not in the liver. Fluorescence (red) is seen penetrating from a blood vessel in the tumor (at 30 min after the injection; left side of bottom panel) into the tumor parenchyma, until all the tumor parenchyma is filled with red fluorescence at 2 h after the injection (center of bottom panel). Only little fluorescence can be seen in the liver 2 h after the injection (right side of bottom panel). Cell nuclei were stained with DAPI, shown in blue. All tested bola vesicles (20 mg/kg) contain CHOL and CHEMS.
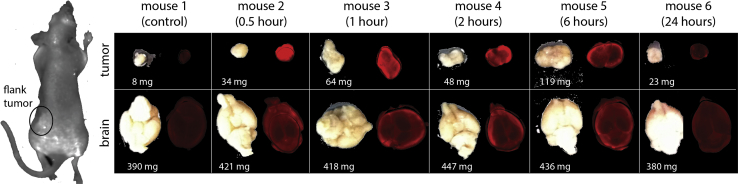
To take advantage of the strengths of both bolas, we examined the ability of GLH-19/GLH-20 vesicles to deliver siRNA to a flank tumor and into the brain. Vesicles were loaded with siRNA tagged with IRDye 700 and injected into the tail vein of mice, which were sacrificed at different time periods after the injection. We then monitored the accumulation of the siRNA-IRDye 700 in the tumor and in the brain at different time points. We found that GLH-19/GLH-20/siRNA-IRDye 700 vesicles successfully delivered the siRNA-IRDye 700 into the implanted flank tumor and into the brain. As shown in Figure 8, the fluorescence of the siRNA-IRDye 700 was seen in both the tumor and in the brain 30 min after the injection of GLH-19/GLH-20/siRNA-IRDye 700 vesicles and remained in these tissues for at least 6 h after the injection.
Figure 8.
Fluorescent Imaging of siRNA-IRDye 700 in the Flank Tumor and in the Brain following Injection of GLH-19/GLH-20/siRNA-IRDye 700 Vesicles into the Tail Vein of Mice
In each time panel, left images are tumor (top) and brain (bottom), and right images show their fluorescence. Fluorescence imaging shows significant accumulation of siRNA-IRDye 700 in the tumor and in the brain at different time points after tail vein injection. All tested bola vesicles (20 mg/kg) contain CHOL and CHEMS.
Discussion
Previous studies32,33 and the present study show that the efficiency of bola vesicles to deliver siRNA into cells is determined by the length of the hydrophobic chain of the bolas, the total charge, the atomic geometry of the head groups, and the assembled conformations, which can be either micelles or vesicles. GLH-58 and GLH-60 that contain relatively short hydrophobic chains formed only micelle conformations. GLH-60 has a total of four positively charged head groups, so that it has strong binding affinity and weak siRNA release. Alternatively, GLH-58 has two positively charged head groups and showed moderate binding affinity along with protection capability against hydrolysis of the associated siRNA by nucleases, transfection capability, and silencing capability.32 However, micelle structures do not penetrate the BBB very well,32 whereas vesicles have been shown to penetrate into the brain and deliver compounds that do not normally penetrate the BBB into the brain.29,30 Delivering siRNA into the brain is important for the treatment of brain tumors and some neurological diseases. Therefore, we examined whether vesicles that have been shown to penetrate into the brain can associate with siRNA and deliver it into the brain and tumors.
Both GLH-19 and GLH-20 have two positively charged head groups with relatively long hydrophobic chains so that they can form either micelle or vesicles, depending on the preparation protocols.29,30 In addition, our present computational and experimental studies showed consistent results arguing that the stability of bola vesicles can be enhanced by the presence of CHOL and CHEMS. Thus, if CHOL and CHEMS were absent, only GLH-19 formed vesicles. However, even GLH-19 vesicles that did not contain CHOL and CHEMS collapsed when they interacted with siRNA. In comparison, when CHOL and CHEMS existed in bola membranes, both GLH-19 and GLH-20 maintained stable vesicle conformations regardless of the association with siRNA.
Our previous MD simulations as well as our MD simulations in the present study showed that GLH-19 head groups have more mobility and extend farther from both the micelle and vesicle surface than does the head group of GLH-20. This difference significantly affected the characteristics of each bola vesicle in terms of stability, binding affinity, protection, transfection, and siRNA release, as summarized below.
Our computational studies and cryo-TEM images indicate that the surface properties of vesicles and micelles are different. Bola head groups in vesicles are uniformly distributed on a flat membrane surface while those of a micelle may be distributed on a rugged micelle surface.32,33 Therefore, bola head groups may have less mobility on a vesicle surface than on the surface of the micelle. The surface differences between micelles and vesicles may explain the moderate binding affinity of siRNA on bola vesicles compared to the strong binding affinity on micelles. In the presence of the detergent Triton X-100, siRNAs were released from GLH-19 vesicles while they were not separated from GLH-19 micelles (compare the data from this study to those described in Kim et al.32). In addition, the uniformly distributed head groups on the vesicle surface may induce a more stable negative ion cloud on its surface than that of the micelle surface. Thus, as it was shown in our experimental and computational studies, siRNAs began to associate with GLH-20 under higher concentrations as compared to the lower concentration of GLH-19 that were needed to associate with siRNA.
The different behaviors of each head group on the surface also explain the different protection efficiencies between GLH-19 and GLH-20 as well as between micelles and vesicles. It was found that siRNA protection by GLH-19 in the form of either a micelle or a vesicle was better than the protection achieved by GLH-20. In addition, the protection of siRNA against the hydrolytic action of nucleases by GLH-20 micelles was worse than that of GLH-20 vesicles. The high mobility and greater extension of the GLH-19 head groups from vesicle and micelle surfaces results in durable siRNA protection. Alternatively, the low mobility of the embedded GLH-20 head groups caused weak siRNA protection. In addition, the rugged surface of GLH-20 micelles may cause weaker siRNA protection than that of GLH-20 vesicles.
The strong binding affinity and efficient siRNA protection by GLH-19 result in a better transfection efficiency than that obtained by GLH-20 in both the micelle and vesicle forms. However, due to the weaker binding affinity of GLH-20, and more efficient release of the siRNA from the bola/siRNA complex, silencing efficiencies after siRNA release were comparable between the two bolas.
The performance of bola vesicles was also examined by in vivo and ex vivo experiments. It was found that a significant amount of siRNA accumulated in the flank implanted tumor, while minor accumulation of siRNA was observed in the liver following the injection of GLH-19 vesicles with fluorescently tagged siRNA. To further explore the delivery capability of bola vesicles, we investigated siRNA delivery with a mixture of two bolas: GLH-19 and GLH-20. We predicted that GLH-19/GLH-20 vesicles will efficiently deliver siRNA into the brain because GLH-19 contributes to the strength of stability, binding affinity, and protection of siRNA against nucleases, while GLH-20 provides the capability of BBB penetration and efficient siRNA release via hydrolysis of the GLH-20 head group by ChE. Our computational studies showed that when the two bolas were added together for vesicle formation, the characteristics of each bola head group are maintained, and no interference occurs due to the interactions between the two different head groups. Our experimental results demonstrated that, indeed, GLH-19/GLH-20 vesicles have strong protection capability and significantly better transfection and silencing efficiencies than those of the commercial transfection agent L2K. Based on these computational and experimental studies, we examined the biodistribution of GLH-19/GLH-20/siRNA-IRDye 700 vesicles and their ability to deliver the associated siRNA into the brain. The bola vesicles cross the BBB by transcytosis, which includes a step of endocytosis followed by exocytosis. In previous papers we showed that the vesicles are endocytosed by endothelial cells from microvessels of the brain (cells that constitute the BBB).30,36 We also demonstrated that proteins that do not cross the BBB are transported into the brain parenchyma when encapsulated in the vesicles.30 There are other examples in the literature of nanoparticles that cross the BBB via transcytosis. A review that discusses these processes can be found in several references.39, 40, 41
In conclusion, we characterized the delivery of siRNA using three bola vesicles, GLH-19, GLH-20, and GLH-19/GLH-20. Each bola vesicle has different characteristics: GLH-19 vesicles have better stability, higher binding affinity, better protection of siRNA against nucleases, and better transfection capability than GLH-20 vesicles, while GLH-20 vesicles release siRNA better due to a weaker binding affinity and the capability of the GLH-20 head groups to be hydrolyzed by ChE.29 Thus, when vesicles were made from a mixture of the two bolas, the characteristics of each bola contribute to the end results, and thus the GLH-19 enhanced the delivery process and the GLH-20 enabled efficient siRNA release, and both bolas together form vesicles capable of crossing the BBB. In addition, our study also showed that the delivery characteristics of bola vesicles are different from those of micelles in terms of binding affinity, protection of siRNA from the action of nucleases, and the resulting biodistributions of the delivered siRNA. Therefore, our studies demonstrate that depending on the delivery purpose, proper delivery agents and their conformation can be chosen according to the target organ and target cells.
Materials and Methods
Synthesis of the Bolas
The bolas, GLH-19 and GLH-20, were synthesized in Ben-Gurion University as previously described.27, 28, 29
Preparation of Bola Vesicles
Ethanol Injection (EI)
This method was typically used for obtaining vesicle suspensions without CHOL and CHEMS. 10 mg of GLH-19 or GLH-20 was weighed in an Eppendorf tube, dissolved in 50 μL of ethanol. The solution of the bolas was transferred to a high-pressure liquid chromatography (HPLC) syringe and quickly injected into in a 5-mL glass vial containing 1 mL of 0.1 mg/mL CF solution in PBS buffer, which was vortexed vigorously. The solution was then bath sonicated for 15 min at room temperature.
Film Hydration and Sonication
This method was typically used for obtaining vesicle suspensions with CHOL and CHEMS. The molar ratio of bolas to the CHEMS and CHOL was 2:1:1, respectively. 10 mg of GLH-19 or GLH-20 was weighed in a 5-mL glass vial; 1.6 mg of CHOL and 2.4 mg of CHEMS were weighed in separate Eppendorf tubes, and 350 μL of chloroform was added to each tube to dissolve the content. Two solutions were transferred to a glass vial containing the bolas and vortexed until a homogeneous solution was obtained. The resulting solution was dried under nitrogen flow to get a uniform thin film spread on the walls of the vial. The film was dried for an additional 12–16 h under vacuum to remove all chloroform residues. To the dried film, 1 mL of solution containing the compound to be encapsulated (e.g., CF when stability of the vesicles was measured) was added and vortexed until the film was dissolved completely. The resulting vesicle solution was sonicated by a probe sonicator for 5 min in an ice-cold bath in cycles as follows: 15-s pulse/10-s rest; the power was adjusted to avoid bubble formation and overheating of the sample.
Agarose Gel Electrophoresis of the Bola/siRNA Vesicles
To determine the binding affinity of the siRNA to the bolas, the bola/siRNA complexes were run at room temperature on 2% agarose gel in the presence of 89 mM Tris-borate (pH 8.3). The assay was performed with several concentrations of the bolas and a fixed concentration of the siRNA. siRNA that was bound to the bolas did not run on the gel, whereas a band of the free siRNA was stained by ethidium bromide and was visualized on the gel by a Hitachi FMBIO II multi-view imager.
Dynamic Light Scattering Experiments
For dynamic light scattering experiments, sample solutions containing bola/siRNA vesicles were used. The samples were measured at 25°C with a Zetasizer Nano (Malvern Instruments, Worcestershire, UK) equipped with a 633-nm laser.
Stability Studies
Vesicle Stability in Solution and after Dilution
When vesicle aggregation in solution occurs, the vesicles lose their stability. Vesicle aggregation was monitored by following the development of turbidity in the vesicle solution. The development of turbidity was measured by following the change in absorbance of the vesicle solution at 620 nm as a function of time.
Vesicle stability was also determined by measuring the change in the amount of CF encapsulation as a function of dilution in diluted PBS buffer. Vesicles were made by EI from 10 mg/mL GLH-19 or GLH-20 in diluted PBS buffer (containing 1:10 NaCl content compared to a regular PBS buffer and designated as PBS/10 buffer), containing 0.1 mg/mL CF. Vesicles were diluted 10-, 20-, 50-, and 100-fold in PBS/10 buffer, kept at room temperature (RT), and encapsulation was measured at 1 and 24 h after the dilution. Encapsulation was calculated from the difference between the fluorescence before and after the addition of Triton X-100 after diluting the vesicles 200-fold in order to reduce the extravesicular CF below its self-quenching concentration.
All stability studies were done in triplicate.
Stability of Associated DNA after Exposure to RQ1 DNase
To study the ability of the vesicles to protect associated nucleic acid against nuclease, DNA duplexes containing one 3′ antisense strand conjugated with Al488 and a 5′ sense strand conjugated with Iowa Black FQ quencher were preincubated with bola (GLH-19, GLH-20, or GLH-19/GLH-20)/siRNA vesicles, and the dequenching of Al488 upon digestion with RQ1 DNase (Promega, Madison, WI, USA) was followed by fluorescence resonance energy transfer measurements.
Cryo-TEM
Quantifoil copper 200 mesh R 3.5/1 grids were washed overnight with acetone. To prepare a frozen, hydrated grid, 2.5 μL of sample was applied to the grid, blotted, and plunged into liquid ethane using Vitrobot III (FEI, Hillsboro, OR, USA). Images were collected at liquid nitrogen temperature (∼100 K) on a JEM-2010F (JEOL, Tokyo, Japan) cryo-TEM equipped with a field emission gun. JEM-2010F was operating at 200 kV and was equipped with a Gatan cryo-holder (model 626) (Gatan, Pleasanton, CA, USA). Images were recorded on DE-12, a 12.6-megapixel (3,072 × 4,096) direct detection device sensor (Direct Electron, San Diego, CA, USA). Samples were imaged at ×13,900 effective magnification targeted at 3–4 μm under focus. We used a total specimen exposure for each image of 30 e−/Å2 over 1 s.
Transfection of MDA-MB-231 Human Breast Cancer Cells with Bola/siRNA Complexes
The siRNA duplexes in the in silico, in vitro, in vivo, and ex vivo experiments were designed to silence EGFP as described in our previous study.32 siRNA transfections were performed with the human breast cancer cell line MDA-MB-231 (that either express or do not express EGFP) using vesicles made from the bolas GLH-19, GLH-20, or a mixture of the two. Upon resuspension in water, the bola vesicles (1 mg/mL) were mixed with siRNAs (100-fold the desired final concentration) at a 1:1 volume ratio. Upon 30 min of incubation at room temperature, the bola/siRNA complexes were diluted 50-fold in Opti-MEM prior to addition to the cells for transfection. Upon a 4-h incubation at 37°C, the media were replaced by regular DMEM for 24 h (transfection efficiency) or 72 h (silencing efficiency).
Fluorescent Light Microscopy
To assess the silencing efficiency, cells were imaged after the transfection with a Nikon 200 TE inverted microscope (Nikon, Melville, NJ, USA). To visualize the uptake of the bola/siRNA particles, experiments were performed with a LSM 710 confocal microscope (Carl Zeiss) and RNAs were labeled with Al546. The cells were fixed with 4% paraformaldehyde and imaged (transfection efficiency) or permeabilized with 0.2% Triton X-100 for 20 min in order to label the early endosome-associated protein EEA1 (with primary [Cell Signaling Technology, Danvers, MA, USA] and Al488-conjugated secondary [Molecular Probes, Eugene, OR, USA] antibodies), and Nikon 200 TE-Al488 imaging was performed with the 488-nm line of an argon laser while Al546 imaging was obtained through excitation with a DPSS 561 laser.
Flow Cytometry
For statistical analysis of the flow cytometry experiments, at least 30,000 MDA-MB-231 cells (Cell Biolabs, San Diego, CA, USA; with or without EGFP) as well as A375 (BRAFV600E) and HMCB (BRAFWT) cells (gifts from Dr. Deborah K. Morrison, NCI-Frederick) were analyzed by FACS analysis with a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA).
BRAFV600E Gene Silencing with Bola/siRNA Complexes
A375 and HMCB cells were plated at 105 cells/35-mm dish and transfected with 100 nM (final concentration) siRNA-Al488, BRAFV600E siRNA (5′-GCUACAGAGAAAUCUCGAUUU-3′), or a non-targeting siRNA pool (Thermo Fisher Scientific, D-001810-10-05). Briefly, the siRNAs were incubated with GLHs for 30 min at 25°C, protected from light. After incubation, the mixture was diluted in 1 mL of Opti-MEM (Thermo Fisher Scientific, #31985088) and directly added to cells previously rinsed with PBS. After 4 h, the mixture was replaced with regular serum-containing media. For siRNA uptake assays, cells were trypsinized after 24 h and analyzed by FACS. For knockdown experiments, whole-cell lysates were generated after 48 h using radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton X-100). Antibodies used for immunoblotting were anti-BRAF (Santa Cruz, sc-5284) and anti-β-actin (Santa Cruz, sc-1616-R).
In Vivo Experiments
All animal in vivo experiments were performed according to the Frederick National Laboratory for Cancer Research (Frederick, MD, USA) Animal Care and Use Committee guidelines. For these experiments, 107 MDA-MB-231 tumor cells were subcutaneously injected in the flank of each athymic nude mouse (Charles River Laboratories, Frederick, MD, USA). Experiments were performed when the tumors reached at least 3 mm in their longest diameter (about 2 weeks after cell injection). Two different fluorescently labeled (with either Al555 or IRDye 700 dyes) duplexes were used. Biodistribution of GLH-19/siRNA-IRDye 700 and GLH-19/siRNA-Al555 was examined at two time points (30 and 120 min post-injection) using four mice, out of which two mice were used to visualize fluorescence in whole isolated organs at 30 and 120 min post-injection, and the other two mice were used for histofluorescence studies of the implanted tumor and of the liver also at 30 and 120 min post-injection. In addition, time-dependent biodistribution of GLH-19/GLH-20/siRNA-IRDye 700 was examined using six mice, one for control and five for five different time points. Images of in vivo experiments were taken by a Nikon 200 TE inverted microscope (Nikon, Melville, NJ, USA). Imaging studies were performed on MDA-MB-231 tumor-bearing athymic nude mice (Charles River Laboratories, Frederick, MD, USA).
MD Simulations
Preparation of Bola Membranes for In Silico Study
The initial molecular structures of GLH-19, GLH-20, CHOL, and CHEMS were built with Discovery Studio Visualizer (Figure S1). The force fields (FFs) and topologies of GLH-19, GLH-20, CHOL, and CHEMS were prepared by the Amber antechamber package42 utilizing the general Amber force field (GAFF),43 which has been developed for small organic molecules. Detailed procedures are discussed in Supplemental Materials and Methods.
Due to the unique structural characteristics of the bolas that were tested in this study, which contained a single hydrophobic chain bound to two positively charged hydrophilic head groups at each end of the hydrophobic chain, these bolas can potentially form a monolayer or a monolayer/bilayer membrane mixture.44 For the in silico studies, we tested three different types of bola membranes: monolayer, bilayer, and the mixture of monolayer and bilayer (monolayer/bilayer). The stability of each membrane was further examined in terms of the presence of CHOL and CHEMS and the association with an siRNA. The list of 31 membrane systems for the in silico studies are summarized in Table S1. The detailed information about membrane patch preparations, MD simulation protocols, and in silico data analysis are provided in Supplemental Materials and Methods.
Author Contributions
T.K. conducted MD simulations. M.V., K.A.A., K.G., M.P., E.H. and J.S. conducted experiments. C.L. and E.H. designed the bolas used in this study and contributed materials. T.K, M.V., K.G., and E.H wrote the paper. B.A.S. conceived of and designed the MD simulations and experiments, as the laboratory chief supervised the experiments, and gave his input as well as edited the manuscript. P.F.J. contributed to design and data interpretation for the BRAFV600E targeting experiments, and J.S. and P.F.J. wrote the associated parts of the text.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, Center for Cancer Research, NCI-Frederick. This work has been funded with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract no. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.02.011.
Supplemental Information
References
- 1.Afonin K.A., Grabow W.W., Walker F.M., Bindewald E., Dobrovolskaia M.A., Shapiro B.A., Jaeger L. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat. Protoc. 2011;6:2022–2034. doi: 10.1038/nprot.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis M.E. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol. Pharm. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 3.Davis M.E., Zuckerman J.E., Choi C.H., Seligson D., Tolcher A., Alabi C.A., Yen Y., Heidel J.D., Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kole R., Krainer A.R., Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pecot C.V., Calin G.A., Coleman R.L., Lopez-Berestein G., Sood A.K. RNA interference in the clinic: challenges and future directions. Nat. Rev. Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkhout B., Sanders R.W. Molecular strategies to design an escape-proof antiviral therapy. Antiviral Res. 2011;92:7–14. doi: 10.1016/j.antiviral.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Grabow W.W., Zakrevsky P., Afonin K.A., Chworos A., Shapiro B.A., Jaeger L. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett. 2011;11:878–887. doi: 10.1021/nl104271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shu Y., Cinier M., Shu D., Guo P. Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods. 2011;54:204–214. doi: 10.1016/j.ymeth.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shukla G.C., Haque F., Tor Y., Wilhelmsson L.M., Toulmé J.J., Isambert H., Guo P., Rossi J.J., Tenenbaum S.A., Shapiro B.A. A boost for the emerging field of RNA nanotechnology. ACS Nano. 2011;5:3405–3418. doi: 10.1021/nn200989r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannon G.J., Rossi J.J. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 12.Bramsen J.B., Kjems J. Development of therapeutic-grade small interfering RNAs by chemical engineering. Front. Genet. 2012;3:154. doi: 10.3389/fgene.2012.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afonin K.A., Kireeva M., Grabow W.W., Kashlev M., Jaeger L., Shapiro B.A. Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs. Nano Lett. 2012;12:5192–5195. doi: 10.1021/nl302302e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams D., Gonzalez-Duarte A., O’Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 15.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 16.Daka A., Peer D. RNAi-based nanomedicines for targeted personalized therapy. Adv. Drug Deliv. Rev. 2012;64:1508–1521. doi: 10.1016/j.addr.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Kim H.J., Kim A., Miyata K., Kataoka K. Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv. Drug Deliv. Rev. 2016;104:61–77. doi: 10.1016/j.addr.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Zheng M., Tao W., Zou Y., Farokhzad O.C., Shi B. Nanotechnology-based strategies for siRNA brain delivery for disease therapy. Trends Biotechnol. 2018;36:562–575. doi: 10.1016/j.tibtech.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Lee J.M., Yoon T.J., Cho Y.S. Recent developments in nanoparticle-based siRNA delivery for cancer therapy. BioMed Res. Int. 2013;2013:782041. doi: 10.1155/2013/782041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cruz-Acuña M., Halman J.R., Afonin K.A., Dobson J., Rinaldi C. Magnetic nanoparticles loaded with functional RNA nanoparticles. Nanoscale. 2018;10:17761–17770. doi: 10.1039/c8nr04254c. [DOI] [PubMed] [Google Scholar]
- 21.Kanasty R., Dorkin J.R., Vegas A., Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 22.Yin H., Kanasty R.L., Eltoukhy A.A., Vegas A.J., Dorkin J.R., Anderson D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 23.Adjei I., Sharma B., Labhasetwar V. Nanoparticles: cellular uptake and cytotoxicity. In: Capco D.G., Chen Y., editors. Nanomaterial. Springer; 2014. pp. 73–91. [DOI] [PubMed] [Google Scholar]
- 24.Ardana A., Whittaker A.K., McMillan N.A.J., Thurecht K.J. Polymeric siRNA delivery vectors: knocking down cancers with polymeric-based gene delivery systems. J. Chem. Technol. Biotechnol. 2015;90:1196–1208. [Google Scholar]
- 25.Dowdy S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017;35:222–229. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- 26.Grinberg S., Kolot V., Linder C., Shaubi E., Kas’yanov V., Deckelbaum R.J., Heldman E. Synthesis of novel cationic bolaamphiphiles from vernonia oil and their aggregated structures. Chem. Phys. Lipids. 2008;153:85–97. doi: 10.1016/j.chemphyslip.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Popov M., Linder C., Deckelbaum R.J., Grinberg S., Hansen I.H., Shaubi E., Waner T., Heldman E. Cationic vesicles from novel bolaamphiphilic compounds. J. Liposome Res. 2010;20:147–159. doi: 10.3109/08982100903218900. [DOI] [PubMed] [Google Scholar]
- 28.Grinberg S., Kipnis N., Linder C., Kolot V., Heldman E. Asymmetric bolaamphiphiles from vernonia oil designed for drug delivery. Eur. J. Lipid Sci. Technol. 2010;112:137–151. [Google Scholar]
- 29.Popov M., Grinberg S., Linder C., Waner T., Levi-Hevroni B., Deckelbaum R.J., Heldman E. Site-directed decapsulation of bolaamphiphilic vesicles with enzymatic cleavable surface groups. J. Control. Release. 2012;160:306–314. doi: 10.1016/j.jconrel.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 30.Dakwar G.R., Abu Hammad I., Popov M., Linder C., Grinberg S., Heldman E., Stepensky D. Delivery of proteins to the brain by bolaamphiphilic nano-sized vesicles. J. Control. Release. 2012;160:315–321. doi: 10.1016/j.jconrel.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 31.Popov M., Abu Hammad I., Bachar T., Grinberg S., Linder C., Stepensky D., Heldman E. Delivery of analgesic peptides to the brain by nano-sized bolaamphiphilic vesicles made of monolayer membranes. Eur. J. Pharm. Biopharm. 2013;85(3 Pt A):381–389. doi: 10.1016/j.ejpb.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Kim T., Afonin K.A., Viard M., Koyfman A.Y., Sparks S., Heldman E., Grinberg S., Linder C., Blumenthal R.P., Shapiro B.A. In silico, in vitro, and in vivo studies indicate the potential use of bolaamphiphiles for therapeutic siRNAs delivery. Mol. Ther. Nucleic Acids. 2013;2:e80. doi: 10.1038/mtna.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta K., Afonin K.A., Viard M., Herrero V., Kasprzak W., Kagiampakis I., Kim T., Koyfman A.Y., Puri A., Stepler M. Bolaamphiphiles as carriers for siRNA delivery: From chemical syntheses to practical applications. J. Control. Release. 2015;213:142–151. doi: 10.1016/j.jconrel.2015.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonin K.A., Viard M., Martins A.N., Lockett S.J., Maciag A.E., Freed E.O., Heldman E., Jaeger L., Blumenthal R., Shapiro B.A. Activation of different split functionalities on re-association of RNA-DNA hybrids. Nat. Nanotechnol. 2013;8:296–304. doi: 10.1038/nnano.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afonin K.A., Viard M., Koyfman A.Y., Martins A.N., Kasprzak W.K., Panigaj M., Desai R., Santhanam A., Grabow W.W., Jaeger L. Multifunctional RNA nanoparticles. Nano Lett. 2014;14:5662–5671. doi: 10.1021/nl502385k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philosof-Mazor L., Dakwar G.R., Popov M., Kolusheva S., Shames A., Linder C., Greenberg S., Heldman E., Stepensky D., Jelinek R. Bolaamphiphilic vesicles encapsulating iron oxide nanoparticles: new vehicles for magnetically targeted drug delivery. Int. J. Pharm. 2013;450:241–249. doi: 10.1016/j.ijpharm.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 37.von Gersdorff K., Sanders N.N., Vandenbroucke R., De Smedt S.C., Wagner E., Ogris M. The internalization route resulting in successful gene expression depends on both cell line and polyethylenimine polyplex type. Mol. Ther. 2006;14:745–753. doi: 10.1016/j.ymthe.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Bucheit A.D., Davies M.A. Emerging insights into resistance to BRAF inhibitors in melanoma. Biochem. Pharmacol. 2014;87:381–389. doi: 10.1016/j.bcp.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 39.de Lange E.C. The physiological characteristics and transcytosis mechanisms of the blood-brain barrier (BBB) Curr. Pharm. Biotechnol. 2012;13:2319–2327. doi: 10.2174/138920112803341860. [DOI] [PubMed] [Google Scholar]
- 40.McCully M., Sanchez-Navarro M., Teixido M., Giralt E. Peptide mediated brain delivery of nano- and submicroparticles: a synergistic approach. Curr. Pharm. Des. 2018;24:1366–1376. doi: 10.2174/1381612824666171201115126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dinda S.C., Pattnaik G. Nanobiotechnology-based drug delivery in brain targeting. Curr. Pharm. Biotechnol. 2013;14:1264–1274. doi: 10.2174/1389201015666140608143719. [DOI] [PubMed] [Google Scholar]
- 42.Wang J., Wang W., Kollman P.A., Case D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 43.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 44.Kaufman Y., Grinberg S., Linder C., Heldman E., Gilron J., Freger V. Fusion of bolaamphiphile micelles: a method to prepare stable supported biomimetic membranes. Langmuir. 2013;29:1152–1161. doi: 10.1021/la304484p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.