Abstract
Neovascular age-related macular degeneration (AMD) is characterized by the formation of choroidal neovascularization, which is responsible for more than 80% of cases of severe vision loss. Ubiquitin protein ligase E3D (UBE3D) gene missense has been proven to be associated with neovascular AMD in the East Asian population based on our previous study. In vivo, we explored the role of ube3d in eye development and the mechanisms underlying the development of neovascular AMD in a zebrafish model. In vitro, we investigated the function and mechanism of ube3d in oxidative damage in human retinal pigment epithelium (hRPE) cells. The ube3d gene was knocked down in zebrafish in our experiments, and rescue of ube3d morphants was also performed. We observed the zebrafish model at the molecular level and functional and morphological changes in vivo. Lentivirus-based gene transfer technology was used to overexpress/knockdown ube3d expression in hRPE cells in vitro. hRPE oxidative damage was induced by tert-butyl hydroperoxide (t-TBH). Cell proliferation and migration were assessed. Quantitative real-time PCR and western blot were used to measure the expression levels of UBE3D and CyclinB1. Abnormal eye development was found in zebrafish in this study, including small eyes, delayed retinal development, delayed retrograde melanosome transport, and reduced dark-induced hyper-locomotor activity under light-off conditions. In addition, increased angiogenesis was observed in ube3d morphants. A negative correlation between UBE3D and CyclinB1 was observed. Low UBE3D expression can promote oxidative damage and inflammatory reactions. UBE3D and autophagy have a synergetic effect on anti-oxidative damage. These findings indicate that ube3d may play an important role in the pathogenesis of AMD by affecting retinal development, oxidative damage, and autophagy.
Keywords: ubiquitin protein ligase E3D, age-related macular degeneration, zebrafish, autophagy, oxidative damage
Introduction
Age-related macular degeneration (AMD), a neurodegenerative disease, is the leading cause of vision loss in elderly persons around the world.1,2 Neovascular AMD is a subtype of advanced AMD that is responsible for more than 80% of cases of severe vision loss.3 Many reports in the literature indicate that oxidative stress and dysfunction of the retinal pigment epithelium (RPE) are associated with the pathogenesis of AMD.4, 5, 6
Drusen formation has been confirmed to be involved in accumulation of abnormal proteins in the retina.7 Two main types of protein degradation pathway exist in human cells: the ubiquitin-proteasome system (UPS) and the lysosomal degradation pathway. The ubiquitin-proteasome proteolytic pathway, a major pathway for protein degradation in cells, plays a critical role in protein metabolism. The UPS is essential for protein degradation in mammalian cells.8 UPS dysfunction can lead to accumulation of abnormal proteins.9 The UPS requires three enzymes: E1, a ubiquitin-activating enzyme; E2, a ubiquitin-conjugating enzyme; and E3, a ubiquitin ligase that is also called substrate recognition factor.10,11 UBE3D, an E3 protein, is also known as ubiquitin-conjugating enzyme E2C-binding protein (UBE2CBP). UBE3D accepts ubiquitin from specific E2 ubiquitin-conjugating enzymes and recognizes the target protein to be degraded. Many types of proteins are degraded by this system, including cyclins and other cell-cycle regulators, transcription factors, and inflammatory factors.8 In addition, the UPS is involved in the removal of damaged or abnormal proteins. Neurodegenerative diseases are characterized by accumulation of aggregates of insoluble proteins. Given the important role of this pathway, unsurprisingly, UPS dysfunction has been implicated in neurodegenerative diseases.12,13
In our previous study, we used whole-exome sequencing to determine the role of ubiquitin-protein ligase E3D (9UBE3D) in age-related macular degeneration in east Asian populations.14 Following the study, we found that Ube3d+/− heterozygous mice expressed decreased levels of the UBE3D protein, and an abnormal amount of pigment granules deposited in the RPE microvilli area in these mice.14 However, the mechanism underlying these abnormalities induced by UBE3D in a neovascular AMD model remained unclear. Our previous experiments revealed that homozygous mice died young; therefore, an appropriate zebrafish model was used instead for further study.
The zebrafish serves as an aquatic vertebrate model system, and its eye is a well-laminated structure similar to the eyes of other vertebrates, including humans. The morphological differentiation of structures in the zebrafish eye has been analyzed using light microscopy (LM) and transmission electron microscopy (TEM).15 Eye morphogenesis in the zebrafish begins at 11.5 h post-fertilization (hpf), and the eyecup is well formed by 24 hpf. By 72 hpf, all of the major retinal cell types and basic synaptic connections are in place. These characteristics render the zebrafish a powerful model organism in human development and disease research.
In this study, in vivo, we characterized the function of ube3d in eye development in zebrafish and explored the mechanisms underlying the involvement of ube3d in neovascular AMD. In vitro, we investigated the function and mechanism of UBE3D in oxidative damage in human RPE (hRPE) cells.
Results
In Vivo
Knockdown of ube3d in Zebrafish Delays Eye Development and Reduces Eye Size
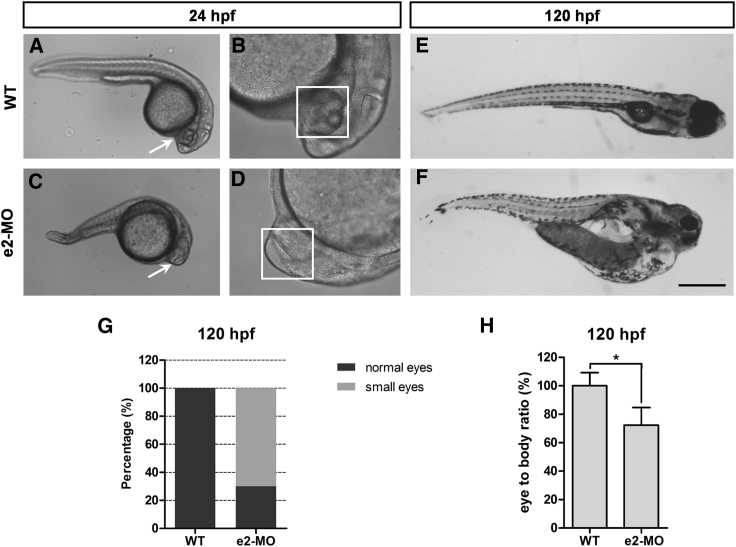
To investigate the role of ube3d in eye development in zebrafish, we analyzed eye phenotypes and measured eye sizes and body lengths in wild-type (WT) larvae and ube3d morphants. As shown in Figure 1, the eyecup was well-formed in WT 24-hpf larvae (Figures 1A and 1B), while eye morphogenesis had only just begun in e2- morpholino oligos (e2-MOs) 24-hpf larvae (Figures 1C and 1D). At 120 hpf, most e2-MO larvae had smaller eyes than WT larvae of the same age. None of the WT larvae and 70% of the e2-MO larvae had small eyes (Figure 1G). Whole-mount in situ hybridization (WISH) showed that ube3d mRNA was specifically expressed in eyes in WT zebrafish (Figure S1). We next measured eye size and body length at 24 hpf, 48 hpf, 72 hpf, and 120 hpf in ube3d morphants and WT larvae. At 120 hpf, the ube3d morphants still had a significantly smaller eye-to-body length ratio and shorter body lengths than the WT larvae (Figures 1E, 1F, and 1H). Ube3d morphants also had smaller eyes at all other time points examined (data not shown). In addition, ube3d knockdown was confirmed in ube3d morphants (Figure S2). These results show that knockdown of ube3d delays zebrafish eye development.
Figure 1.
Knockdown of ube3d Delays Zebrafish Eye Development and Reduces Eye Size
(A) Live images of WT 24-hpf larvae. (B Enlargement of (A) with the 3.2 magnification. (C) Live images of e2-MO 24-hpf larvae. (D) Enlargement of (C) with the 3.2 magnification. (E) Live images of WT 120-hpf larvae. (F) Live images of e2-MO 120-hpf larvae. (G) At 120 hpf, the percentage of small eyes in e2-MO larvae was significantly higher than the percentage in WT larvae. (H) At 120 hpf, eye size in e2-MO larvae was significantly smaller than eye size in WT larvae. The data are presented as the mean ± SD. ∗p < 0.05. Scale bars represent 400 μm (A and C), 125 μm (B and D), and 500 μm (E and F).
Rescue of ube3d Morphants
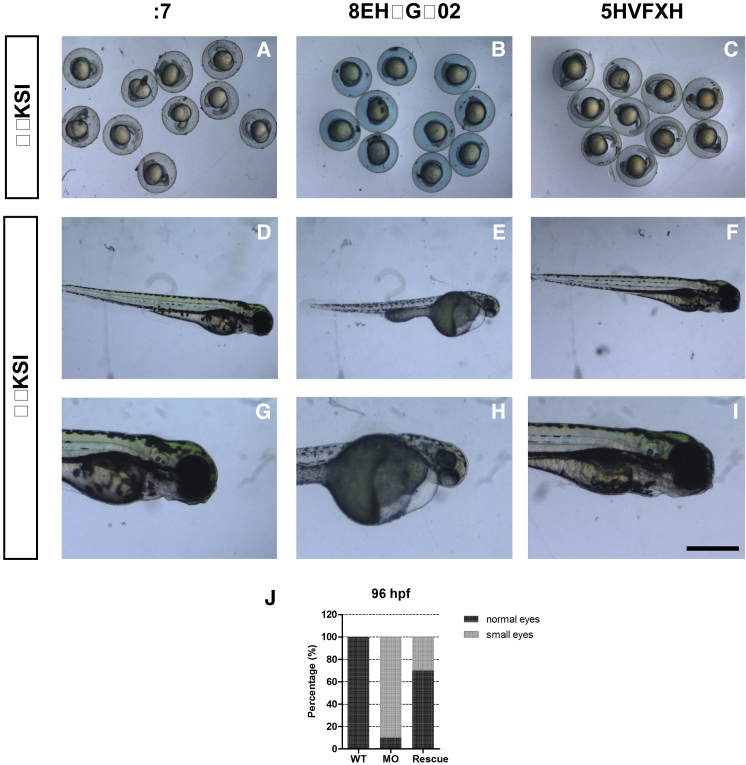
To provide further evidence that the phenotype observed in Figure 1 is caused by ube3d knockdown, we performed the above-mentioned rescue experiment and found that the ube3d MO embryos were partially rescued by coinjection with human UBE3D mRNA (Figure 2).
Figure 2.
Rescue of ube3d Morphants
(A–C) (A) Live images of 24 hpf WT; (B) Live images of 24 hpfMO; (C) Live images of rescue 24-hpf larvae. (D–I) (D and G) Live images of 96 hpf WT; (E and H) Live images of 96 hpf MO; (F and I) Live images of Rescue 96-hpf larvae. (G) Enlargement of (D), (H) Enlargement of (E), (I) Enlargement of (F). (J) At 96 hpf, the ube3d MO embryos were partially rescued by coinjection with human ube3d mRNA, and the percentage of small eyes in the rescued larvae was significantly lower than the percentage in MO larvae.
Knockdown of ube3d in Zebrafish Causes Increased Cell Death in Eyes
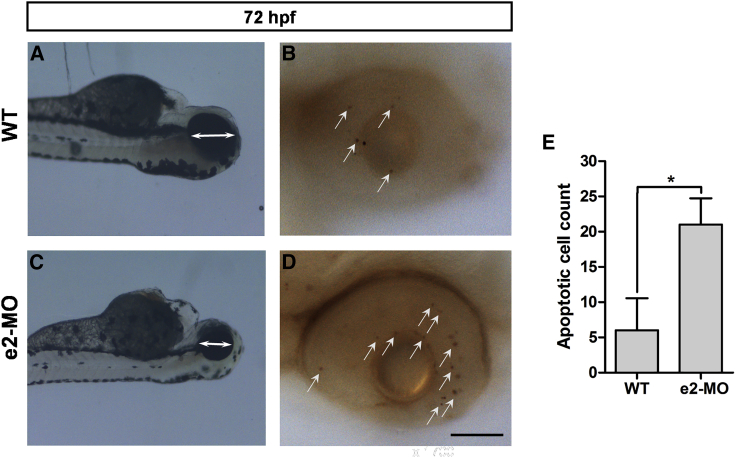
To evaluate whether apoptosis contributed to the small size of the eyes observed in the e2-MO zebrafish, we used terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining to detect apoptotic cells. TUNEL staining revealed a higher proportion of apoptotic cells in the eyes of e2-MO 72-hpf larvae (Figures 3C and 3D) than that in WT 72-hpf larvae (Figures 3A and 3B). A graphical representation has been provided to demonstrate that the number of apoptotic cells in e2-MO 72-hpf larvae (21.0 ± 2.2) was significantly higher than the number in WT larvae (6.0 ± 2.6; p < 0.05; Figure 3E).
Figure 3.
Knockdown of ube3d Causes Increases Cell Death in the Eye
(A) TUNEL staining revealed few apoptotic cells (arrows) in the eyes of WT 72-hpf larvae. (B) Enlargement of (A). (C) TUNEL staining revealed an increase in apoptotic cells (arrows) in the eyes of e2-MO 72-hpf larvae. (D) Enlargement of (C). (E) A graphical representation demonstrating that the number of apoptotic cells in e2-MO 72-hpf larvae (21.0 ± 2.2) was significantly higher than the number in WT larvae (6.0 ± 2.6). The data are presented as the mean ± SD. ∗p < 0.05. Scale bars represent 250 μm (A and C) and 70 μm (B and D).
Knockdown of ube3d in Zebrafish Results in Retinal Degeneration in Adult Zebrafish
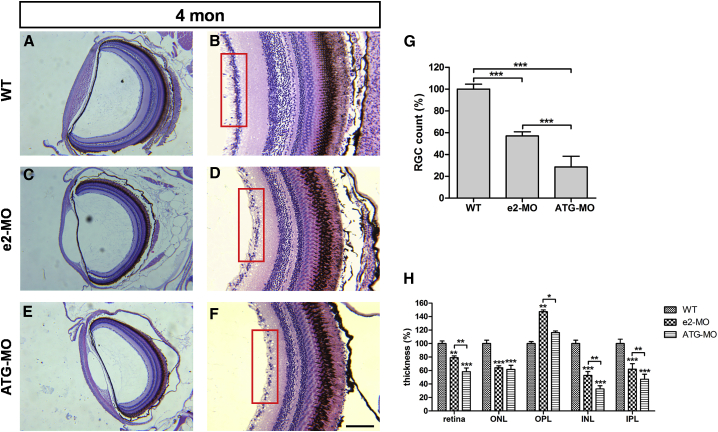
To investigate the role of ube3d in the eyes of adult zebrafish, we analyzed retinal structures using H&E staining in transverse retinal paraffin sections of eyes obtained from WT zebrafish and e2-MO and ATG-MO ube3d morphants at 4 months. As shown in Figure 4, the number of retinal ganglion cells was significantly lower in e2-MO and ATG-MO ube3d morphants than that in WT zebrafish (Figures 4A–4G). In addition, all of the layers of the retina in the e2-MO and ATG-MO ube3d morphants were significantly thinner than the layers in the WT zebrafish (Figures 4A–4F and 4H).
Figure 4.
Knockdown of ube3d Results in Retinal Degeneration in Adult Zebrafish
(A) Hematoxylin and eosin (H&E) staining of transverse retinal paraffin sections through WT zebrafish eyes at 4 months old. (B) enlargement of (A). (C) H&E staining of transverse retinal paraffin sections through e2-MO ube3d morphant eyes at 4 months old. (D) Enlargement of (C). (E) H&E staining of transverse retinal paraffin sections through ATG-MO ube3d morphant eyes at 4 months old. (F) enlargement of (E). (G) A graphical representation demonstrating that the number of retinal ganglion cells was significantly lower in e2-MO and ATG-MO ube3d morphants than the number in WT zebrafish. (H) All layers of the retina in e2-MO and ATG-MO ube3d morphants were significantly thinner than the corresponding layers in WT zebrafish. The red boxes in (B), (D), and (F) indicate the ganglion cell layer. The data are presented as the mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars represent 100 μm (A, C, and E) and 70 μm (B, D, and F).
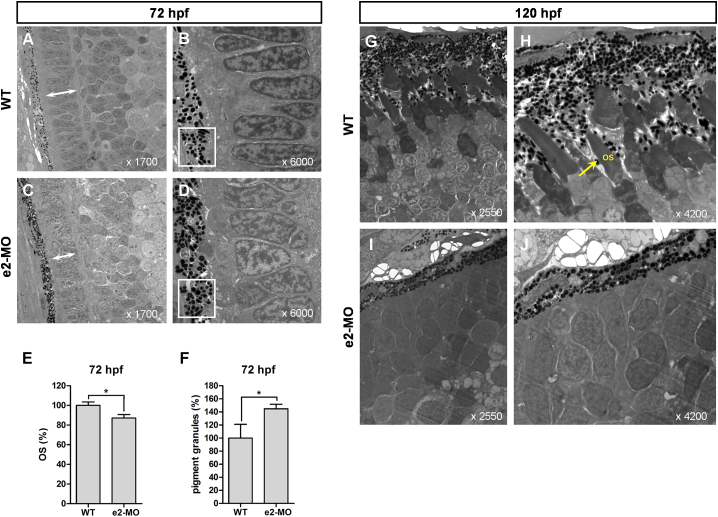
Knockdown of ube3d in Zebrafish Results in a Shorter Photoreceptor Outer Segment Layer and Increased Deposition of Pigment Granules
We used TEM to analyze the morphological differentiation of the zebrafish eye. At 72 hpf, TEM of transverse sections cut along the dorsal-ventral axis of the eye revealed shorter photoreceptor outer segments (OSs) and a larger number of deposited pigment granules in the photoreceptor OS layer in ube3d morphants than those in WT larvae (Figures 5A–5F). These differences were statistically significant (Figures 5E and 5F). At 120 hpf, photoreceptor OSs were present in WT larvae (Figures 5G and 5H), but not in ube3d morphants (Figures 5I and 5J).
Figure 5.
Knockdown of ube3d Results in More Deposited Pigment Granules and an Abnormal Photoreceptor-Outer Segment Layer
(A) Transmission electron micrographs of transverse sections cut along the dorsal-ventral axis of WT zebrafish eyes at 72 hpf (with ×1,700 and ×6,000 magnification, as indicated). (B) Enlargement of (A). (C) Transmission electron micrographs of transverse sections cut along the dorsal-ventral axis of e2-MO ube3d morphant eyes at 72 hpf (with ×1,700 and ×6,000 magnification, as indicated). (D) Enlargement of (C). (E) Graphical representation demonstrating that OS (arrow) lengths were shorter in the ube3d morphants than those in the WT larvae. (F) Graphical representation demonstrating that more pigment granules were deposited in the photoreceptor OS layer in ube3d morphants than were deposited in WT larvae. (G) Photoreceptor OSs (arrow) were present in WT 120-hpf larvae (with ×2,550 and ×4,200 magnification, as indicated). (H) Enlargement of (G). (I) Photoreceptor OSs were not observed in the ube3d morphants at 120 hpf (with ×1,700 and ×6,000 magnification, as indicated). (J) Enlargement of (I). The data are presented as the mean ± SD. ∗p < 0.05.
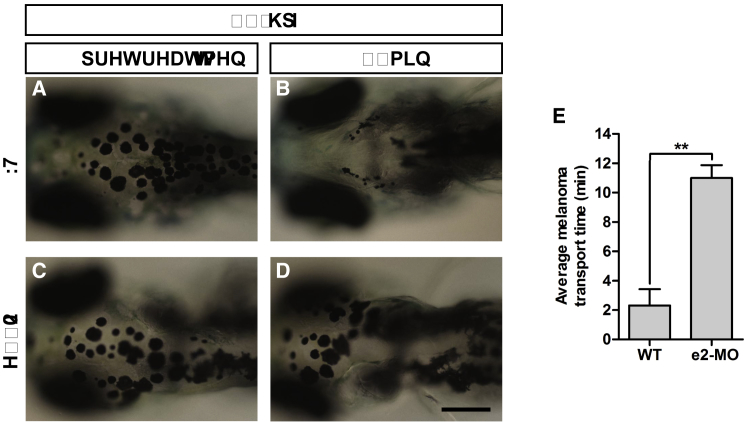
Knockdown of ube3d in Zebrafish Delays Retrograde Intracellular Transport
To induce maximum melanophore dispersion, we dark-adapted 120-hpf WT and MO larvae, and epinephrine was then used to induce melanosome transport. Following exposure to epinephrine, WT larvae rapidly retracted their melanosomes (Figures 6A, 6B, and 6E) (t = 2.3 min), whereas ube3d morphants exhibited a significant delay in melanosome retraction (Figures 6C–6E; t = 11.0 min; p < 0.01).
Figure 6.
Knockdown of ube3d Results in Delayed Epinephrine-Induced Melanosome Retraction
(A and B) WT 120-hpf larvae prior to treatment (A) and at 4 min after exposure to epinephrine (B). (C and D) e2-MO 120-hpf larvae prior to treatment (C) and at 4 min after exposure to epinephrine (D). (E) A graphical representation demonstrating that epinephrine-induced melanosome retrograde trafficking was significantly slower in e2-MO 120-hpf larvae (11.0 ± 0.5 min) than trafficking in WT larvae (2.32 ± 0.64 min). The data are presented as the mean ± SD. ∗∗p < 0.01.
Knockdown of ube3d in Zebrafish Decreases Dark-Induced Hyperlocomotor Activity under Light-Off Conditions
Behavioral tests demonstrated that ube3d morphants exhibited aberrant locomotor responses during the light-off phase (Figure S3), including reductions in the total distance traveled and the total time spent moving. These findings strongly indicate that ube3d plays an important role in visual functions and that ube3d dysfunction can cause abnormal activity.
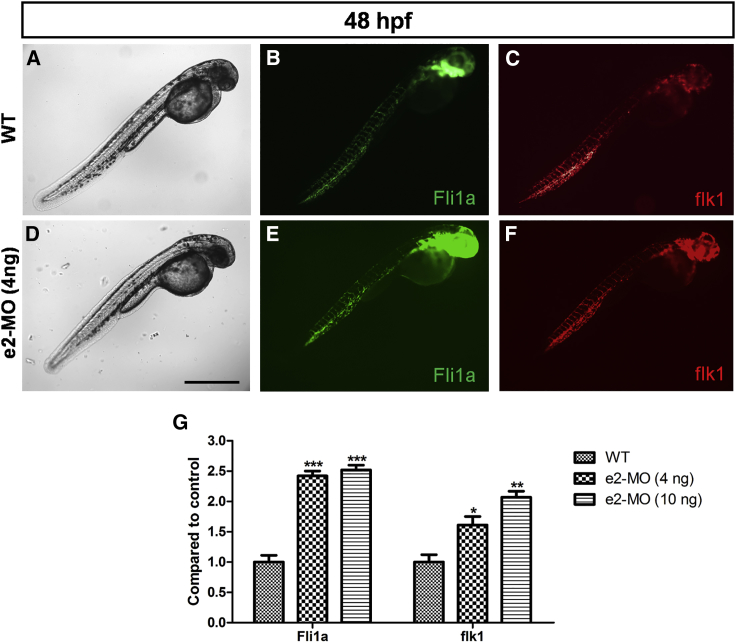
Knockdown of ube3d in Zebrafish Promotes Angiogenesis
To compare the distribution pattern of blood vessels between WT zebrafish and ube3d morphants, we used Tg(flk:mCherry, flila:EGFP) zebrafish as a control. As shown in Figure 7, Fli1a and flk1 expression levels were significantly higher in the e2-MO ube3d morphants than those in the controls.
Figure 7.
Knockdown of ube3d Promotes Angiogenesis
(A–C) Live images (A), Fli1a expression (B), and flk1 expression (C) in WT 48-hpf larvae. (D–F) Live images (D), Fli1a expression (E), and flk1 expression (F) in e2-MO (4 ng) 48-hpf larvae. (G) The expression levels of Fli1a and flk1 were significantly increased in e2-MO 48-hpf larvae. The data are presented as the mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bar represents 500 μm.
In Vitro
Identification of Recombinant Plasmid UBE3D
The positive clones were identified after PCR amplification. The detected sequence was identical to the known human UBE3D sequence in GenBank (Gene ID 90025). A western blot revealed a 32-kDa band in cell extracts, which was in accordance with the expected size of an UBE3D-Flag confluency protein (28 kDa–36 kDa). These results indicate that the UBE3D recombinant plasmid was successfully expressed in hRPE cells and suggested that the cells were successfully transduced with lentivirus.
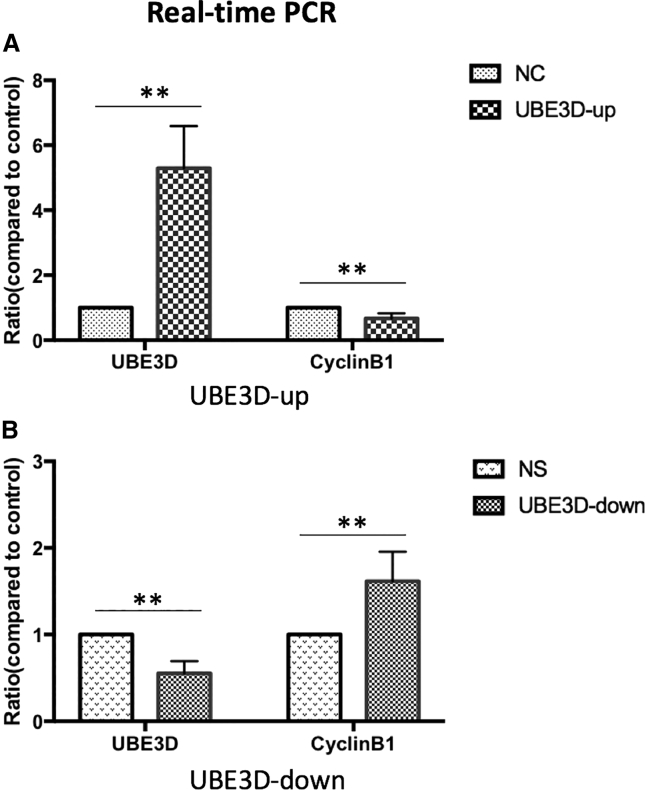
Overexpression/Low Expression of UBE3D by Stable Transfection of UBE3D
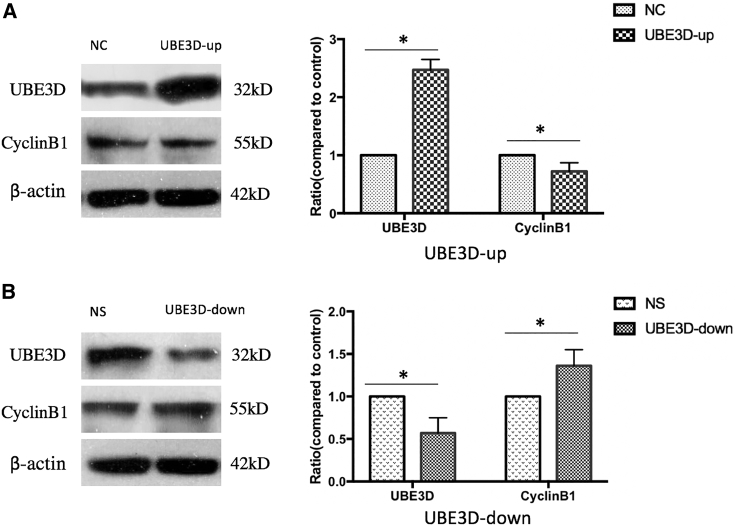
Total mRNA and protein extracts were prepared from lentiviral vector control (NC), overexpression of UBE3D-transfected (UBE3D-up), lentiviral nonspecific control (NS), and low expression of UBE3D-transfected (UBE3D-down) hRPE cells. The UBE3D mRNA and protein expression levels in these cells were determined by real-time RT-PCR and western blot assays, respectively. Real-time RT-PCR demonstrated that the mRNA levels of UBE3D protein expression in the transfectants overexpressing UBE3D were increased compared with those in the lentiviral vector control (NC) transfectants, which was consistent with the increased UBE3D protein expression (p < 0.01; Figure 8A). In contrast, compared with the lentiviral nonspecific control (NS) transfectants, the levels of UBE3D protein expression in the transfectants containing the UBE3D-down vector were decreased (p < 0.01; Figure 8B). Western blots showed consistent results (p < 0.05; Figures 9A and 9B). These results indicate that stable transfection upregulated or downregulated UBE3D expression in hRPE cells.
Figure 8.
Effect of UBE3D on CyclinB1 Protein Expression in hRPE Cells as Determined by Real-Time RT-PCR
mRNA expression levels of UBE3D and CyclinB1 in hRPE cells transfected with overexpression/low expression of UBE3D vectors. Compared with the control groups, the UBE3D mRNA levels were increased, while CyclinB1 levels were reduced in UBE3D-overexpressing cells (∗∗p < 0.01; A). Additionally, the UBE3D mRNA levels were decreased, while CyclinB1 levels were increased in low-UBE3D-expressing cells (∗∗p < 0.01; B).
Figure 9.
Effect of UBE3D on CyclinB1 Protein Expression in hRPE Cells as Determined by Western Blots
Protein expression of UBE3D and CyclinB1 in hRPE cells transfected with overexpression/low expression of UBE3D vectors. Compared with the control groups, the UBE3D protein levels were increased, while CyclinB1 levels were reduced in UBE3D-overexpressing cells (∗p < 0.05; A). The UBE3D protein levels were decreased, while CyclinB1 levels were increased in low-UBE3D-expressing cells (∗p < 0.05; B).
Effect of UBE3D on CyclinB1 Protein Expression
Real-time RT-PCR demonstrated that CyclinB1 mRNA expression levels exhibited an opposite pattern compared to those of UBE3D (p < 0.05; Figures 8A and 8B). Western blots showed consistent results (p < 0.05; Figures 9A and 9B). CyclinB1 is the product of UBE3D. A negative correlation between UBE3D and CyclinB1 expression was observed.
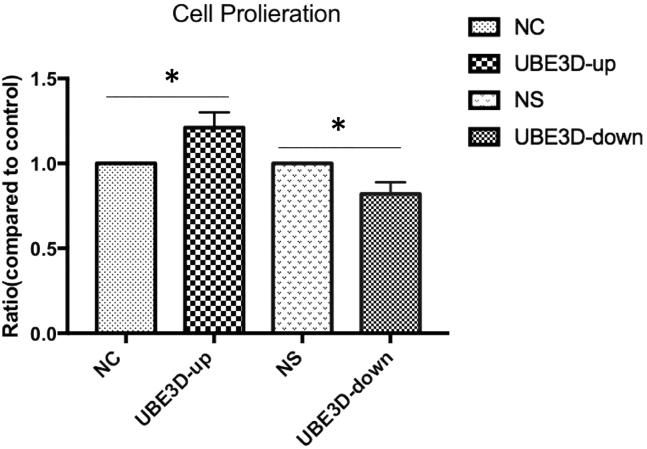
UBE3D Regulates hRPE Cell Proliferation and Migration
To determine the effect of UBE3D-induced hRPE cell proliferation, we quantified the metabolic activities of UBE3D-up, NC, UBE3D-down, and NS cells using the cell counting kit-8 (CCK-8) assay. UBE3D-up cell proliferation was enhanced by 21% compared with NC cell proliferation (p < 0.05, Figure 10). In contrast, UBE3D-down cell proliferation was increased by 18% compared with NS cell proliferation (p < 0.05, Figure 10).
Figure 10.
Effect of UBE3D on hRPE Cell Proliferation
Proliferation of hRPE cells was measured with a CCK-8 assay. The values are listed as the mean ± SD of at least three independent experiments. Compared with the control groups, hRPE cell proliferation was enhanced in the UBE3D-overexpressing group, and hRPE cell proliferation was induced in the low-UBE3D-expression group (∗p < 0.05).
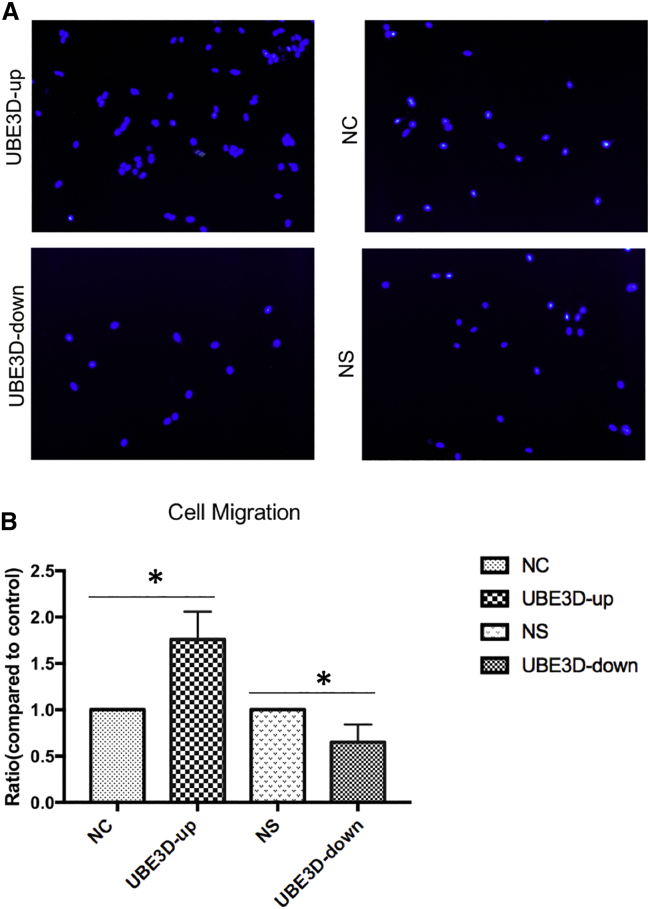
To explore the role of UBE3D in the migration of hRPE cells, we used a modified Boyden chamber in which the cells migrated through a porous membrane. The mean counts of migrating cells in the UBE3D-up group were significantly higher than those in the NC group (p < 0.05, Figure 11). In contrast, the mean counts of migrating cells in the UBE3D-down group were significantly lower than those in the NS group (p < 0.05, Figure 11).
Figure 11.
Effect of UBE3D on the Migration of hRPE Cells
(A) shows RPE through Boyden upper compartment (DAPI staining, RPE-colored nuclei through the compartment as seen under fluorescence microscopy). (B) Statistical results. The migratory activities of cell lines were estimated based on the numbers of cells that had migrated through the filter of the chamber. The numbers of migrating cells in the UBE3D-overexpressing group were greater than those observed in the lentiviral vector control group. The numbers of migrating cells in the low-UBE3D-expressing group were lower than those observed in the lentiviral nonspecific control group (∗p < 0.05).
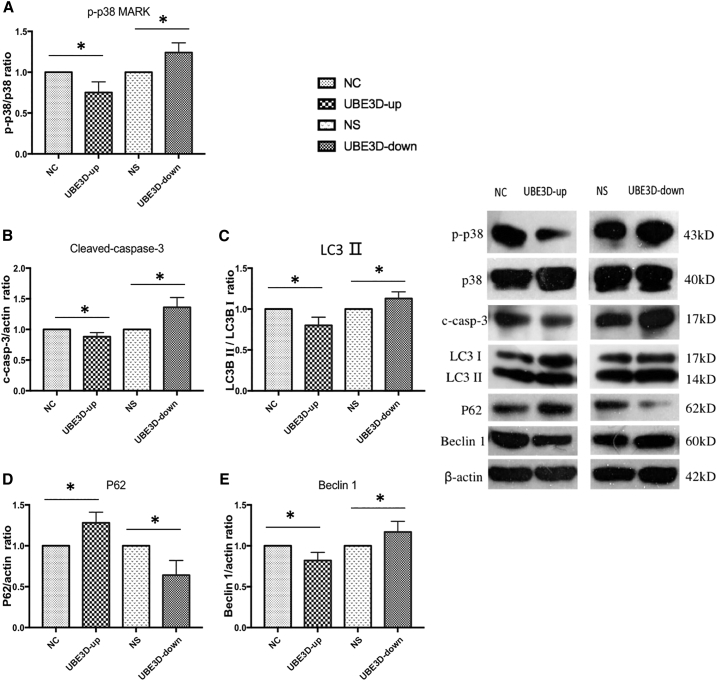
Effects of UBE3D on Caspase-3, p38MARK, LC3, P62, and Beclin1 Protein Expression
To investigate a possible mechanism of UBE3D in oxidative damage in hRPE cells, we performed western blot to assess the protein levels of cleaved-caspase-3, p-p38MARK, LC3, P62, and Beclin1. We observed significant downregulation of cleaved-caspase-3, p-p38MARK, LC3II, and Beclin1 in the UBE3D-up group compared with the corresponding observations in the NC group (p < 0.05, Figures 12A–12C and 12E). In contrast, the levels of cleaved-caspase-3, p-p38MARK, LC3II, and Beclin1 were upregulated in the UBE3D-down group compared with those in the NS group (p < 0.05, Figures 12A–12C and 12E). A negative correlation between LC3II and P62 expression was observed (Figure 12D).
Figure 12.
UBE3D Regulated p-p38MARK, Cleaved-Caspase-3, LC3, P62, and Beclin1 Protein Expression in hRPE Cells
The right panel shows representative blot images. (A–E) Results of the statistical analysis of the western blot data. As shown in this figure, p-p38MARK (A), cleaved-caspase-3 (B), LC3II (C), and Beclin1 (E) expression levels were downregulated in the UBE3D-up group (∗p < 0.05), while P62 (D) expression levels were upregulated (∗p < 0.05). In contrast, p-p38MARK (A), cleaved-caspase-3 (B), LC3II (C), and Beclin1 (E) expression levels were upregulated in the UBE3D-down group (∗p < 0.05), while P62 (E) expression levels were downregulated (∗p < 0.05).
Discussion
Accumulation of abnormal proteins and dysregulation of angiogenesis in the retina have been associated with the formation of drusen and choroidal neovascularization (CNV), which are hallmarks of AMD.16,17 The major role of the UPS is to prevent accumulation of abnormal proteins.8,17 Therefore, dysfunction in the UPS may contribute to the development of AMD. In this study, we established a ube3d-knockdown model in zebrafish to study the roles of ube3d, an important component of the UPS, during retinal development and normal visual functions in zebrafish.
The zebrafish retina is a well-laminated structure that is similar in many ways to the retinas of other vertebrates. The morphological differentiation of the retina is a complex process that involves cellular differentiation and regulation of synaptic connectivity. Previous reports have indicated that the UPS plays a crucial role in retinal differentiation and development in mammals.18, 19, 20 Genetic screens in Drosophila have shown that mutations in the genes encoding a ubiquitin ligase (E3) and ubiquitin-specific protease enzymes can lead to axon guidance defects, including defects in the regulation of synaptic connectivity.21 Other reports have shown that inhibiting the UPS in developing Xenopus retinal ganglion cells (RGCs) decreased terminal arborization, which is the basis of synaptic connectivity, in vivo.18 In addition, several transcription factors and signaling molecules known to be involved in normal retinal differentiation are degraded by the UPS.17 For example, microphthalmia-associated transcription factor (MITF) plays a crucial role in melanin biosynthesis in RPE cells, and the UPS regulates RPE differentiation by targeting MITF for proteasomal degradation.20 Retinal development is a complex process including both normal cellular differentiation and normal cell apoptosis. The apoptotic process is highly regulated by the UPS. Some members of the inhibitors of apoptosis family belong to the E3 family, and these factors block apoptosis by inactivating pro-apoptotic caspases by targeting them for ubiquitination.22 In agreement with these reports, we found that knockdown of ube3d in zebrafish delayed eye development, reduced eye size, caused apoptosis, and resulted in retinal degeneration. Ube3d is likely involved in regulating the levels of the transcription factors and signaling molecules that control retinal differentiation and development. In our previous study, homozygous mice all died young, and abnormalities in eye development in Ube3d+/- heterozygous mice was not observed. This evidence strongly indicates that ube3d is important during mouse development.
In the vertebrate eye, RPE cells lie next to photoreceptor OSs, which are the outermost cells in the retina. Interactions between these two cell types are essential for functional differentiation in the retina.23, 24, 25 Light collection is an essential task of the retina, especially for the light-sensitive photoreceptor OSs. Because they are exposed to high levels of light throughout the day, many photoreceptor OSs are damaged as a result of photo-oxidative damage. To maintain normal functions, the damaged photoreceptor OSs are constantly being removed by phagocytosis by cells in the RPE layer.26,27 A functional RPE is therefore required to digest photoreceptor OS membranes by taking them up into phagosomes, indicating that the length of a photoreceptor OS is constantly being regulated by coordination between the RPE layer and photoreceptors.27 The UPS has been shown to be involved in many cellular processes, including regulation of immune responses and inflammation. Because the RPE is a major retinal source of inflammation, impairment of the UPS may contribute to RPE dysfunction, which may result in photoreceptor OSs becoming abnormally elongated.28 In the present study, we found that knockdown of ube3d in zebrafish led to an increase in the number of deposited pigment granules and an abnormal photoreceptor outer segment layer. These data are consistent with those in our previous study showing that more pigment granules were deposited in the RPE microvilli in UBE3D+/- heterozygous mice than in WT mice, indicating that dysfunction of ubiquitin protein ligase (E3) results in the accumulation of extracellular deposits.14 Such dysfunction was also associated with abnormal development of photoreceptor OSs and an increase in the number of deposited pigment granules, which resulted in reduced light sensitivity of photoreceptors and decreased dark-induced hyperlocomotor activity under the light-off condition.
Melanosome transport is widely used as a model to analyze intracellular transport. In mammals, melanosomes are produced within skin melanocytes, RPE cells, and other cell types throughout the body. Retrograde melanosome transport toward the microtubule minus-ends at the cell center is accomplished by dynein and dynein-associated proteins, and this process can be used to evaluate melanosome motility within pigmented cells.29, 30, 31 In RPE cells, melanosome motility is achieved by RPE microvilli. In our previous study, we reported that more pigment granules were deposited in the RPE microvilli of Ube3d+/– heterozygous mice than in those of WT mice.14 In the current study, we observed that knockdown of ube3d in zebrafish significantly delayed melanosome retraction, further indicating the ube3d may be involved in RPE microvilli functions.
Angiogenesis, the process that results in the formation of new capillaries, plays a crucial role in several human diseases, including retinal diseases. Inhibitors of angiogenesis have been widely used in clinical settings as a novel therapeutic strategy.32,33 An increasing number of studies indicate that an organized distribution pattern of blood vessels in the retina is required for normal retinal development. Abnormal angiogenesis has been observed in AMD,33,34 and deregulation of vascular endothelial growth factor (VEGF) expression has been shown to contribute to abnormal angiogenesis. A previous in vitro study showed that impairment of the UPS resulted in increased expression and secretion of pro-angiogenic factors, including VEGF, in RPE cells.35 In the current study, we used the Tg(flk:mCherry, flila:EGFP) line of zebrafish in experiments to determine the function of ube3d in angiogenesis. Friend leukemia integration 1a (Fli1a) is a downstream target of E 26 transforming specific family variant 2 (Etv2), which is essential to the initiation of vascular morphogenesis in early zebrafish embryos.36 Various forms of VEGF bind to various VEGF receptors (VEGFRs) to induce angiogenesis, and the VEGF-VEGFR2 pathway is particularly important for the regulation of angiogenesis. Flk1 is the gene that encodes VEGFR-2 (also known as KDR/flk-1).37 In the data presented in this study, we show that knockdown of ube3d increased the expression levels of Fli1a and flk1, which resulted in an increase in angiogenesis. These results further confirm the association between ube3d and neovascular AMD, a hallmark of which is angiogenesis. However, we did not obtain positive results in our experiments aiming to induce angiogenesis in Ube3d+/– heterozygous mice in our previous study. We propose two reasons for this discrepancy: first, Ube3d+/– mice were used, which exhibited normal retinal vessels, a second, the Ube3d+/– mice (3 months of age) in our experiments may not have been sufficiently old to develop abnormal angiogenesis under the experimental conditions.
To demonstrate a potential link between ube3d and AMD, in vivo studies are required to reveal the mechanisms underlying this relationship. Our experiments at both the gene level and protein level show a negative correlation between UBE3D and CyclinB1 (Figures 8 and 9). We verified that ubiquitin ligase (E3) could specifically identify CyclinB1, which was undergoing ubiquitination and then degradation. CyclinB1 is a key cell-cycle regulator in the progression of mitosis, and the ubiquitin-protein degradation system plays an important role in mitosis regulation.38 Identifying specific E3 ligase substrates and correcting the pathological state of E3 ligase activation may become medical intervention strategies for AMD.
Oxidative damage to cellular components is known to be largely involved in the pathogenesis of AMD.39,40 In this study, after induction of oxidative damage, proliferation (Figure 10), and migration (Figure 11) abilities were better in the UBE3D-up group compared with those in the NC group. As expected, proliferation and migration abilities became worse in the UBE3D-down group, indicating that hRPE cells in the UBE3D-down group were more sensitive to oxidative damage. Thus, UBE3D may play a role in maintaining the normal physiologic functions and homeostatic functions of hRPE cells.
Western blot analysis (Figure 12) showed that the protein levels of cleaved-caspase-3 and p-p38 were downregulated in the UBE3D-up group, while opposite results were observed in the UBE3D-down group, indicating that ubiquitin proteasome dysfunction was involved in the pathological course of oxidative damage. One study found oxidative damage and ubiquitin-proteasome system disorder in macular drusen and retinal tissue biopsies from AMD patients.41 Ubiquitin is present throughout the retina in the RPE. E1 and E2 activities of the ubiquitin conjugation system have been reported to be increased in the human lens and RPE cells in response to mild oxidative stress.42 The pathogenesis of age-related macular degeneration (AMD) essentially involves chronic oxidative stress, increased accumulation of lipofuscin in RPE cells, and extracellular drusen formation, as well as chronic inflammation. The capacity to prevent the accumulation of cellular cytotoxic protein aggregates is decreased in senescent cells, which may evoke lipofuscin accumulation in lysosomes in post-mitotic RPE cells. This presence of lipofuscin decreases lysosomal enzyme activity and impairs autophagic clearance of damaged proteins, which should be removed from cells. Proteasomes are another crucial proteolytic machine that degrade cellular proteins damaged by oxidative stress.43
In this study, the expression levels of autophagy-related protein also changed. The protein levels of LC3II and Beclin1 were downregulated in the UBE3D-up group, while opposite results were observed in the UBE3D-down group, suggesting interactions between the UPS and autophagy. A negative correlation was found between LC3II and P62 expression. P62 is an encoding ubiquitin binding protein for SQSTM1 and is involved in both UPS and autophagy protein degradation processes.44 The UPS and autophagy-lysosome pathway (ALP) are the two most important mechanisms that normally repair or remove abnormal proteins. Whereas the ubiquitin-proteasome system is involved in the rapid degradation of proteins, autophagy pathways can selectively remove protein aggregates and damaged or excess organelles. Although autophagy has long been viewed as a random cytoplasmic degradation system, the involvement of ubiquitin as a specificity factor for selective autophagy is rapidly emerging.45
In conclusion, we show that ube3d is essential for eye development and the maintenance of the normal functions of the eye in zebrafish. Dysfunction in ube3d may contribute to the development of neovascular AMD. Further, we identified a negative correlation between UBE3D and CyclinB1. The UBE3D gene was involved in the pathogenesis of age-related macular degeneration. Low UBE3D expression can promote oxidative damage and inflammatory reactions. UBE3D and autophagy may have a synergetic effect on anti-oxidative damage.
Materials and Methods
In Vivo
Zebrafish Care and Ethics Statement
The Tübingen strain of zebrafish and Tg(flk:mCherry, flila:EGFP) zebrafish were raised at ∼28.5°C under standard conditions with a 14-h light and 10-h dark cycle. All of the experiments were performed in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research and the guidelines of the Institutional Animal Care and Use Committee (IACUC) of Peking University. The procedures were approved by the Animal Care and Use Committee of Peking University People’s Hospital (2011-72, Beijing, China).
WISH of ube3d
WISH is a process that allows detection of the expression and localization of mRNA in fixed embryos. WISH was performed essentially as previously described.46 NBT/BCIP (50 mg/mL; Promega) was used as an alkaline phosphatase substrate. To obtain the probe for in situ hybridization, we amplified ube3d cDNA by PCR using the primer pairs 5′-AGATGTGAAGATCGAATCCAAAC-3′ and 5′-ACATCCACAACATCTCTGAGCTT-3′ and then cloned into pEASY-T3 (TransGen). The plasmid was then linearized with ApaI, and in vitro transcription was performed using the SP6 RNA polymerase. The localization of ube3d mRNA was detected in WT zebrafish (Figure S1).
Zebrafish Injections for Morpholino Knockdown and Rescue
The following antisense MOs were designed against ube3d (Ensembl: ENSDART00000038036): E2I2 MO, 5′-ATCTTAGCGTGTACCTGTTGGCTGC-3′ and ATG MO, 5′-CATCTGTGTTCGCAGTCTCTTCCAT-3′. They were obtained from Gene Tools (Corvallis, OR, USA). We injected 10 ng of the MOs diluted with phenol red tracer into one-cell-stage embryos. For the rescue experiments, capped human full-length ube3d mRNA was synthesized using the mMessage mMachine T7 kit according to the manufacturer’s instructions (AM1344; Ambion, Grand Island, NY, USA). 400 picograms of UBE3D mRNA were coinjected with the morpholinos into one- to two-cell-stage embryos.
RT-PCR
Total RNA was extracted from splice-blocking morphants (5 embryos/stage) at 72 hpf and synthesized into cDNA. The effect of ube3d-E2I2-MO on mRNA splicing was then analyzed using the following primers, which amplified the product of either normal splicing or exon 2 skipping: primer pair e1 (5′-TTTATTGAGCTCAGACAGAAGCTG-3′) and e3 (5′-GCCCGTGTCAGACTCTCATC-3′). The expression of β-actin was analyzed as an internal control.
The Ube3d ATG-MOs target sequence and a full-length GFP cDNA were amplified using PCR with the forward primer 5′-cggatccCCAATGGAAGAGACTGCGAACACA GATGTGAGCAAGGGCGAGGAGCTG-3′ and the reverse primer 5′-GGACGAGCTGTACAAGTAAtctagag-3′. Then, the ube3d ATG-MO-GFP cDNA was digested and ligated into the pCS2vector between its BamHI and XbaI cloning sites. Ube3d ATG-MO-GFP mRNA was synthesized using an mMessage mMachine kit (Ambion) from a pCS2-GDNF vector that was linearized with NotI, transcribed using SP6, ethanol precipitated, and resuspended in sterile distilled water for injection at a concentration of 100 ng/nL. To test the specificity of the ATG-MO, we generated a modified pCS2-GFP containing the MO target sequences for the ube3d ATG-MO directly upstream of the corresponding start codon (Figure S2).
Quantification of Eye/Body Size
To compare the eye-to-body ratio between WT and morphant zebrafish embryos, we measured the diameters of zebrafish eyes and body lengths using the software ImageJ (National Institutes of Health, Bethesda, MD, USA) in images of whole embryos collected using an Axioimager Z1 fluorescence microscope (Zeiss) and an AxioCam MRm digital camera. Eye-to-body length ratios were determined by collecting one image showing the area with the widest diameter from each morphant or WT embryo.
Whole-Mount TUNEL Assay
Embryos (n = 20/groups) were fixed overnight at 72 hpf in 4% paraformaldehyde, dehydrated in a series of phosphate-buffered saline (PBS)/ethanol solutions, and then incubated in 100% acetone at −20°C. Then, the embryos were rehydrated and permeabilized by incubating them in a solution of proteinase K in PBS with 0.1% Tween-20 (PBST). TUNEL-positive cells were detected using an In Situ Cell Death Detection Kit, POD (Roche Applied Science, Rotkreuz, Switzerland), according to the manufacturer’s instructions.
Histopathological Observation
At 4 months post-injection, the zebrafish were euthanized with a lethal dose of tricaine methanesulfonate, and their eyes were removed and dissected. Paraffin-embedded sections were then prepared from whole eyes. Serial sections (5 μm) were stained using a conventional hematoxylin and eosin (H&E) protocol, and the slides were then observed using a microscope (Olympus, Japan).
TEM
Zebrafish control animals and zebrafish subjected to MO knockdown for 72 h, 120 h, or 4 months were fixed using 3% glutaraldehyde in 0.1 M PBS (pH 7.0) for 2 h at 4°C. The zebrafish were then post-fixed in 1% osmic acid in the same buffer, dehydrated in ethanol, and embedded in epoxy resin. Thin sections (1,000 nm) were cut and then stained with toluidine blue. Finally, thinner sections (70 nm) were stained with uranium and lead prior to examination using a transmission electron microscope (FEI Company, Hillsboro, OR, USA).
Melanosome Transport Assay
After an overnight dark-adaptation period, control and MO-knockdown zebrafish (120 hpf) were exposed to epinephrine by adding this agent to the embryonic medium to obtain a final concentration of 500 μg/mL, as previously described.47 Melanosome retraction was continuously monitored under a bright-field microscope, and the endpoint was achieved when all melanosomes in the head and trunk were perinuclear.
Behavioral Observation
EthoVision-XT software (Noldous Information Technology, Netherlands) was used to track the location of the zebrafish to assay general locomotor activity. In the present study, 144-hpf WT and MO zebrafish (n = 6/group) were placed in 24-well plates with fresh zebrafish system water. During recording of locomotor activity, each zebrafish was kept in one well (area = 2 cm2) for a total of 50 min as follows: after allowing 10 min for adaptation to the environment, measurements were performed for the next 40 min, which consisted of two 20-min light/dark cycles (light on for 10 min and light off for 10 min). Measurements included the total distance traveled, the velocity of travel and the duration of movements. All zebrafish were tested between 10:00 and 11:00 a.m.
Imaging
All micrographs were obtained under the same conditions and saved as TIFF files. The images were processed using graphics software (Adobe Photoshop CS5, Adobe Systems, San Jose, CA, USA).
In Vitro
Cell Culture and Lentivirus Transduction
We obtained hRPE cells from the American Type Culture Collection (ARPE-19, CRL-2302; ATCC, Manassas, VA, USA). The cells were cultured in a 1:1 mixture of Dulbecco’s Modified Eagle’s Medium and F12 (GIBCO, Grand Island, NY) supplemented with 10% FBS at 37°C in 5% CO2 and 95% humidity. When the hRPE cells were approximately 50% confluent in fresh serum-free medium, they were transiently transduced with control lentivirus, UBE3D-up expression lentivirus or UBE3D-down expression lentivirus at MOI (multiplicity of infection) of 10. The cells were further cultured in DMEM with 10% FBS after infection for 4 h. When more than 90% of the transduced cells were found to strongly express GFP under fluorescence microscopy, the hRPE cells were plated in six-well culture dishes and used for experiments at 80% to 90% confluence, and the cells were incubated in DMEM with 1% FBS for 24 h before tert-butyl hydroperoxide (t-BHP, 500 μM, Sigma-Aldrich) was added to the wells. Cells were collected at the 4-h time-point after t-BHP treatment.
RNA Isolation and Relative Quantitative Real-Time PCR
Total RNA was isolated from hRPE cells using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Reverse transcriptase reactions were performed using a RevertAid First Strand cDNA Synthesis Kit with oligo-dT primer (Fermentas, Pittsburgh, PA, USA). Real-time PCR reactions were performed with SYBR Green PCR mix (Thermo, Pittsburgh, PA, USA) using an ABI7300 real-time PCR system (Applied Biosystems, Life Technologies, Foster City, CA, USA) with 10 μL reaction mixtures containing 1 μL of cDNA, 3.5 μL of sterilized water, 5 μL of SYBR Green Real-Time PCR Master Mix, and 0.5 μL of primer (10 μM).
The UBE3D and CyclinB1 amplification signals were normalized to the housekeeping gene human glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and relative multiples of changes in mRNA expression were determined by calculating the fold change = 2-ΔΔcq. Each experiment was repeated five times. The primers used for real-time PCR are shown in Table 1.
Table 1.
Gene Subtype Oligonucleotide Primers
| Gene Subtype | Primer Sequence (5′-3′) | Size (bp) |
|---|---|---|
| Human UBE3D | F: TTCCCCTTGTTGGAAAACACA | 138 |
| R: TGGCTGGTAGAGGACCTTGAC | ||
| Human CyclinB1 | F: TTGGGGACATTGGTAACAAAGTC | 126 |
| R: ATAGGCTCAGGCGAAAGTTTTT | ||
| Human GAPDH | F: GAGTCCACTGGCGTCTTCAC | 100 |
| R: GTTCACACCCATGACGAACA |
F, forward; R, reverse.
In Vitro Cell Proliferation Assay
The cell proliferation assay was performed as described previously.48 The hRPE cells were incubated in 96-well plates. CCK-8 (Dojindo, Japan) assays were performed according to the manufacturer’s instructions. The absorbance at 450 nm was measured using an ELISA plate reader (Finstruments Multiskan Models 347; MTX Lab Systems, Vienna, VA, USA). Each experiment was performed in five wells and repeated at least three times.
Cell Migration Assay in hRPE ells
The hRPE cell migration study was performed using Transwells (Cat#3422; Corning, Tewksbury, MA, USA) as described previously.49 Briefly, 5 × 104 cells were placed in the upper chamber in a final volume of 200 μL of serum-free medium. The DMEM/F12 medium with 10% FBS was placed in the bottom chamber at a final volume of 600 μL. All migration assays were conducted at 37°C for 6 h, and the cells were then fixed in 4% paraformaldehyde (PFA) and stained with 4,6-diamidino-2-phenylindole (DAPI; Roche Diagnostics, Indiana- polis, IN, USA). The remaining cells were wiped away with a cotton swab, and the membrane was imaged under fluorescence microscopy (Zeiss Axiophot, Thornwood, NY, USA). The hRPE cells in five random fields of view were counted. Each experiment was repeated three times.
Protein Extraction and Western Blot Analysis
The hRPR cells were washed three times with ice-cold PBS (4°C, pH 7.4) for 5 min at room temperature and prepared using a protein extraction kit and a protease inhibitor kit (Pierce, Rockford, IL). After centrifugation, the supernatant was collected, and the protein content of each lysate was measured with a bicinchoninic acid (BCA) protein assay kit (Pierce) according to the manufacturer’s instructions. Equal amounts of protein (40 μg) were separated by a 12% sodium dodecyl sulfate (SDS) polyacrylamide gel and transferred onto a 0.22-μm polyvinylidene fluoride (PVDF) membrane (Millipore). The primary antibodies used to probe the membranes included anti-UBE3D (1:500, PAB21883, Abnova, USA), anti-CyclinB1 (1:1,000,# 4138, CST, USA), anti-caspase-3 (1:500, ab13847, Abcam, Cambridge, MA, USA), anti-p38 mitogen-activated protein kinases (p38MARK) (1:1,000, #8690, CST, USA), anti-phosphorylated p38 MAPK (p-p38MARK) (1:1,000, #4511, CST, USA), anti-P62 (1:2,000, PM045, MBL, Japan), anti-light chain3 (LC3) (1:1,000, #2775, CST, USA), anti-Beclin1 (1:2,000, ab207612, Abcam, Cambridge, MA, USA) and anti-β-actin (1:2,000, #4970 CST, USA). The membranes were washed and incubated with peroxidase-conjugated secondary antibodies (1:5,000, Boster, China). The proteins were visualized with enhanced chemiluminescence western blot detection reagents (Millipore).
Statistical Analysis
Data analyses were performed using SPSS software, version 22.0 (SPSS, Chicago, IL, USA). All data are presented as the means ± standard deviation (SD). Differences between groups were evaluated using the Mann-Whitney U test. A p value less than 0.05 was considered statistically significant (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 in the figures).
Author Contributions
Conceived and designed the experiments: H.X., Q.Z., L.H., and X.L. Performed the experiments: H.X. and Q.Z. Analyzed the data: H.X., Q.Z., Y.S., Y.B., X.M., and L.H. Contributed reagents/materials/analysis tools: H.X., Q.Z., L.H., B.Z., Y.Q., J.Z., Q.H., W.D., L.Z., H.X., P.Z., and B.W. Wrote the paper: H.X., Q.Z., L.H., and X.L.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant number 81470649, 81670870), the National Basic Research Program of China (973 Program, grant number 2011CB510200, 2012CB945101), the Beijing Nova Program (Z161100004916058), Science and Technology Innovation Project of Chinese Academy of Medical Sciences (2019-RC-HL-019), and Huaxia Translational Medicine Fund for Young Scholars (grant number 2017-C-001). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.02.010.
Contributor Information
Lvzhen Huang, Email: huanglvzhen@126.com.
Xiaoxin Li, Email: drlixiaoxin@163.com.
Supplemental Information
References
- 1.Resnikoff S., Pascolini D., Etya’ale D., Kocur I., Pararajasegaram R., Pokharel G.P., Mariotti S.P. Global data on visual impairment in the year 2002. Bull. World Health Organ. 2004;82:844–851. [PMC free article] [PubMed] [Google Scholar]
- 2.Pascolini D., Mariotti S.P., Pokharel G.P., Pararajasegaram R., Etya’ale D., Négrel A.D., Resnikoff S. 2002 global update of available data on visual impairment: a compilation of population-based prevalence studies. Ophthalmic Epidemiol. 2004;11:67–115. doi: 10.1076/opep.11.2.67.28158. [DOI] [PubMed] [Google Scholar]
- 3.Rosenfeld P.J., Brown D.M., Heier J.S., Boyer D.S., Kaiser P.K., Chung C.Y., Kim R.Y., MARINA Study Group Ranibizumab for neovascular age-related macular degeneration. N. Engl. J. Med. 2006;355:1419–1431. doi: 10.1056/NEJMoa054481. [DOI] [PubMed] [Google Scholar]
- 4.Boulton M., Róanowska M., Wess T. Ageing of the retinal pigment epithelium: implications for transplantation. Graefes Arch. Clin. Exp. Ophthalmol. 2004;242:76–84. doi: 10.1007/s00417-003-0812-8. [DOI] [PubMed] [Google Scholar]
- 5.Zarbin M.A. Current concepts in the pathogenesis of age-related macular degeneration. Arch. Ophthalmol. 2004;122:598–614. doi: 10.1001/archopht.122.4.598. [DOI] [PubMed] [Google Scholar]
- 6.Zhou J., Cai B., Jang Y.P., Pachydaki S., Schmidt A.M., Sparrow J.R. Mechanisms for the induction of HNE- MDA- and AGE-adducts, RAGE and VEGF in retinal pigment epithelial cells. Exp. Eye Res. 2005;80:567–580. doi: 10.1016/j.exer.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Ambati J., Atkinson J.P., Gelfand B.D. Immunology of age-related macular degeneration. Nat. Rev. Immunol. 2013;13:438–451. doi: 10.1038/nri3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doherty F.J., Dawson S., Mayer R.J. The ubiquitin-proteasome pathway of intracellular proteolysis. Essays Biochem. 2002;38:51–63. doi: 10.1042/bse0380051. [DOI] [PubMed] [Google Scholar]
- 9.Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., Suzuki T. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 10.Haas A.L., Siepmann T.J. Pathways of ubiquitin conjugation. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- 11.van Wijk S.J.L., Timmers H.T.M. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 2010;24:981–993. doi: 10.1096/fj.09-136259. [DOI] [PubMed] [Google Scholar]
- 12.Lennox G., Lowe J., Morrell K., Landon M., Mayer R.J. Ubiquitin is a component of neurofibrillary tangles in a variety of neurodegenerative diseases. Neurosci. Lett. 1988;94:211–217. doi: 10.1016/0304-3940(88)90297-2. [DOI] [PubMed] [Google Scholar]
- 13.Lam Y.A., Pickart C.M., Alban A., Landon M., Jamieson C., Ramage R., Mayer R.J., Layfield R. Inhibition of the ubiquitin-proteasome system in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2000;97:9902–9906. doi: 10.1073/pnas.170173897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang L.Z., Li Y.J., Xie X.F., Zhang J.J., Cheng C.Y., Yamashiro K., Chen L.J., Ma X.Y., Cheung C.M., Wang Y.S. Whole-exome sequencing implicates UBE3D in age-related macular degeneration in East Asian populations. Nat. Commun. 2015;6:6687. doi: 10.1038/ncomms7687. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt E.A., Dowling J.E. Early retinal development in the zebrafish, Danio rerio: light and electron microscopic analyses. J. Comp. Neurol. 1999;404:515–536. [PubMed] [Google Scholar]
- 16.Crabb J.W., Miyagi M., Gu X., Shadrach K., West K.A., Sakaguchi H., Kamei M., Hasan A., Yan L., Rayborn M.E. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc. Natl. Acad. Sci. USA. 2002;99:14682–14687. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang F., Taylor A. Roles for the ubiquitin-proteasome pathway in protein quality control and signaling in the retina: implications in the pathogenesis of age-related macular degeneration. Mol. Aspects Med. 2012;33:446–466. doi: 10.1016/j.mam.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drinjakovic J., Jung H., Campbell D.S., Strochlic L., Dwivedy A., Holt C.E. E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron. 2010;65:341–357. doi: 10.1016/j.neuron.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galy A., Néron B., Planque N., Saule S., Eychène A. Activated MAPK/ERK kinase (MEK-1) induces transdifferentiation of pigmented epithelium into neural retina. Dev. Biol. 2002;248:251–264. doi: 10.1006/dbio.2002.0736. [DOI] [PubMed] [Google Scholar]
- 20.Xu W., Gong L., Haddad M.M., Bischof O., Campisi J., Yeh E.T.H., Medrano E.E. Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzyme hUBC9. Exp. Cell Res. 2000;255:135–143. doi: 10.1006/excr.2000.4803. [DOI] [PubMed] [Google Scholar]
- 21.Muralidhar M.G., Thomas J.B. The Drosophila bendless gene encodes a neural protein related to ubiquitin-conjugating enzymes. Neuron. 1993;11:253–266. doi: 10.1016/0896-6273(93)90182-q. [DOI] [PubMed] [Google Scholar]
- 22.Wilson R., Goyal L., Ditzel M., Zachariou A., Baker D.A., Agapite J., Steller H., Meier P. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat. Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- 23.Marmorstein A.D., Finnemann S.C., Bonilha V.L., Rodriguez-Boulan E. Morphogenesis of the retinal pigment epithelium: toward understanding retinal degenerative diseases. Ann. N Y Acad. Sci. 1998;857:1–12. doi: 10.1111/j.1749-6632.1998.tb10102.x. [DOI] [PubMed] [Google Scholar]
- 24.Rizzolo L.J. Polarity and the development of the outer blood-retinal barrier. Histol. Histopathol. 1997;12:1057–1067. [PubMed] [Google Scholar]
- 25.Lamb T.D., Collin S.P., Pugh E.N., Jr. Evolution of the vertebrate eye: opsins, photoreceptors, retina and eye cup. Nat. Rev. Neurosci. 2007;8:960–976. doi: 10.1038/nrn2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaVail M.M. Outer segment disc shedding and phagocytosis in the outer retina. Trans. Ophthalmol. Soc. U. K. 1983;103:397–404. [PubMed] [Google Scholar]
- 27.Kevany B.M., Palczewski K. Phagocytosis of retinal rod and cone photoreceptors. Physiology (Bethesda) 2010;25:8–15. doi: 10.1152/physiol.00038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z., Qin T., Zhou J., Taylor A., Sparrow J.R., Shang F. Impairment of the ubiquitin-proteasome pathway in RPE alters the expression of inflammation related genes. Adv. Exp. Med. Biol. 2014;801:237–250. doi: 10.1007/978-1-4614-3209-8_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barral D.C., Seabra M.C. The melanosome as a model to study organelle motility in mammals. Pigment Cell Res. 2004;17:111–118. doi: 10.1111/j.1600-0749.2004.00138.x. [DOI] [PubMed] [Google Scholar]
- 30.Hume A.N., Seabra M.C. Melanosomes on the move: a model to understand organelle dynamics. Biochem. Soc. Trans. 2011;39:1191–1196. doi: 10.1042/BST0391191. [DOI] [PubMed] [Google Scholar]
- 31.Kushimoto T., Valencia J.C., Costin G.E., Toyofuku K., Watabe H., Yasumoto K., Rouzaud F., Vieira W.D., Hearing V.J. The Seiji memorial lecture: the melanosome: an ideal model to study cellular differentiation. Pigment Cell Res. 2003;16:237–244. doi: 10.1034/j.1600-0749.2003.00034.x. [DOI] [PubMed] [Google Scholar]
- 32.Li X., Hu Y., Sun X., Zhang J., Zhang M., Neovascular Age-Related Macular Degeneration Treatment Trial Using Bevacizumab (NATTB) Bevacizumab for neovascular age-related macular degeneration in China. Ophthalmology. 2012;119:2087–2093. doi: 10.1016/j.ophtha.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 33.Figurska M., Robaszkiewicz J., Wierzbowska J. Safety of ranibizumab therapy in wet AMD and the role of vascular endothelial growth factors in physiological angiogenesis. Klin. Oczna. 2010;112:147–150. [PubMed] [Google Scholar]
- 34.Arjamaa O., Nikinmaa M., Salminen A., Kaarniranta K. Regulatory role of HIF-1alpha in the pathogenesis of age-related macular degeneration (AMD) Ageing Res. Rev. 2009;8:349–358. doi: 10.1016/j.arr.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Fernandes A.F., Guo W., Zhang X., Gallagher M., Ivan M., Taylor A., Pereira P., Shang F. Proteasome-dependent regulation of signal transduction in retinal pigment epithelial cells. Exp. Eye Res. 2006;83:1472–1481. doi: 10.1016/j.exer.2006.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abedin M.J., Nguyen A., Jiang N., Perry C.E., Shelton J.M., Watson D.K., Ferdous A. Fli1 acts downstream of Etv2 to govern cell survival and vascular homeostasis via positive autoregulation. Circ. Res. 2014;114:1690–1699. doi: 10.1161/CIRCRESAHA.1134303145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neufeld G., Cohen T., Gengrinovitch S., Poltorak Z. Vascular endothelial growth factor (VEGF) and its receptors. FASEB J. 1999;13:9–22. [PubMed] [Google Scholar]
- 38.Castro A., Bernis C., Vigneron S., Labbé J.C., Lorca T. The anaphase-promoting complex: a key factor in the regulation of cell cycle. Oncogene. 2005;24:314–325. doi: 10.1038/sj.onc.1207973. [DOI] [PubMed] [Google Scholar]
- 39.Cai J., Nelson K.C., Wu M., Sternberg P., Jr., Jones D.P. Oxidative damage and protection of the RPE. Prog. Retin. Eye Res. 2000;19:205–221. doi: 10.1016/s1350-9462(99)00009-9. [DOI] [PubMed] [Google Scholar]
- 40.Winkler B.S., Boulton M.E., Gottsch J.D., Sternberg P. Oxidative damage and age-related macular degeneration. Mol. Vis. 1999;5:32. [PMC free article] [PubMed] [Google Scholar]
- 41.Campello L., Esteve-Rudd J., Cuenca N., Martín-Nieto J. The ubiquitin-proteasome system in retinal health and disease. Mol. Neurobiol. 2013;47:790–810. doi: 10.1007/s12035-012-8391-5. [DOI] [PubMed] [Google Scholar]
- 42.Shang F., Gong X., Palmer H.J., Nowell T.R., Jr., Taylor A. Age-related decline in ubiquitin conjugation in response to oxidative stress in the lens. Exp. Eye Res. 1997;64:21–30. doi: 10.1006/exer.1996.0176. [DOI] [PubMed] [Google Scholar]
- 43.Kaarniranta K., Hyttinen J., Ryhanen T., Viiri J., Paimela T., Toropainen E., Sorri I., Salminen A. Mechanisms of protein aggregation in the retinal pigment epithelial cells. Front. Biosci. (Elite Ed.) 2010;2:1374–1384. doi: 10.2741/e198. [DOI] [PubMed] [Google Scholar]
- 44.Viiri J., Hyttinen J.M.T., Ryhänen T., Rilla K., Paimela T., Kuusisto E., Siitonen A., Urtti A., Salminen A., Kaarniranta K. p62/sequestosome 1 as a regulator of proteasome inhibitor-induced autophagy in human retinal pigment epithelial cells. Mol. Vis. 2010;16:1399–1414. [PMC free article] [PubMed] [Google Scholar]
- 45.Geisler S., Holmström K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 46.Thisse C., Thisse B., Schilling T.F., Postlethwait J.H. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 47.Yen H.J., Tayeh M.K., Mullins R.F., Stone E.M., Sheffield V.C., Slusarski D.C. Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer’s vesicle cilia function. Hum. Mol. Genet. 2006;15:667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- 48.Bai Y., Yu W., Han N., Yang F., Sun Y., Zhang L., Zhao M., Huang L., Zhou A., Wang F., Li X. Effects of semaphorin 3A on retinal pigment epithelial cell activity. Invest. Ophthalmol. Vis. Sci. 2013;54:6628–6638. doi: 10.1167/iovs.13-12625. [DOI] [PubMed] [Google Scholar]
- 49.Huang L., Xu Y., Yu W., Li Y., Chu L., Dong J., Li X. Effect of Robo1 on retinal pigment epithelial cells and experimental proliferative vitreoretinopathy. Invest. Ophthalmol. Vis. Sci. 2010;51:3193–3204. doi: 10.1167/iovs.09-3779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.