Abstract
Background
CDGSH iron sulfur domain-containing protein 1 (CISD-1) belongs to the CISD protein family that is evolutionary conserved across different species. In mammals, CISD-1 protein has been implicated in diseases such as cancers and diabetes. As a tractable model organism to study disease-associated proteins, we employed Caenorhabditis elegans in this study with an aim to establish a model for interrogating the functional relevance of CISD-1 in human metabolic conditions.
Methods
We first bioinformatically identified the human Cisd-1 homologue in worms. We then employed N2 wild-type and cisd-1(tm4993) mutant to investigate the consequences of CISD-1 loss-of-function on: 1) the expression pattern of CISD-1, 2) mitochondrial morphology pattern, 3) mitochondrial function and bioenergetics, and 4) the effects of anti-diabetes drugs.
Results
We first identified C. elegans W02B12.15 gene as the human Cisd-1 homologous gene, and pinpointed the localization of CISD-1 to the outer membrane of mitochondria. As compared with the N2 wild-type worm, cisd-1(tm4993) mutant exhibited a higher proportion of hyperfused form of mitochondria. This structural abnormality was associated with the generation of higher levels of ROS and mitochondrial superoxide but lower ATP. These physiological changes in mutants did not result in discernable effects on animal motility and lifespan. Moreover, the amount of glucose in N2 wild-type worms treated with troglitazone and pioglitazone, derivatives of TZD, was reduced to a comparable level as in the mutant animals.
Conclusions
By focusing on the Cisd-1 gene, our study established a C. elegans genetic system suitable for modeling human diabetes-related diseases.
Keywords: C. elegans, Diabetes, Glucose consumption, Lifespan, Iron-sulfur containing protein, ROS
At a glance of commentary
Scientific background on the subject
Several lines of evidence have shown that alternation of CISD-1 is associated with human diseases, such as cancer and diabetes. However, animal models for a rapid screening drugs targeting to CISD-1 are less developed. C. elegans is thus used to interrogate the functional relevance of CISD-1 in human metabolic conditions.
What this study adds to the field
Using bioinformatic analyses, we found C. elegans W02G12.15 is the human Cisd-1 gene homologue. Furthermore, the wild-type worms treated with anti-diabetic drugs behave similar to cisd-1 mutants. We recommend C. elegans can be a good model for screening drugs against CISD-1 associated diseases.
Iron-sulfur-containing proteins are characterized by the presence of iron-sulfur clusters, which harbors sulfide-linked di-iron [Fe2S2], tri-iron [Fe3S4] and tetra-iron [Fe4S4] centers [1]. These proteins are versatile and evolutionarily conserved in prokaryotes and eukaryotes, and play key roles in the oxidation-reduction of mitochondrial electron transport, such as NADH dehydrogenase and coenzyme Q-cytochrome c [2]. Furthermore, iron-sulfur proteins are also implicated in the regulation of iron storage [3], gene expression and enzyme activity [4], [5], as exemplified respectively by ferredoxins, Redox-sensitive transcriptional activator (SoxR) and glutamine phosphoribosylpyrophosphate amidotransferase (GPAT). Assembly of [Fe–S] clusters is required for biosynthesis of multiple proteins in both prokaryotes and eukaryotes. In eukaryotes, [Fe–S] protein maturation takes place primarily in the mitochondria [6]. Defects in [Fe–S] protein biosynthetic machineries or [Fe–S] proteins themselves are the causes of many neurodegenerative and hematological disorders [7], reinforcing their biological significance.
Recently, a family of [Fe–S] proteins called CDGSH iron sulfur domain-containing protein (CISD) has been identified in many organisms [8]. They are classified into two groups: one containing a single CDGSH sequence, such as CISD-1 (mitoNEET) and CISD-2 (NAF-1), and the other having two CDGSH sequences, such as CISD-3 (MiNT). CISD-3 is a monomeric protein that resides in the mitochondrial matrix and regulates labile iron and reactive oxygen species, particularly in cancer cells [9]. CISD-2 is reportedly the genetic basis of human Wolfram syndrome 2, a rare autosomal neurodegenerative disorder [10]. Phenotypes of the Cisd-2-knockout mouse model are consistent with the notion that CISD-2 is important for aging-associated mitochondria integrity [11], [12]. CISD-1 was originally called mitoNEET due to a NEET (Asn-Glu-Glu-Thr) sequence that is presumably targeted by thiazolidinedione (TZD), an anti-diabetic drug [13], clinical derivatives of which also include pioglitazone, troglitazone, and rosiglitazone [14]. The biological function of CISD-1 is likely to modulate glucose metabolism or cellular bioenergetics, given that functional loss of this gene is related to obesity, diabetes, cancer, and neurodegenerative disease [15], [16], [17], [18], [19]. Mouse studies strengthen this metabolic role – CISD-1 transgenic mice are prone to obesity and insulin insensitivity, whereas loss-of-function mice conversely exhibit lower body weights and lipid storage [20]. Consequently, in addition to TZD, many drugs targeting CISD-1 have recently been developed for the treatment of diseases with dysfunction mitochondria [21], [22], [23].
Free-living soil nematode, Caenorhabditis elegans, because of its simplicity and short life cycle, has been used as a model organism for studying human diseases, with notable examples of glucose-6-phosphate dehydrogenase deficiency and Alzheimer's disease [24], [25]. To further exploit this system for studying CISD-1-associated metabolic diseases, we first identified in this study the C. elegans W02B12.15 gene as the most phylogenetically close counterpart to the human Cisd-1 gene. We then further defined that cisd-1 gene is critical for maintaining mitochondria integrity. In addition, abnormally higher levels of ROS and superoxide were observed in the cisd-1(tm4993) mutants, which conversely produced 55% less ATP. However, these physiological alterations had no effect on cisd-1(tm4993) mutant motility and lifespan. Interestingly, the levels of ATP production and glucose in troglitazone-treated N2 worms appeared to be comparable to those in cisd-1(tm4993) mutants, suggesting that this genetic system of C. elegans is a suitable model for studying human diabetes-related diseases and the consequent exploration of drugs against these diseases.
Materials and methods
C. elegans strains and maintenance
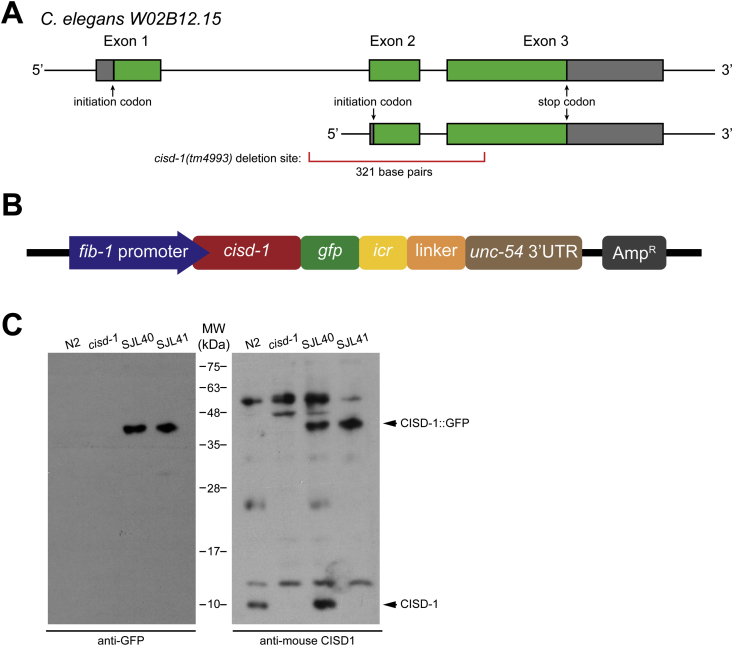
C. elegans were cultured and maintained following the standard procedure [26] on the normal growth medium (NGM)plates with E. coli (OP50 strain) food source. The cisd-1(tm4993) mutant [27], which was created by the TMP/UV method, has a 321-base pairs deletion from the nucleotide positions 74 to 424. Two additional worm strains, SJL40 [Pfib-1:: cisd-1:: gfp:: 3′ UTRunc-54] and SJL41 [cisd-1; Pfib-1:: cisd-1:: gfp:: 3′ UTRunc-54], were generated by microinjection and crossing in this study [28].
Plasmid constructions
Full-length W02B12.15b gene fragment, including three introns and two exons, was amplified from the genomic DNA by PCR and sub-cloned via NheI and AgeI sites into Pfib-1:: fib-1:: gfp:: 3′ UTRunc-54 [29], between the fib-1 promoter and gfp sequence. The primers used for cloning the entire genomic locus of W02B12.15b are:
Forward, 5′-CAATTTGCTAGCAAAAATGACCATCGCTGGA-3, Reverse, 5′-CGAGACACCGGTAGCTTCTTTTCGGATTTG-3′.
Mitochondria isolation
One day-old adult worms were collected and washed twice with M9 buffer, then suspended in ice-cold 0.1 M NaCl. Ice-cold sucrose was added to the worms to a final concentration of 30%. The worms were then centrifuged at 1,100 g for 5 min at 4 °C. Worms floating to the top were collected with a glass Pasteur pipette. After washes with 0.1 M NaCl, worms were resuspended with worm lysis buffer with protease inhibitor cocktail and transferred to the ice-cold beads beater chamber containing the beads about one-half volume of the worm pellet. Grinding proceeded with four pulses of 1 min each interspersed with 1 min interval on ice. The recovered supernatant and the rinsing of the beads were pooled and homogenized by hand in a glass homogenizer. The lysate was centrifuged at 2,000 g for 10 min at 4 °C, with the pellet subsequently resuspended in cold worm lysis buffer and centrifuged again at 15,000 g for 30 min at 4 °C. The pellet was resuspended in worm lysis buffer and layered onto a sucrose gradient of 1 M–2 M. After centrifugation at 80,000 g for 90 min at 4 °C, the intact mitochondria appeared as a brown band in the middle of the gradient, which was collected, diluted with 4 volumes of cold worm lysis buffer, and then centrifuged again at 30,000 g for 30 min at 4 °C. The purified mitochondria were gently resuspended with a small amount of cold worm lysis buffer.
Mitochondria sub-fractionation
Further fractionation of mitochondria was performed as described previously [30], [31]. Fifty μg of mitochondria were digested with 100 μg/ml proteinase K in isolation buffer for 30 min. The reaction was inhibited by adding 5 mM phenylmethylsulfonyl fluoride (PMSF) for 10 min on ice. Fifty micrograms of mitochondria were treated with 0.1 M sodium carbonate (pH = 11.5) on ice for 30 min. The mixtures were centrifuged at 55,000 g for 30 min, with the supernatant collected as mitochondria soluble fraction and pellet as mitochondria membrane fraction.
Western blot analysis
Young adult worms were lysed in sodium dodecyl sulfate (SDS) sample buffer by boiling. The protein extracts were loaded on the 10% or 15% SDS-polyacrylamide gel, electrophoresed and transferred to a polyvinylidene difluoride (PVDF) membrane. After blocking with 5% w/w skimmed milk powder in TBST, the membranes were incubated with primary antibodies against MitoNEET (Santa Cruz, sc-138,987), green fluorescence protein (GFP) (Santa Cruz, sc-9996), HSP60 (ENZO, ADI-SPA-807-E), FIB-1 (Novus, NB300-269), or NDUFS3 (Abcam, ab14711), and subsequently with species-specific secondary horseradish peroxidase (HRP)-conjugated IgG. The signals were developed with ECL (Thermo, 34,095) according to the manufacturer's instructions. The images were acquired with BioRad ChemiDoc™ Touch Imaging system.
Mitotracker Red staining
One-hundred μl of Mitotracker Red FM stock solution (100 μM, Invitrogen, M7512) were mixed with 500 μl heat-killed OP50 and seeded on agar plates. The L4 stage worms were grown on Mitotracker Red-containing plates at 20 °C for 12 h. When the worms reached young adult stage, they were transferred to clean OP50 agar plates for another 2 h at 20 °C to remove intestinal accumulation. Worms were then washed with M9 buffer, followed by mounting onto 2% agarose pads. Images of each strain were acquired with an Olympus FV10i confocal laser-scanning microscope. Morphologies of mitochondria were classified based on a previous report [32].
Mitochondrial ROS and superoxide determination
Around 100 L4 worms were collected in microcentrifuge tubes. After three washes with M9 buffer, worms were resuspended in 100 μl PBST containing 1 mM PMSF, and then subjected to freeze- and -thaw cycle and sonicated at 30% amplitude for 3 cycles, each of which consists of 15 s on and 30 s off, on ice. Worm lysates were incubated with CM-H2DCFDA (Thermo; C6827) in PBS at final concentration of 100 μM for 1 h in 96-well plates. The ROS-related fluorescence levels were measured using SpectraMax M2 Microplate Reader at excitation wavelength of 493 nm and emission wavelength of 522 nm. For superoxide detection, around 100 L4 worms were collected in microcentrifuge tubes. After three washes with M9 buffer, worms were stained with MitoSOX Red (Thermo; M36008) in PBS at final concentration of 20 μM for 1 h in the dark. Worms were transferred to an NGM plate for an hour to clear their intestines of residual dye. The fluorescence signals were measured using SpectraMax M2 Microplate Reader at excitation wavelength of 510 nm and emission wavelength of 580 nm. Data were normalized by protein concentration of worm total lysates.
ATP measurement
Worm growth was synchronized at the L4 stage at 20 °C, washed with M9 buffer, and frozen in the liquid nitrogen. The samples were immersed in the boiling water 15 min before analysis. The ATP levels were measured using the ATP determination kit (Molecular Probes; A22066) and the ATP levels were normalized by protein concentration of respective samples, as determined using the Bio-Rad protein assay dye (cat#: 500-0006).
Lifespan study
All worms were synchronized and maintained at 20 °C until they developed into young adults. Approximately 60 young adult worms were transferred onto NGM plates freshly seeded with bacteria OP50, at the density of 10 worms per plate. The worms were transferred to new plates every day until worms stopped laying eggs. Worms were inspected for cessation of crawling or touch response. The survival curves and the statistical log rank tests of each condition were analyzed by using GraphPad prism 6.
Swimming and crawling assay
One-day young adults were collected, washed and then transferred to a new NGM plate. For the crawling behavior, worms were recorded for 15 s and analyzed by the WrmTrck plugin of ImageJ. For the swimming behavior, worms were transferred to a 24-well plate with PBS and recorded for 15 s. The body bends were analyzed by the WrmTrck plugin of ImageJ.
Drug treatment and glucose level determination
Synchronized L1 larvae were transferred to NGM supplied with metformin (25 μM), troglitazone (100 μM) or pioglitazone (100 μM). Worms were collected once adulthood is reached. The glucose levels were determined by using Glucose (HK) Assay Kit (Sigma; GAHK20) and normalized by protein concentration of respective samples.
Results
Phylogenetic sequence analysis reveals that W02B12.15 is the C. elegans cisd-1 gene
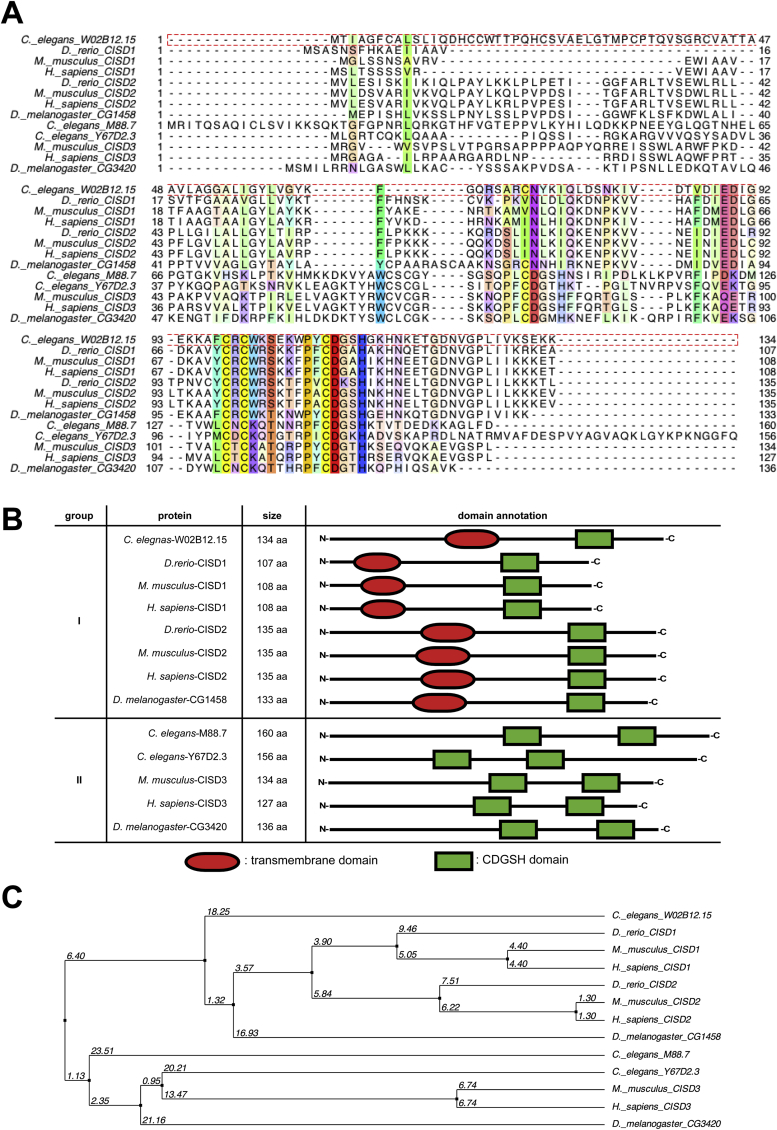
To pinpoint the human Cisd-1 gene homologue in C. elegans, we performed BLAST analysis of human CISD-1 and CISD-2 amino acid sequences against the wormbase data, and uncovered a gene annotated as W02B12.15 with the highest score of alignment [Fig. 1A]. The W02B12.15 encoded protein possesses one CDGSH domain close to the N-terminus and a C-terminal transmembrane domain, an organization analogous to human CISD-1 and CISD-2 [Fig. 1B]. As shown by the phylogenetic tree of CISD homologues encoded by the H. sapiens, M. musculus, D. rerio, D. melanogaster and C. elegans genomes, W02B12.15 was closest to the Cisd-1 clade [Fig. 1C] and thus designated hereafter as cisd-1. Interestingly, no cisd-2 homologue was detected in C. elegans, while there were two cisd-3 homologous genes (M88.7 and Y67D2.3) [8] [Fig. 1B, C].
Fig. 1.
Phylogenetic tree analysis of W02B12.15. (A) Amino acid sequence alignment of CISD proteins among C. elegans, D. rerio, D. melanogaster, M. musculus and H. sapiens. The single letters denote amino acids, of which highly conserved residues are colored. Numbers indicate the position of amino acids. (B) Schematic view of the transmembrane and CDGSH domains of CDGSH-containing proteins and their conservation among C. elegans, D. rerio, D. melanogaster, M. musculus and H. sapiens. The domain architectures were analyzed by SMART (a Simple Modular Architecture Research Tool). (C) C. elegans W02B12.15 gene is evolutionarily close to the Cisd-1 clades. Neighbor joining phylogenetic tree analysis of the CDGSH domain-containing proteins from different model organisms were calculated based on identity (%).
Based on the sequence characterization, C. elegans cisd-1 putatively encodes two alternative protein isoforms, with lengths of 134 or 103 amino acids depending on the choice of initiation codon [Fig. 2A]. To verify its endogenous expression pattern, we generated transgenic worm strains expressing cisd-1 fused with the green fluorescence protein (GFP) gene (strain SJL40 in N2 wild-type background and SJL41 in cisd-1(tm4993) mutant background; [Fig. 2B]) and performed Western blot analysis. Both anti-GFP and anti-mouse CISD-1 antibodies were reactive to a band around 37 kDa, the presumed size of CISD-1::GFP fusion, in both SJL40 and SJL41 but not the control stains of N2 wild-type and cisd-1(tm4993) animals [Fig. 2C, left panel]. Immunoblotting results using the anti-mouse CISD-1 antibodies in N2 wild-type and SJL40 worms revealed a 10-kDa signal that corresponds to the endogenous CISD-1 [Fig. 2C, right panel, lanes 1 and 3]. Based on these results, we inferred that the cisd-1 gene expresses a protein product of 134 amino acids.
Fig. 2.
Characterization of protein encoded by W02B12.15. (A) Genomic structure of W02B12.15 and two possible transcripts encoded by the gene. The deletion allele in the mutant cisd-1(tm4993) is indicated, from nucleotide positions 424 to 745. (B) Scheme of the construct Pfib-1::cisd-1::gfp::icr::linker::3′UTRunc-54-AMPR used for generation of transgenic worms (cguIs14) (strain SJL40). (C) Western blot analysis of four strains of worms - wild-type (N2), cisd-1(tm4993), SJL40 (transgenic worm), and SJL41 (transgenic worm in tm4993 background) - using anti-GFP (left) and anti-mouse CISD-1 (right) antibodies.
CISD-1 is an outer membrane protein of mitochondria
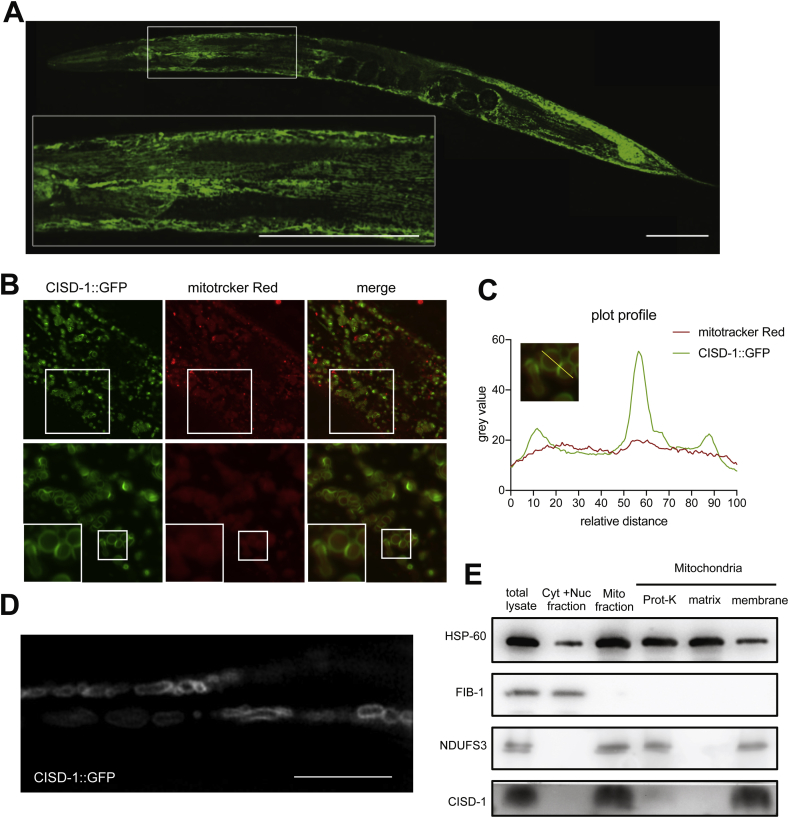
To study the subcellular localization of the CISD-1 protein, we examined the transgenic worms (SJL40) expressing CISD-1::GFP using fluorescence microscopy. Fluorescence signals evidenced a thread- and rod-like structure in low-magnification images [Fig. 3A], while images of higher magnification indicated that GFP is co-localized with the Mitotracker Red dye to the periphery of mitochondria [Fig. 3B–D], suggesting a spatial association with mitochondrial outer membrane. To further confirm this spatial distribution via biochemical means, mitochondria were fractionated from disrupted N2 wild-type worms, treated with or without proteinase K, and subjected to protein expression profiling by Western blot. As control, the FIB-1 expression was detected in the cytosol plus nuclear fraction but not in mitochondria fraction [Fig. 3E, the second row, lane 2 vs. lane 3]. In the mitochondria fraction, the expression of NDUFS3 and HSP60, known respectively as inner membrane and matrix resident proteins, were resistant to proteinase K treatment [Fig. 3E, lane 4]. By contrast, CISD-1 signal was detectable only in intact mitochondria [Fig. 3E, lane 3] but absent in the protease-treated sample [Fig. 3E, lane 4]. When the mitochondrial membrane pellet was extracted with sodium carbonate, CISD-1 was recovered in the pellet, indicating that it is an integral membrane protein as NDUFS3 [Fig. 3E, lanes 5 and 6]. These results thus demonstrated that CISD-1 is a mitochondrial protein with an outer membrane distribution.
Fig. 3.
C. elegans CISD-1 is an outer membrane protein of mitochondria. (A) Fluorescence images of the expression pattern of CISD-1::GFP, with a high magnification image showing the body wall muscle region. Scale bar, 100 μm. (B) The Pfib-1::CISD-1::GFP transgenic worms (cguIs14, strain SJL40) were stained with mitochondria specific dye, Mitotracker Red FM, and shown in different magnifications. The upper panels show partial co-localization of GFP with Mitotracker under low magnification. The bottom panels represent magnified images of the box regions in the upper panels, with inserts at lower left denoting further magnification of boxed regions. (C) The images from (B) were analyzed by plot profile through the yellow line. (D) High-resolution image of the tail region in SJL40 was acquired by 3D structured illumination microscopy. Scale bar, 1.5 μm. (E) Biochemical characterization of CISD-1's mitochondrial localization. Worms were separated into cytosol, nuclear, and mitochondrial fractions. Isolated mitochondria were treated with protease K (Prot-K), or separated into soluble fraction (matrix) or membrane fraction using sodium carbonate. Western blot analyses of the extracts were done by using anti-mitoNEET, anti-NDUFS3, anti-FIB-1 and anti-HSP60 antibodies, as indicated.
Mutant of cisd-1 engenders aberrant mitochondrial morphology
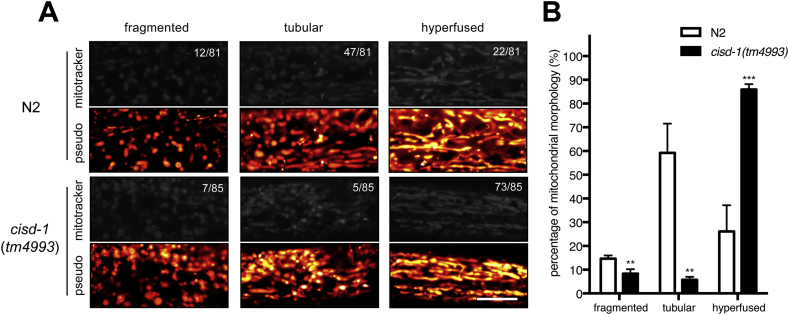
The role of CISD-1 in mitochondria was first examined by measuring morphological changes of mitochondria in a cisd-1(tm4993) mutant, which has a 321-bp deletion within the gene locus [Fig. 2A]. Based on an earlier report [32], mitochondrial morphology could be classified structurally as fragmented, tubular, and hyperfused shape [Fig. 4A]. Among 81 N2 wild-type worms and 85 cisd-1(tm4993) mutant worms collected for this analysis, the mitochondrial morphology appeared in the majority of N2 (about 59%) was of the tubular shape, in contrast to hyperfused morphology exhibited predominantly by the mutant worms (85%) [Fig. 4B]. These observations implied that CISD-1 is important for maintaining mitochondria in the proper tubular form.
Fig. 4.
Mutation of cisd-1 compromises mitochondrial morphology. (A) Representative images and pseud-color images of mitochondrial organization in the body wall muscles of wild-type (N2) (n = 81) and cisd-1(tm4993) mutants (n = 85). Mitochondria morphologies are classified into 3 structural types: fragmented, tubular, and hyperfused. The numbers indicate fractions of worms with the respective mitochondrial forms. Scale bars, 10 m.(B) The quantitative results of (A) showing the percentage of various mitochondrial morphologies in wild-type (N2) and cisd-1(tm4993) (N = 3). **p < 0.01; ***p < 0.001. Error bars represent SEMs.
Elevation of ROS, mitochondrial superoxide and lower ATP production in cisd-1 (tm4993) mutants
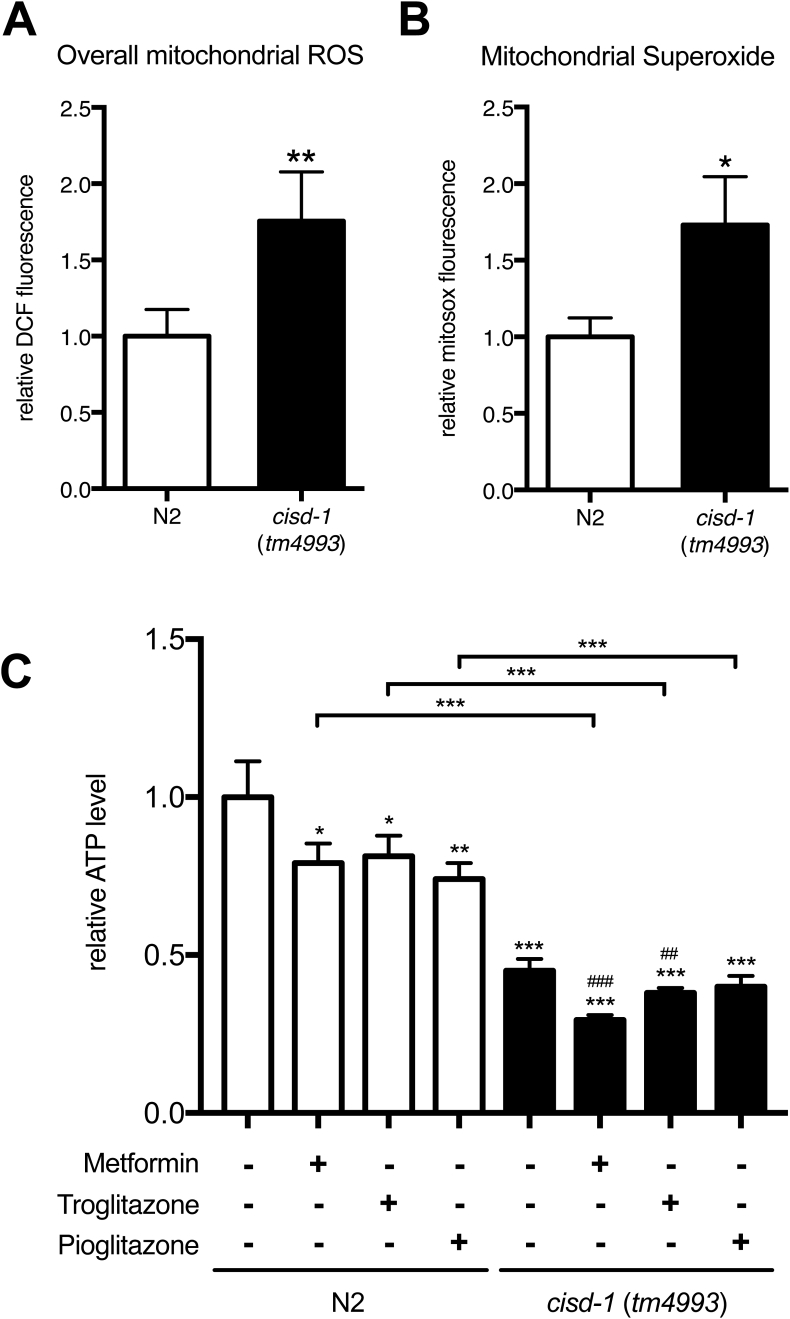
Aberrant mitochondrial organization observed in the cisd-1(tm4993) mutant could be correlated with altered function. To test this possibility, we assessed changes in mitochondrial reactive oxygen species (ROS), superoxide and ATP production. There were 1.5-fold elevation in the overall levels of both mitochondrial ROS and superoxide in cisd-1(tm4993) mutant worms compared with N2 wild-type worms [Fig. 5A, B]. In addition, the ATP production level was about 55% lower in cisd-1(tm4993) mutants [Fig. 5C, lane 1 vs. lane 5]. Viewed together, these biochemical changes indicate a key role of CISD-1 in mitochondrial structure and bioenergetics.
Fig. 5.
The cisd-1(tm4993) mutant exhibits elevated mitochondrial ROS and superoxide but lower ATP production. (A) The overall mitochondrial ROS level was measured by using ROS indicator CM-H2DCFDA in wild-type (N2) and cisd-1(tm4993) (N = 6). (B) The mitochondrial superoxide level was measured by using mitochondrial superoxide indicator, MitoSOX Red, in wild-type (N2) and cisd-1(tm4993) (N = 6). (C) The relative ATP level was analyzed in wild-type (N2) and cisd-1(tm4993) (N = 5). *p < 0.05; **p < 0.01; ***p < 0.001. ##p < 0.01 and ###p < 0.001 denote comparison with the untreated mutant. Error bars represent SEMs.
Motility and lifespan are normal in cisd-1(tm4993) worms
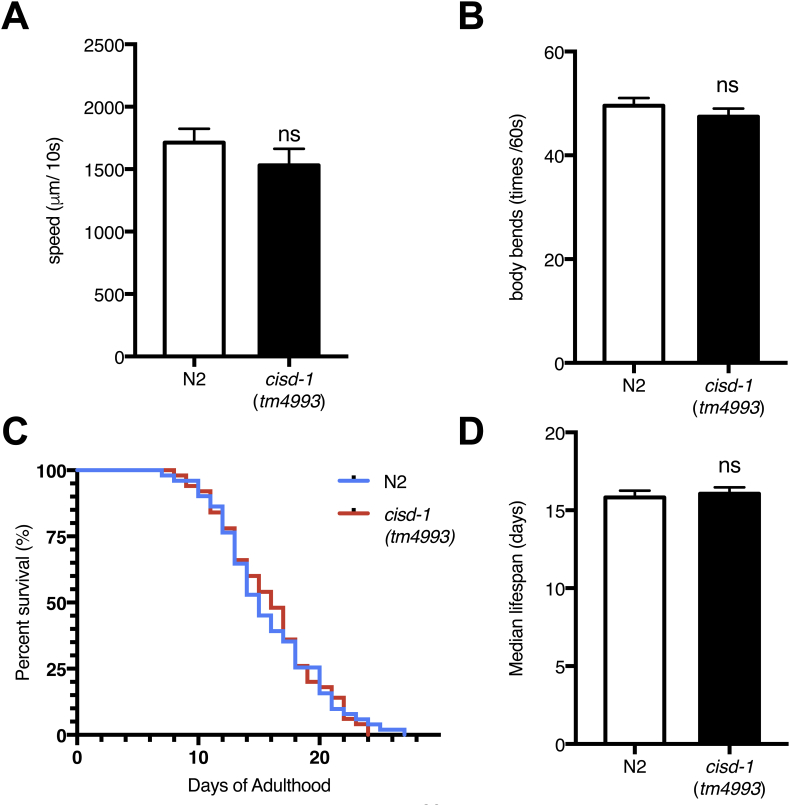
We then speculated that the impaired mitochondrial bioenergetics upon loss of cisd-1 might affect energy-dependent processes such as motility and lifespan. However, comparison between cisd-1(tm4993) mutants and N2 wild-type worms did not reveal apparent differences in movement speed on agar plates and body bends in liquid media [Fig. 6A, B]. In addition, lifespan studies showed no apparent difference between cisd-1(tm4993) mutant and N2 wild-type worms [Fig. 6C]; the median lifespan was about 10 days for both N2 wild-type worms and cisd-1(tm4993) mutant worms [Fig. 6D]. Despite the reduction in ATP production, we speculated that the energy required for preserving normal motility and lifespan in cisd-1(tm4993) mutants might be compensated by minimizing energetically costly processes, such as the lipid and carbohydrate anabolic pathways.
Fig. 6.
The motility and lifespan remain normal in cisd-1(tm4993) mutant. (A) The crawling behavior assay was performed on the normal growth medium for 10 s by video recording (n = 33 at N2 and n = 27 at tm4993). The moving speed was automatically calculated by the WrmTrck plugin in Image J. (B) The body bending behavior was analyzed by counting worm body bends per 15 s (n = 12 at N2 and n = 11 at tm4993). (C) and (D) Survival curves and median lifespan of wild-type (n = 51), cisd-1(tm4993) (n = 50) at 20 °C. *p < 0.05; **p < 0.01; ***p < 0.001. Error bars represent SEMs.
Effects of three anti-diabetic drugs on N2 wild-type and cisd-1(tm4993) worms
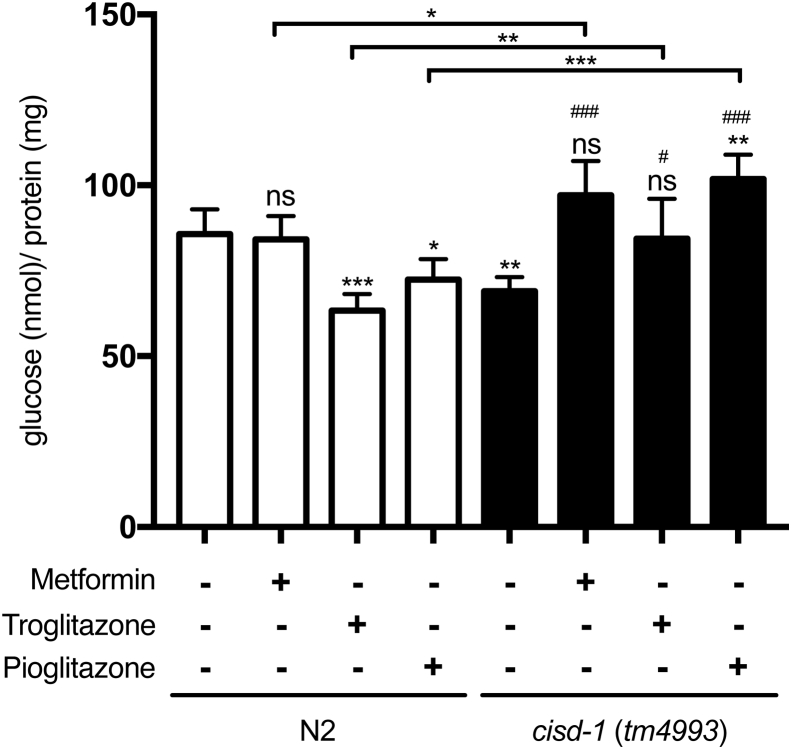
To test whether the metabolic phenotypes of cisd-1(tm4993) worms could be recapitulated in N2 wild-type worms treated with TZD and other anti-diabetic drugs, we analyzed the mitochondria morphology, ATP production and glucose levels in both N2 wild-type and cisd-1(tm4993) mutant worms with or without metformin, troglitazone and pioglitazone treatment. Our results first showed that mitochondria morphology in the N2 wild-type worms were altered by troglitazone in such a manner that 77% of the animal exhibited the hyperfused form (Supplementary Fig. 1), a proportion that was similarly observed in cisd-1(tm4993) worms. ATP production was also decreased in N2 wild-type worms treated with troglitazone [Fig. 5C, lane 2]. In the absence of drugs, the basal level of glucose concentration in cisd-1(tm4993) worms was 30% lower than that of N2 wild-type worms [Fig. 7, lane 5]. Interestingly however, upon treatment with troglitazone and pioglitazone, N2 wild-type worms exhibited respectively 26% and 15% decline in glucose level [Fig. 7, lanes 2–4 vs. lane 1]. Unexpectedly, these anti-diabetic drug treatments all increased to different extents the glucose levels in cisd-1(tm4993) worms: 40% with metformin, 22% with troglitazone and 47% with pioglitazone [Fig. 7, lanes 6–8]. Nevertheless, the notion that basal ATP production and glucose level in cisd-1(tm4993) worms were comparable to those in N2 wild-type worms treated with troglitazone [Fig. 7, lane 5 vs. lane 3] indicates that genetic defects in cisd-1 gene phenocopy the pharmacological targeting of CISD-1 in C. elegans.
Fig. 7.
The glucose level of wild-type worms and mutant worms treated with or without anti-diabetic drugs. The glucose level was analyzed for both wild-type (N2) and cisd-1(tm4993) treated with metformin, troglitazone or pioglitazone (N = 5). *p < 0.05; **p < 0.01; ***p < 0.001. #p < 0.05 and ###p < 0.001 denote comparison with the untreated mutant. Error bars represent SEMs.
Discussion
By sequence analysis, we have identified in this study three members of the C. elegans CISD family - W02B12.15, M88.7 and Y67D2.3 genes [Fig. 1A, B]. Among them, W02B12.15 is the human homolog of CISD-1, while M88.7 and Y67D2.3 correspond to CISD-3. Since mice and humans encode three distinct members of the family (cisd-1, cisd-2 and cisd-3) but worms lack cisd-2, we hypothesized that mammals gained cisd-2 probably through a duplication of cisd-1 gene along the path of evolution. C. elegans CISD-1 therefore represents the ancient type I CISD-1 protein in the animal kingdom with the conserved distribution in mitochondria outer membrane [Fig. 2]. Based on its molecular characterization and mitochondria localization, CISD-1 likely acts to maintain inter-compartment iron balance and also the proper function of electron transport chain complex. This notion was further strengthened by our study, which demonstrated that its absence triggered mitochondria morphology alteration [Fig. 4], ROS and superoxide increase [Fig. 5], and compromised ATP production [Fig. 5].
One unresolved issue of our study is the seemingly normal longevity and motility of the cisd-1(tm4993) worms in spite of the perturbations in mitochondria ROS and superoxide, as well as the ATP production. Numerous studies have shown that mitochondrial dysfunction, such as diminished oxidative capacity, increase in ROS, or aberrant morphology, are highly correlated with animal aging [33], [34], [35]. Interestingly, enlarged mitochondria typically promote sufficient ATP production, which is linked to diverse longevity pathways [36]. However, a comparable lifespan as wild-type exhibited by the mutant worms [Fig. 6] is somewhat unexpected, given that a nuo-6 mutant, which lacks a complex I subunit of the electron transport chain, was found to have an extended lifespan owing to protective stress responses elicited by the intrinsic apoptosis pathway [37]. While a recent study has indicated that loss of CISD-1 triggers apoptosis in germ cells [27], it still remains unknown whether this could be a compensatory mechanism exploited by the cisd-1(tm4993) mutants to maintain lifespan.
One important discovery of our study is the functional relevance of CISD-1 in maintaining basal glucose level [Fig. 7]. However, how a defect in the mitochondria-associated protein could contribute to glucose homeostasis is currently unknown. One possible explanation is that glycolysis rate might be enhanced in the event of ATP deficit to replenish energy shortage. Nevertheless, among three anti-diabetic drugs tested in this study, metformin and TZDs are known to target energy-related pathways that converge on the AMPK signaling [38], [39], [40], [41]. Both metformin and TZDs have been shown to inhibit respiratory complex I, and TZDs additionally activate peroxisome proliferator-activated receptor gamma to enhance adiponectin release. Interestingly, CISD-1 is also a target of TZDs and was shown in this study to be a critical factor in mitochondrial energy output. The current study raises the possibility that CISD-1 may be a viable target for averting pathological conditions related to bioenergetics imbalance, as CISD-1 inhibition manifests no overt phenotype in the presence of functional AMPK.
Conclusion
In this study, we demonstrated that the C. elegans W02B12.15 gene is the human cisd-1 homolog. The cisd-1(tm4993) mutant exhibited hyperfused mitochondria accompanied by elevation of ROS and superoxide but lower ATP production. However, no defects in movement speed, body bending and lifespan were discernibly observed in cisd-1(tm4993) mutants. Furthermore, treatment of the wild-type worms with glucose-reducing TZD derivatives equalized the glucose concentration to that of the cisd-1(tm4993) mutants. We thus recommend C. elegans is a good model to study human cisd-1 related diseases and to screen drugs against CISD-1-associated diseases.
Conflicts of interest
All authors declare they have no conflicts of interest.
Acknowledgements
We thank Professor Simon Silver for critically commenting and editing of the manuscript. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by grants from the Ministry of Science and Technology of Taiwan (MOST104-2320-B-182-029-MY3 and MOST105-2314-B-182-061-MY4 to BC-MT), Chang Gung Memorial Hospital (CMRPD3E0153, CMRPD1F0442, CMRPD1H0021 and BMRP960 to BC-MT, and BMRP742 to SJL), and the Ministry of Education (EMRPD1C0051 to SJL).
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bj.2019.07.009.
Contributor Information
Bertrand Chin-Ming Tan, Email: btan@mail.cgu.edu.tw.
Szecheng J. Lo, Email: losj@mail.cgu.edu.tw.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Analyses of mitochondrial morphology changes in N2 worms treated with troglitazone. (A) Representative images and pseud-color images of mitochondrial organization in the body wall muscles of in N2 worms and cisd-1(tm4993) treated with troglitazone. Mitochondria morphologies are classified into 3 structural types as in Fig. 4. Scale bars, 10 m. (B) The quantitative results of (A) showing the percentage of various mitochondrial morphologies (N = 3). *p<0.05. **p < 0.01. ***p < 0.001. Error bars represent SEMs.
References
- 1.Beinert H. Iron-sulfur proteins: ancient structures, still full of surprises. J Biol Inorg Chem. 2000;5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- 2.Rieske J.S. Composition, structure, and function of complex III of the respiratory chain. Biochim Biophys Acta. 1976;456:195–247. doi: 10.1016/0304-4173(76)90012-4. [DOI] [PubMed] [Google Scholar]
- 3.Fidai I., Wachnowsky C., Cowan J.A. Mapping cellular Fe-S cluster uptake and exchange reactions - divergent pathways for iron-sulfur cluster delivery to human ferredoxins. Metallomics. 2016;8:1283–1293. doi: 10.1039/c6mt00193a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson D.C., Dean D.R., Smith A.D., Johnson M.K. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 5.Tong W.H., Rouault T. Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J. 2000;19:5692–5700. doi: 10.1093/emboj/19.21.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lill R., Hoffmann B., Molik S., Pierik A.J., Rietzschel N., Stehling O. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta. 2012;1823:1491–1508. doi: 10.1016/j.bbamcr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Stehling O., Wilbrecht C., Lill R. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie. 2014;100:61–77. doi: 10.1016/j.biochi.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Inupakutika M.A., Sengupta S., Nechushtai R., Jennings P.A., Onuchic J.N., Azad R.K. Phylogenetic analysis of eukaryotic NEET proteins uncovers a link between a key gene duplication event and the evolution of vertebrates. Sci Rep. 2017;7:42571. doi: 10.1038/srep42571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipper C.H., Karmi O., Sohn Y.S., Darash-Yahana M., Lammert H., Song L. Structure of the human monomeric NEET protein MiNT and its role in regulating iron and reactive oxygen species in cancer cells. Proc Natl Acad Sci U S A. 2018;115:272–277. doi: 10.1073/pnas.1715842115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amr S., Heisey C., Zhang M., Xia X.J., Shows K.H., Ajlouni K. A homozygous mutation in a novel zinc-finger protein, ERIS, is responsible for Wolfram syndrome 2. Am J Hum Genet. 2007;81:673–683. doi: 10.1086/520961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y.F., Kao C.H., Chen Y.T., Wang C.H., Wu C.Y., Tsai C.Y. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev. 2009;23:1183–1194. doi: 10.1101/gad.1779509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y.F., Kao C.H., Kirby R., Tsai T.F. Cisd2 mediates mitochondrial integrity and life span in mammals. Autophagy. 2009;5:1043–1045. doi: 10.4161/auto.5.7.9351. [DOI] [PubMed] [Google Scholar]
- 13.Colca J.R., McDonald W.G., Waldon D.J., Leone J.W., Lull J.M., Bannow C.A. Identification of a novel mitochondrial protein ("mitoNEET") cross-linked specifically by a thiazolidinedione photoprobe. Am J Physiol Endocrinol Metab. 2004;286:E252–E260. doi: 10.1152/ajpendo.00424.2003. [DOI] [PubMed] [Google Scholar]
- 14.Lebovitz H.E. Differentiating members of the thiazolidinedione class: a focus on safety. Diabetes Metab Res Rev. 2002;18(Suppl 2):S23–S29. doi: 10.1002/dmrr.252. [DOI] [PubMed] [Google Scholar]
- 15.Sohn Y.S., Tamir S., Song L., Michaeli D., Matouk I., Conlan A.R. NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proc Natl Acad Sci U S A. 2013;110:14676–14681. doi: 10.1073/pnas.1313198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittler R., Darash-Yahana M., Sohn Y.S., Bai F., Song L., Cabantchik I.Z. NEET proteins: a new link between iron metabolism, ROS and cancer. Antioxidants Redox Signal. 2019;30:1083–1095. doi: 10.1089/ars.2018.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusminski C.M., Chen S., Ye R., Sun K., Wang Q.A., Spurgin S.B. MitoNEET-parkin effects in pancreatic alpha- and beta-cells, cellular survival, and intrainsular cross talk. Diabetes. 2016;65:1534–1555. doi: 10.2337/db15-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kusminski C.M., Park J., Scherer P.E. MitoNEET-mediated effects on browning of white adipose tissue. Nat Commun. 2014;5:3962. doi: 10.1038/ncomms4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geldenhuys W.J., Benkovic S.A., Lin L., Yonutas H.M., Crish S.D., Sullivan P.G. MitoNEET (CISD1) knockout mice show signs of striatal mitochondrial dysfunction and a Parkinson's disease phenotype. ACS Chem Neurosci. 2017;8:2759–2765. doi: 10.1021/acschemneuro.7b00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusminski C.M., Holland W.L., Sun K., Park J., Spurgin S.B., Lin Y. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat Med. 2012;18:1539–1549. doi: 10.1038/nm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geldenhuys W.J., Yonutas H.M., Morris D.L., Sullivan P.G., Darvesh A.S., Leeper T.C. Identification of small molecules that bind to the mitochondrial protein mitoNEET. Bioorg Med Chem Lett. 2016;26:5350–5353. doi: 10.1016/j.bmcl.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabchevsky A.G., Patel S.P., Sullivan P.G. Targeting mitoNEET with pioglitazone for therapeutic neuroprotection after spinal cord injury. Neural Regen Res. 2017;12:1807–1808. doi: 10.4103/1673-5374.219040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi T., Yamamoto M., Amikura K., Kato K., Serizawa T., Serizawa K. A novel MitoNEET ligand, TT01001, improves diabetes and ameliorates mitochondrial function in db/db mice. J Pharmacol Exp Ther. 2015;352:338–345. doi: 10.1124/jpet.114.220673. [DOI] [PubMed] [Google Scholar]
- 24.Kaletta T., Hengartner M.O. Finding function in novel targets: C. elegans as a model organism. Nat Rev Drug Discov. 2006;5:387–398. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- 25.Yang H.C., Chen T.L., Wu Y.H., Cheng K.P., Lin Y.H., Cheng M.L. Glucose 6-phosphate dehydrogenase deficiency enhances germ cell apoptosis and causes defective embryogenesis in Caenorhabditis elegans. Cell Death Dis. 2013;4:e616. doi: 10.1038/cddis.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King S.D., Gray C.F., Song L., Nechushtai R., Gumienny T.L., Mittler R. The cisd gene family regulates physiological germline apoptosis through ced-13 and the canonical cell death pathway in Caenorhabditis elegans. Cell Death Differ. 2019;26:162–178. doi: 10.1038/s41418-018-0108-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mello C., Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 29.Lee L.W., Lee C.C., Huang C.R., Lo S.J. The nucleolus of Caenorhabditis elegans. J Biomed Biotechnol. 2012;2012:601274. doi: 10.1155/2012/601274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diekert K., de Kroon A.I., Kispal G., Lill R. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 2001;65:37–51. doi: 10.1016/s0091-679x(01)65003-9. [DOI] [PubMed] [Google Scholar]
- 31.Pallotti F., Lenaz G. Isolation and subfractionation of mitochondria from animal cells and tissue culture lines. Methods Cell Biol. 2001;65:1–35. doi: 10.1016/s0091-679x(01)65002-7. [DOI] [PubMed] [Google Scholar]
- 32.Rolland S.G., Motori E., Memar N., Hench J., Frank S., Winklhofer K.F. Impaired complex IV activity in response to loss of LRPPRC function can be compensated by mitochondrial hyperfusion. Proc Natl Acad Sci U S A. 2013;110:E2967–E2976. doi: 10.1073/pnas.1303872110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Hekimi S. Mitochondrial dysfunction and longevity in animals: untangling the knot. Science. 2015;350:1204–1207. doi: 10.1126/science.aac4357. [DOI] [PubMed] [Google Scholar]
- 34.Kauppila T.E.S., Kauppila J.H.K., Larsson N.G. Mammalian mitochondria and aging: an update. Cell Metabol. 2017;25:57–71. doi: 10.1016/j.cmet.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 35.Cedikova M., Pitule P., Kripnerova M., Markova M., Kuncova J. Multiple roles of mitochondria in aging processes. Physiol Res. 2016;65:S519–S531. doi: 10.33549/physiolres.933538. [DOI] [PubMed] [Google Scholar]
- 36.Chaudhari S.N., Kipreos E.T. Increased mitochondrial fusion allows the survival of older animals in diverse C. elegans longevity pathways. Nat Commun. 2017;8:182. doi: 10.1038/s41467-017-00274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang H.W., Pisano S., Chaturbedi A., Chen J., Gordon S., Baruah A. Transcription factors CEP-1/p53 and CEH-23 collaborate with AAK-2/AMPK to modulate longevity in Caenorhabditis elegans. Aging Cell. 2017;16:814–824. doi: 10.1111/acel.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehmann J.M., Moore L.B., Smith-Oliver T.A., Wilkison W.O., Willson T.M., Kliewer S.A. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 39.Fryer L.G., Parbu-Patel A., Carling D. The Anti-diabetic drugs rosiglitazone and metformin stimulate AMP-activated protein kinase through distinct signaling pathways. J Biol Chem. 2002;277:25226–25232. doi: 10.1074/jbc.M202489200. [DOI] [PubMed] [Google Scholar]
- 40.Brunmair B., Staniek K., Gras F., Scharf N., Althaym A., Clara R. Thiazolidinediones, like metformin, inhibit respiratory complex I: a common mechanism contributing to their antidiabetic actions? Diabetes. 2004;53:1052–1059. doi: 10.2337/diabetes.53.4.1052. [DOI] [PubMed] [Google Scholar]
- 41.Hardie D.G., Schaffer B.E., Brunet A. AMPK: an energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol. 2016;26:190–201. doi: 10.1016/j.tcb.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analyses of mitochondrial morphology changes in N2 worms treated with troglitazone. (A) Representative images and pseud-color images of mitochondrial organization in the body wall muscles of in N2 worms and cisd-1(tm4993) treated with troglitazone. Mitochondria morphologies are classified into 3 structural types as in Fig. 4. Scale bars, 10 m. (B) The quantitative results of (A) showing the percentage of various mitochondrial morphologies (N = 3). *p<0.05. **p < 0.01. ***p < 0.001. Error bars represent SEMs.