Abstract
Prokaryotes have developed an adaptive immune system called Clustered regularly interspaced short palindromic repeats (CRISPR) to combat attacks by foreign mobile genetic elements (MGEs) such as plasmids and phages. In the past decade, the widely characterized CRISPR-Cas9 enzyme has been redesigned to trigger a genome editing revolution. Class II type V CRISPR-Cas12a is a new RNA guided endonuclease that has been recently harnessed as an alternative genome editing tool, which is emerging as a powerful molecular scissor to consider in the genome editing application landscape. In this review, we aim to provide a mechanistic insight into the working mechanism of Cas12a, comparing it with Cas9, and eventually provide an overview of its current applications in genome editing and biotechnology applications.
Keywords: CRISPR-Cas12a, RNA guided endonucleases, crRNA biogenesis, Indiscriminate ssDNAse, Endonuclease recycling, Genome editing
Genome editing is a type of genetic engineering where a DNA is inserted, deleted or replaced in the genome of a living organism. The application of this technology has revolutionized various research areas ranging from biomedicine to biotechnology or synthetic biology [[1], [2], [3]]. A key point to initiate the editing is the need to generate a double strand break (DSB) in the DNA at a specific locus in the genome. To achieve this precise DSB researchers have developed engineered nucleases, also termed “molecular scissors”. Previous efforts have focused in the molecular understanding and redesign of different protein templates, such as homing endonucleases, zinc finger nucleases (ZFN) and TALEN. These tools have shown their utility in different genome editing applications [[4], [5], [6]] including the correction of mutations involving monogenic diseases [7,8]. However, the engineering of new DNA specificities in these protein scaffolds is cumbersome. Therefore, the development of the versatile CRISPR-Cas systems (Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR associated proteins) as molecular scissors, where a simple exchange of the RNA guide sequence is enough to redesign the nuclease specificity has paved the way for a revolution in the life sciences [[9], [10], [11]].
CRISPR repeats are associated with Cas proteins constituting an adaptive immune system in bacteria and archaea protecting them from foreign mobile genetic elements [12]. The discovery of CRISPR-Cas revealed the capability of bacteria and archaea to acquire and integrate genetic elements into its own genome, demonstrating the exchange of information between the environment and prokaryotic genomes [12]. The genetic record of previous attacks by foreign nucleic acids is stored in the CRISPR arrays. These arrays are made of short and conserved repetitive sequences called repeats which are strategically placed between unique sequences called spacers. They are inserted by specialized Cas proteins into the CRISPR array during infections by invading nucleic acids [[13], [14], [15], [16]]. The adaptive immunity by prokaryotes against foreign MGEs is achieved through the formation of RNA-guided endonucleases, which constitute the effector complexes and are able to detect secondary infection by a foreign DNA that was previously incorporated into the CRISPR array [10].
The CRISPR-Cas systems are classified into two classes (Classes 1 and 2) that are subdivided into six types (types I through VI). Class 1 (types I, III and IV) systems use multiple Cas proteins in their CRISPR ribonucleoprotein effector nucleases and Class 2 systems (types II, V and VI) use a single Cas protein [17]. Class 1 CRISPR-Cas systems are most commonly found in bacteria and archaea, and comprise ∼90% of all identified CRISPR-Cas loci. The Class 2 CRISPR-Cas systems, comprising the remaining ∼10%, exists almost exclusively in bacteria [18], and assemble a ribonucleoprotein complex, consisting of a CRISPR RNA (crRNA) and a Cas protein [10]. The crRNA contains information to target a specific DNA sequence [19]. These multidomain effector proteins achieve interference by complementarity between the crRNA and the target sequence after recognition of the PAM (Protospacer Adjacent Motif) sequence, which is adjacent to the target DNA [20]. These ribonucleoprotein complexes have been redesigned for precise genome editing by providing a crRNA with a redesigned guide sequence, which is complementary to the sequence of the targeted DNA [21,22]. The most widely characterised CRISPR-Cas system is the type II subtype II-A that is found in Streptococcus pyogenes (Sp), which uses the protein SpCas9, Cas9 was the first Cas-protein engineered for use in gene editing [9]. Class 2 type V is further classified into 4 subtypes (V-A, V–B, V–C, V–U). At present, V–C and V–U remain widely uncharacterised and no structural information on these systems is available [23]. V-A encodes the protein Cas12a (also known as Cpf1) and recently several high resolution structures of Cas12a have provided an insight into its working mechanism [[24], [25], [26]].
This system, involving RNA-guided interference, has been harnessed into a versatile biotechnological tool for genome editing [[9], [10], [11]], whereby a simple exchange of the RNA guide sequence can be employed to re-engineer nuclease specificity, leading to a revolution in the life sciences. Cas9, belonging to Class 2 Type II CRISPR-Cas interference system, is the more extensively used tool for genome editing. However, the overwhelming efficiency in genome sequencing of different organisms has generated a large amount of data helping the identification of new systems whose use in genome editing is currently being explored. Among them there are new members of Class 2, such as Cas12a [18,23,27].
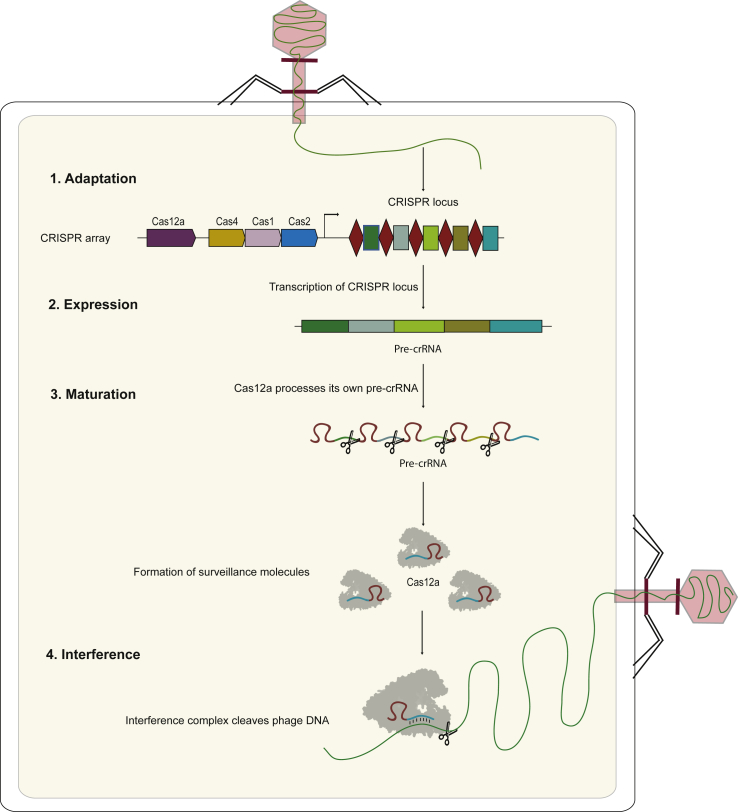
CRISPR-Cas immunity involves three major sequential steps: adaptation, expression/maturation and interference (Fig. 1), each step needs specific Cas proteins encoded by the cas genes near the CRISPR array, together with other accessory proteins [2,28,29]. The CRISPR-Cas adaptation stage involves the identification and extraction of the protospacer from the invading DNA/RNA and its subsequent incorporation into the CRISPR array. Both these functions are performed by the versatile Cas1-Cas2 adaptation complex. The identification of the protospacer starts with the recognition of the PAM by the adaptation complex [28], subsequently the spacer (sequence adjacent to the PAM) is integrated into the CRISPR array and the conserved repeat sequence is duplicated. The PAM sequence is excluded from the CRISPR array and is one of the first recognition motifs used for identifying target nucleic acids for degradation [16,24,28]. During the expression/maturation stage, the CRISPR array is transcribed into a long pre-CRISPR RNA (pre-crRNA) molecule. The pre-crRNA is processed into shorter crRNA molecules each containing a spacer and a part of the repeat sequence. Finally, interference can occur, after the crRNA forms a complex with the effector protein, forming a functional RNA guided endonuclease. This endonuclease is guided by the crRNA, which after PAM recognition hybridizes with the target DNA through its spacer sequence, and eventually, cuts the target DNA sequence. In this review, we describe the structural and functional features of Cas12a, a cousin of Cas9 belonging to the Class 2 Type V CRISPR-Cas system, which has been repurposed into an alternative and promising gene editing tool based on its substantial differences with Cas9 [17,30].
Fig. 1.
Stages of CRISPR-Cas immunity- Adaptation, Expression/Maturation, Interference.
Cas12a crRNA biogenesis
RNA sequencing of small RNA molecules extracted from Francisella novicida U112 culture containing Cas12a-based CRISPR loci revealed that mature crRNAs for Cas12a are 42–44 nt in length, with the first 19/20 nt corresponding to the repeat sequence and the remaining 23-25 nt to the spacer sequence [31]. In type II CRISPR systems, the maturation of crRNA is done by host housekeeping protein RNase III together with the trans-activating crRNA (tracrRNA), which is base paired with the pre-crRNA, in presence of Cas9 [32,33]. In contrast, it has been shown that Cas12a processes its own pre-crRNA into mature crRNAs, without the requirement of a tracrRNA, making it a unique effector protein with both endoribonuclease and endonuclease activities [34].After the pre-crRNA has been transcribed during the expression stage, Cas12a cuts it 4 nt upstream of the hairpin structures formed by the CRISPR repeats, producing intermediate crRNA molecules which undergo further processing in vivo into mature crRNAs.
Cas12a domain organisation and ribonucleoprotein complex assembly
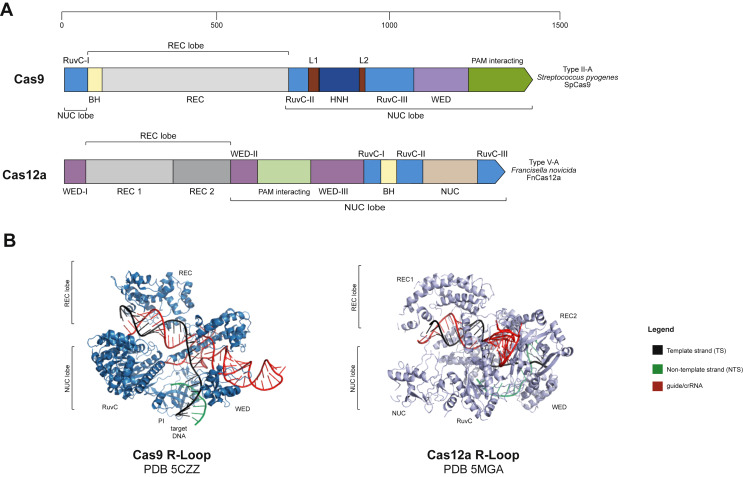
Type II (Cas9) and type V (Cas12a) CRISPR-Cas systems possess a characteristic Ruv-C like nuclease domain (Fig. 2A), which has been shown to be related to IS605 family transposon encoded TnpB proteins [18]. Crystallographic and cryo-EM data [24,31,35,36] reveal that Cas12a adopts a bilobed structure formed by the REC and Nuc lobes (Fig. 2B). The REC lobe is comprised of REC1 and REC2 domains, and the Nuc lobe is comprised of the RuvC, the PAM-interacting (PI) and the WED domains, and additionally, the bridge helix (BH). The RuvC endonuclease domain of this effector protein is made up of three discontinuous parts (RuvC I-III). The RNase site for processing its own crRNA is situated in the WED-III subdomain, and the DNase site is located in the interface between the RuvC and the Nuc domains. These structural studies have also shown that the only the 5’ repeat region of the crRNA is involved in the assembly of the binary complex. The 19/20 nt repeat region forms a pseudoknot structure through intramolecular base pairing. The crRNA is stabilized through interactions with the WED, RuvC and REC2 domains of the endonuclease, as well as two hydrated Mg2+ ions [[24], [25], [26],35]. This binary interference complex is then responsible for recognizing and degrading foreign DNA.
Fig. 2.
Domain organisation and R-loop complex of Ca9 and Cas12a.
PAM recognition
PAM recognition is a critical initial step in identifying a prospective DNA molecule for degradation since the PAM allows the CRISPR-Cas systems to distinguish their own genomic DNA from invading nucleic acids [37]. Cas12a employs a multistep quality control mechanism to ensure the accurate and precise recognition of target spacer sequences. The WED II-III, REC1 and PAM-interacting domains are responsible for PAM recognition and for initiating the hybridization of the DNA target with the crRNA. After recognition of the dsDNA by WED and REC1 domains, the conserved loop-lysine helix-loop (LKL) region in the PI domain, containing three conserved lysines (K667, K671, K677 in FnCas12a), inserts the helix into the PAM duplex with assistance from two conserved prolines in the LKL region. Structural studies show the helix is inserted at an angle of 45° with respect to the dsDNA longitudinal axis, promoting the unwinding of the helical dsDNA. The critical positioning of the three conserved lysines on the dsDNA initiates the uncoupling of the Watson–Crick interaction between the base pairs of the dsDNA after the PAM. The target dsDNA unzipping allows the hybridization of the crRNA with the strand containing the PAM, the ‘target strand (TS), while the uncoupled DNA strand, non-target strand (NTS), is conducted towards the DNase site by the PAM-interacting domain [24,26,35]. Cas12a has been shown to efficiently target spacer sequences following 5’T-rich PAM sequence. The PAM for LbCas12a and AsCas12a has a sequence of 5′-TTTN-3' and for FnCas12a a sequence of 5′-TTN-3′ and is situated upstream of the 5'end of the non-target strand [26,31,34]. It has also been shown that in addition to the canonical 5′-TTTN-3′ PAM, Cas12a also exhibits relaxed PAM recognition for suboptimal C-containing PAM sequences by forming altered interactions with the targeted DNA duplex [38].
DNA unzipping, propagation and cleavage
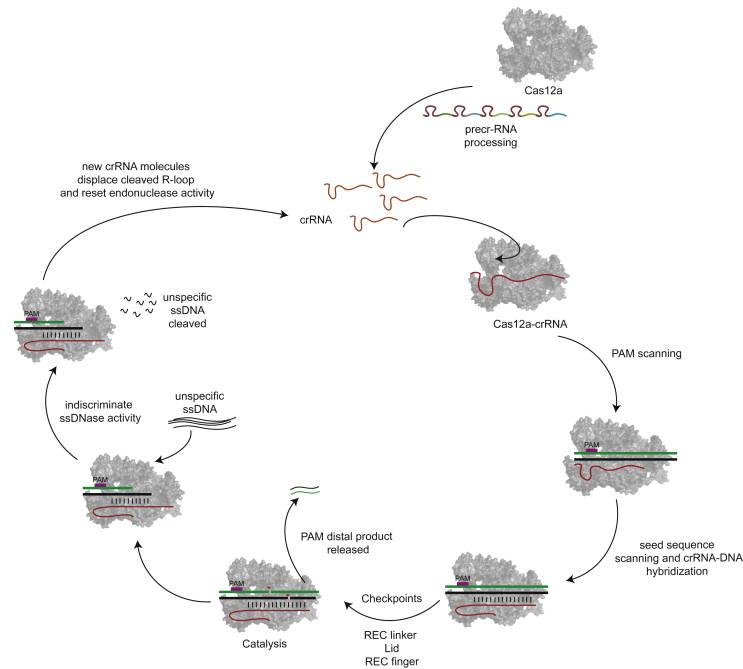
Once the crRNA-DNA hybrid R-loop starts forming, the enzyme then looks for a seed sequence of 3-5 nt on the PAM proximal end, the next check point in the correct identification of the target (Fig. 3). It has been reported that mismatches in the seed sequence results in the loss of cleavage activity [31,34]. Presence of the seed sequences promotes further hybridization of the crRNA-target DNA. Structural studies have shown that TS and NTS follow different pathways to the nuclease site [24], with several residues in the PI domain undergoing conformational changes and adopting a ‘rail’ shape to accommodate the nt-strand and eventually guiding it to the catalytic site. This structure also shows the presence of a barrier, the septum, to prevent the re-annealing of the dsDNA.
Fig. 3.
Model mapping the catalytic pathway of CRISPR-Cas12a.
A recent cryo-EM analysis on the intermediate catalysis products of Cas12a [36] revealed that the enzyme utilizes a further three-checkpoint control to sense the hybridization between the crRNA and the DNA. Three regions within the enzyme, the ‘REC linker’, the ‘lid’ and the ‘REC finger’ sequentially scan the hybrid through conformational changes and only when all three checkpoints are able to recognize the hybrid, the enzyme is in a conformation competent for catalysis. The endonuclease produces a staggered cut on a PAM distal site on the DNA with a 5 nt overhang on the target strand [31,36], and the PAM distal end of the cleaved product is then released from the complex [39].
Cleavage in the t-strand of the DNA by Cas12a produces a 5′-phosphorylated product [35]. In order for both DNA strands to be cut, they must enter the catalytic site with a 5′-3′ polarity. Structural studies reveal that the NTS is positioned to enter the RuvC-Nuc pocket with the 5′-3′ polarity, while the TS has the reverse polarity. An smFRET analysis suggests that Cas12a has to undergo conformational changes in the distal part of the REC and NUC lobes in order to allow the TS enter the nuclease site with the correct polarity [36]. This could explain why the NTS appears to be hydrolyzed faster than the TS. Therefore, the cleavage of the NTS is a consequence of the proper positioning of this strand in the RuvC-Nuc catalytic pocket rather than a requirement to initiate the cleavage reaction. After both strands have been cleaved, the PAM distal end of the cleavage product dissociates from the complex, but the PAM proximal site remains associated to Cas12a forming a cleaved R-loop [24,39].
Indiscriminate ssDNA cleavage
Besides high-specific dsDNA cleavage, Cas12a has also been shown to exhibit indiscriminate ssDNA degradation activity upon activation with a ssDNA complementary to the crRNA guide. This activity is displayed by all Cas12a orthologs and degrades any available ssDNA molecule into single/double nucleotides [40]. Comparisons of the structures of Cas12a before, during and after cleavage reveal the structural changes that result in such an indiscriminate activity. The lid region, which is involved in the checkpoints for accurate target recognition [36] is responsible for this action. Before the crRNA-DNA hybrid is formed, the lid occludes the cleft where the catalytic residues reside. Upon formation of the hybrid, the lid changes conformation to form an α helix, thus interacting with the crRNA of the hybrid assembly, thus dissociating the polar interactions and making available the catalytic pocket. In the R-loop structure after cleavage [24], this region appears disordered indicating that the catalytic site is accessible after the distal part of the dsDNA substrate dissociates from the complex. Therefore, the catalytic cleft is open and able to sever ssDNA indiscriminately. This molecular mechanism would explain how ssDNA molecules are degraded by Cas12a after being activated by the presence of the RNA-DNA hybrid [24,35,36]. In addition, recent studies have reported non-specific nicking of target sequences bearing mismatches in distal regions of the target DNA [41], suggesting that this could be a problem for potential applications.
Cas12a endonuclease recycling
Unspecific ssDNA degradation presents a potential harmful situation for the host cell since it could hinder basic cellular processes such as replication, transcription and DNA repair. This poses an important question: how could we eliminate this harmful indiscriminate activity? As the cell cannot allow an indiscriminate ssDNA degradation unleashed. The answer to these problems can be found in the bacterial genomes encoding Cas12a. Use of a conserved sequence of the crRNA for a database search disclosed that different bacteria encode a single copy of the Cas12a gene, whereas they encode multiple copies (up to 68) of the crRNA [18]. If the transcription rates are presumed to be similar, at any given time, the concentration of various crRNAs in the cell would be multiple times higher than that of the enzyme. It has been experimentally shown that with sufficient concentration of a new crRNA molecule, it is able to displace the cleaved R-loop from the enzyme, with the help of accessory host proteins, forming a new interference complex (Fig. 3). This shows that Cas12a can revert the active conformation to shut down unspecific activity by displacing the cleaved R-loop with a new crRNA. In doing so, it reverts back to a conformation where the molecular ‘lid’ forms polar interactions again to make the catalytic pocket inaccessible [36]. By doing so, not only does the endonuclease shut down the indiscriminate ssDNase activity, but also recycles its catalytic activity towards other target DNAs.
Cas12a vs Cas9
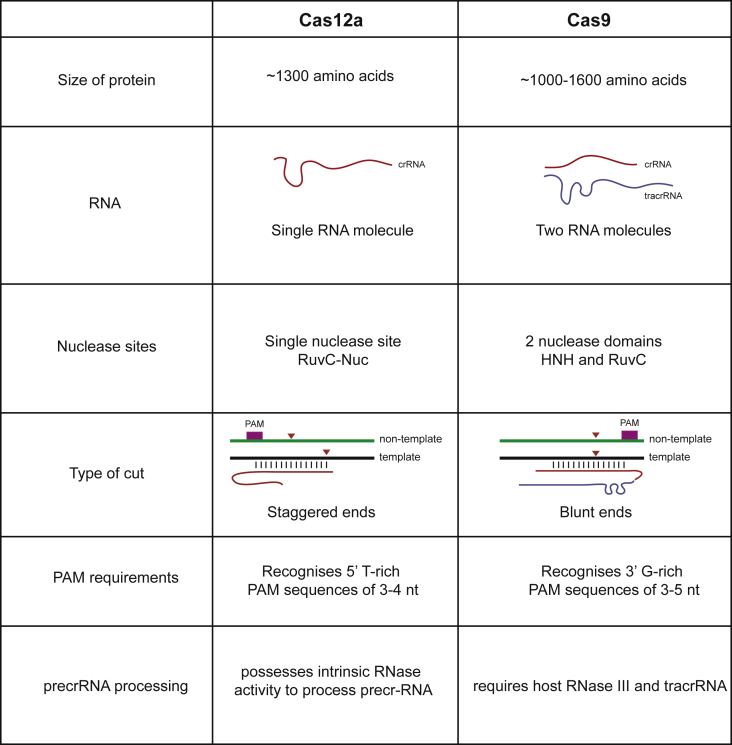
Cas12a and Cas9 have striking functional similarities despite having evolved through independent pathways (Fig. 2, Fig. 4), with similar sizes (1368 amino acids for SpCas9; 1307 for FnCas12a). They are both multidomain effector proteins and adopt a bilobed architecture when in complex with their respective RNAs. Cas9 requires two RNA molecules: tracrRNA and a crRNA, whereas Cas12a requires only a single RNA molecule, the crRNA. Cas9 possesses two nuclease sites HNH and RuvC domains, while Cas12a possesses only one nuclease site in the RuvC domain. Additionally, Cas12a also possesses an RNA processing site [1,9,24,31,34,37]. There are distinct differences in the mechanisms employed by the two proteins when it comes to RNA processing, PAM recognition, target DNA binding and eventually catalysis.
Fig. 4.
Cas12a vs Cas9.
After the CRISPR array has been transcribed into a long pre-crRNA molecule, it is processed into mature crRNAs before it can form an RNP with the endonuclease. In the case of Cas9, the tracrRNA (encoded close to the CRISPR locus) first needs to hybridize with the pre-crRNA, and then this hybrid RNA–RNA duplex structure is recognized by Cas9, following which the host RNase III cleaves the duplex, leaving a ∼75 nt long tracrRNA and a 39-42 nt long crRNA, which then forms the RNP complex responsible for recognizing and degrading the target DNA [33,42,43]. In contrast, Cas12a does not require a tracrRNA or RNase III, since the protein processes its own crRNA in its ribonuclease catalytic site [44]. In type-V CRISPR locus of F. novicida, the spacers are 27–32bp sequences interspersed by 36bp repeat sequences in between the spacers. In the entire CRISPR array transcript, which is the pre-crRNA, the repeat derived sequences form pseudoknots, which are recognized by Cas12a. Following the recognition, the pre-crRNA is cleaved forming ∼43 nt mature crRNA [25,26,31,34,35,45]. Logistically, Cas12a presents a more minimalistic system than Cas9.
For Cas9 targeted DNA sequences, the PAM is situated downstream of the spacer sequence on the non-template strand, and is recognized by the PI domain, which is primed for identifying a 5′-NGG-3′ PAM [43,46]. In contrast, Cas12a recognizes A-T rich sequences, with the PAM, typically 5′-TTTV-3′, located upstream of the spacer. Upon PAM recognition, the target DNA is unzipped and hybridization of the RNA-DNA takes place. For both enzymes exists a crucial seed sequence next to the PAM to determine the specificity of target DNA binding. The seed sequence for Cas9 is about ∼10 nt whereas for Cas12a it is about ∼5–6 nt [9,31,34,35]. When the hybridization of the DNA with the RNA is complete, Cas9 cleaves the template strand and the non-template strands in the catalytic sites located in the HNH and the RuvC domains respectively, producing a blunt DSB, with the cleavage site being 3 base pairs upstream from the PAM sequence [9]. However, in the case of Cas12a due to the presence of a single nuclease site, the strands of the DNA are cut in the same nuclease site. Since it has been shown that the two strands follow different pathways to reach the catalytic site, explaining the staggered DSB produced by Cas12a [24,31].
Cleavage fidelity is an important issue for many of the nucleases used in genome editing applications [47]. An optimal tool must introduce modifications just on the target site, leaving the rest of the genome unmodified in order to avoid undesired changes in other sites of the genome with unpredictable consequences. Therefore, specificity and the resulting cleavage products are key in genome modification applications. A recent comparison of different Cas12a and Cas9 from different species using nuclease digestion and deep sequencing (NucleaSeq) in vitro, revealed that both enzymes share similar types of specificities and tolerate similar mismatches [48], in contrast to in vivo reports that show the lower off-target effects of Cas12a [49,50]. This apparent contradiction may be related to different recognition and cleavage kinetics, but also to a possible different behaviour of these cutters on a chromatin context, thus posing the question whether in vitro or in vivo approaches should be pursued for nuclease redesign efforts.
CRISPR-Cas12a mediated genome editing
Application of CRISPR-Cas systems as molecular tools for genome editing exploits their ability to produce a double strand break (DSB) at a specific genomic locus, and depends entirely on the host cell DNA repair machinery to fix the lesion produced by these systems. The repair mechanisms can be either of the following processes: homology-directed repair (HDR) or non-homologous end joining (NHEJ). HDR utilizes a template DNA that is homologous to the break site (an unbroken sister chromatid or a homologous chromosome) to repair the DSB, whereas NHEJ is based on direct joining of broken ends of the DSB, making NHEJ the more error prone mechanism of the two. HDR can thus be used to supply exogenous template DNA to implement a user defined change in the host genome. NHEJ can be applied for gene disruption whereas HDR allows for the scope of introducing new genetic information or direct correction of the sequence at a specific locus.
At the center of CRISPR mediated genome engineering today is Cas9, with applications including, but not limited to, gene knockout and precise genome editing. Despite the rapid advances in genome editing by Cas9, it still presents challenges owing to the possibility of off-target effects and difficulty of delivering the ribonucleoprotein particle [18]. Cas12a, owing to its substantial differences with Cas9, presents an alternate molecular genome editing tool. The use of Cas12a in genome editing for various cell types has been probed in several studies up to date. Comparative studies of gene repression by catalytically dead Cas9 from S. pyogenes (SpdCas9) and catalytically dead Cas12a from Eubacterium eligens (EedCas12a) revealed that the latter displays a higher gene repression in the template strand of the target DNA than SpdCas9 [51]. It was also shown that the pre-crRNA processing activity of Cas12a makes it an attractive candidate for multiplex gene regulation, which is cumbersome when attempted with Cas9 [52]. This auto-processing of its own crRNA has been used to modify multiple genetic elements simultaneously generating constitutive, conditional, inducible, orthogonal and multiplexed genome engineering of endogenous targets using multiple CRISPR RNAs delivered on a single plasmid [53].
The viability of this approach has been further established by other studies, in which multiplex gene regulation by Cas12a was successfully observed in bacteria, plants, as well as in mammalian cells [52,[54], [55], [56]]. Cas12a can also serve as a solution in cell types where use of Cas9 is toxic, such as in some industrial strains of Streptomyces [54].
Targeted mutagenesis in plants can also be achieved through co-expression of Cas12a and its cognate crRNA in vivo, as was shown in rice. Additionally, it was also shown that the mutagenesis was more efficient through the use of pre-crRNAs with full-length direct repeat sequences than with mature crRNAs [57]. Efficient mutagenesis through delivery of the pre-assembled ribonucleoprotein (RNP) particle was also observed in soybean and wild tobacco. The RNP was assembled from recombinantly expressed Cas12a and in vitro transcribed or chemically synthesized crRNAs [58].
Successful gene editing of mammalian cells using Cas12a include correction of mutations causing Duchenne muscular dystrophy (DMD) in patient derived induced pluripotent stem cells (iPSCs) and in mdx mice, a popular model for studying DMD. Dystrophin expression was reinstated in iPSCs after Cas12a-mediated gene editing, while in the mdx mice, corrections in the pathophysiological hallmarks of muscular dystrophy were observed [59]. Delivery of the adenovirus vector with an AsCas12a expression cassette yielded successful mutations in primary human hepatocytes from humanized mice with chimeric liver [60]. Cas12a-mediated genome editing was also used to engineer rat models that mimic human atherosclerosis and this system may have potential applications in understanding early stage atherosclerosis [61].
All of the above studies how Cas12a can be engineered for various applications. Despite the numerous recent advances in the application of Cas12a, there remain vast avenues of unexplored potential of Cas12a in terms of therapeutics and diagnostics.
Cas12a applications in bioengineering
Currently, a vast effort is ongoing to redesign all these tools for biomedical and biotechnological applications. However, recent studies have envisioned the possibility of using CRISPR-Cas nucleases in bioengineering of smart materials, for example hydrogels [62] These water-filled polymers are encapsulated by DNA. In a recent study, Cas12a has been used to specifically degrade the DNA scaffold of DNA hydrogels, thus opening the possibility that this smart cutter can be turned into a programmable device to deliver the cargo of DNA encaged hydrogels in a determined location at a certain time. The cleavage properties of Cas12a make it an ideal candidate to promote controlled delivery of the cargo. Although, application of these approaches and their combinations can be now envisioned by many researchers, the range of possibilities in different areas is so large that it is beyond our imagination.
Conclusions
In this review we have sought to offer a condensed overview of the functionality of CRISPR-Cas12a, discussing the structural and functional features of the different stages of the reaction pathway leading up to the catalysis of the target DNA, and eventually discussing the applications of Cas12a in brief. Cas12a employs a multi-checkpoint mechanism to ensure precise targeting of DNA, which is a desirable property in a genome editing tool in order to have low off target effects. Although it has been shown that the indiscriminate ssDNA degradation of Cas12a could be shut down through the recruitment of a new crRNA molecule, it could still potentially harm the host cell targeted for genome modification. Modulation of this activity is necessary to achieve higher regulation and control of Cas12a catalysis, and in turn to achieve a more robust genome editing tool. In this direction structural information has been used to redesign Cas12a obtaining variants without ssDNA unspecific activity, thus severing only dsDNA specifically [36].
Currently, Cas9 and Cas12a, are the sole members of the CRISPR family that have been utilized for genome editing. Owing to their significant similarities and differences, just these two endonucleases between themselves have made the applications of CRISPR highly versatile. Cas12a, in some cases, offers certain advantages over Cas9, for example in its capability to be used for multiplex genome editing and production of staggered DSB, which promotes HDR instead of NHEJ. Significant research also is ongoing to engineer artificial variants of Cas9 and Cas12a to recognize different PAM than the wild type proteins, which will facilitate the targeting of a wider library of genomes.
The rapid advent of the CRISPR-Cas technology for genome manipulation has been revolutionary for life sciences. Despite the vast application areas of this technology, the current state of the art of the CRISPR molecular tools (Cas9 or Cas12a) suffers from one important drawback: dependence on host cell DNA repair machinery. Both Cas9 and Cas12a based technology produce a double strand break (DSB) in the target DNA, and this break is then repaired by endogenous DNA repair machinery with or without the presence of a template. Although these tools have been successfully utilized to obtain precise insertion of DNA into the targeted genomic loci, their efficiency differs from cell type to cell type [[63], [64], [65]]. DNA repair through HDR is also related to active cell division, which makes these tools ineffective in cell types that are not actively dividing, such as neurons. Recent studies characterizing CRISPR-associated transposase (CAST), which comprises Tn7-like transposase subunits and a CRISPR effector from type V–K, could pave the way to new avenues of gene editing using CRISPR systems since these systems are self-sufficient in precise DNA insertion and do not depend on endogenous cell DNA repair machinery [66,67]. However, a large ongoing research is aiming to tailor both Cas9 and Cas12a further to ensure precise DNA insertion into the targeted genome. Even apart from this apparent drawback, both these tools have a vast range of applicability and ongoing efforts are striving to produce improved and more robust engineered genome editing tools.
Conflicts of interest
GM declares that he is an inventor in a Cas12a patent application.
Acknowledgements
The Novo Nordisk Foundation Center for Protein Research is supported financially by the Novo Nordisk Foundation (grant NNF14CC0001). Our work is also supported by the cryoEM (grant NNF0024386), cryoNET (grant NNF17SA0030214), and Distinguished Investigator (NNF18OC0055061) grants to GM, who is a member of the Integrative Structural Biology Cluster (ISBUC) at the University of Copenhagen. We thank Stefano Stella for advice during the preparation of the manuscript.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Doudna J.A., Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 2.Wright A.V., Nuñez J.K., Doudna J.A. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R., Horvath P. A decade of discovery: CRISPR functions and applications. Nat Microbiol. 2017;2:17092. doi: 10.1038/nmicrobiol.2017.92. [DOI] [PubMed] [Google Scholar]
- 4.Leitao A.L., Costa M.C., Enguita F.J. Applications of genome editing by programmable nucleases to the metabolic engineering of secondary metabolites. J Biotechnol. 2017;241:50–60. doi: 10.1016/j.jbiotec.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Knipping F., Osborn M.J., Petri K., Tolar J., Glimm H., von Kalle C. Genome-wide specificity of highly efficient TALENs and CRISPR/Cas9 for T cell receptor modification. Mol Ther - Methods Clin Dev. 2017;4:213–224. doi: 10.1016/j.omtm.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandrasegaran S., Carroll D. Origins of programmable nucleases for genome engineering. J Mol Biol. 2016;428:963–989. doi: 10.1016/j.jmb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redondo P., Prieto J., Munoz I.G., Alibes A., Stricher F., Serrano L. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 9.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Chapentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N. Multiplex genome engineering using CRISPR/cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mojica F.J.M., Díez-Villaseñor C., García-Martínez J., Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 13.Nuñez J.K., Kranzusch P.J., Noeske J., Wright A.V., Davies C.W., Doudna J.A. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat Struct Mol Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nuñez J.K., Harrington L.B., Kranzusch P.J., Engelman A.N., Doudna J.A. Foreign DNA capture during CRISPR–Cas adaptive immunity. Nature. 2015;527:535–538. doi: 10.1038/nature15760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J.J. CRISPR-Cas: adapting to change. Science. 2017;356:eaal5056. doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 16.Mojica F.J.M., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 17.Nishimasu H., Nureki O. Structures and mechanisms of CRISPR RNA-guided effector nucleases. Curr Opin Struct Biol. 2017;43:68–78. doi: 10.1016/j.sbi.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 18.Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W. Diversity and evolution of class 2 CRISPR–Cas systems. Nat Rev Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath P., Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 20.Stella S., Alcón, Montoya G. Class2 CRISPR-cas RNA-guided endonucleases: Swiss army knives of genome editing. Nat Struct Mol Biol. 2017;24:882–892. doi: 10.1038/nsmb.3486. [DOI] [PubMed] [Google Scholar]
- 21.Liu L., Chen P., Wang M., Li X., Wang J., Yin M. C2c1-sgRNA complex structure reveals RNA-guided DNA cleavage mechanism. Mol Cell. 2017;65:310–322. doi: 10.1016/j.molcel.2016.11.040. [DOI] [PubMed] [Google Scholar]
- 22.Yang H., Gao P., Rajashankar K.R., Patel D.J. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-cas endonuclease. Cell. 2017;167:1814–1828. doi: 10.1016/j.cell.2016.11.053. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shmakov S., Abudayyeh O.O.O., Makarova K.S.S., Wolf Y.I.I., Gootenberg J.S.S., Semenova E. Discovery and functional characterization of diverse class 2 CRISPR-cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stella S., Alcón P., Montoya G. Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature. 2017;546:559–563. doi: 10.1038/nature22398. [DOI] [PubMed] [Google Scholar]
- 25.Dong D., Ren K., Qiu X., Zheng J., Guo M., Guan X. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016;532:522–526. doi: 10.1038/nature17944. [DOI] [PubMed] [Google Scholar]
- 26.Yamano T., Nishimasu H., Zetsche B., Hirano H., Slaymaker I.M., Li Y. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016;165:949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amitai G., Sorek R. CRISPR–Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol. 2016;14:67–76. doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]
- 29.van der Oost J., Westra E.R., Jackson R.N., Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stella S., Alcón P., Montoya G. Class 2 CRISPR–Cas RNA-guided endonucleases: Swiss Army knives of genome editing. Nat Struct Mol Biol. 2017;24:882–892. doi: 10.1038/nsmb.3486. [DOI] [PubMed] [Google Scholar]
- 31.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charpentier E., Richter H., van der Oost J., White M.F. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev. 2015;39:428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fonfara I., Richter H., Bratovič M., Le Rhun A., Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 35.Swarts D.C., van der Oost J., Jinek M. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-cas12a. Mol Cell. 2017;66:221–233. doi: 10.1016/j.molcel.2017.03.016. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stella S., Mesa P., Thomsen J., Paul B., Alcón P., Jensen S.B. Conformational activation promotes CRISPR-cas12a catalysis and resetting of the endonuclease activity. Cell. 2018;175:1856–1871. doi: 10.1016/j.cell.2018.10.045. e21. [DOI] [PubMed] [Google Scholar]
- 37.Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamano T., Zetsche B., Ishitani R., Zhang F., Nishimasu H., Nureki O. Structural basis for the canonical and non-canonical PAM recognition by CRISPR-cpf1. Mol Cell. 2017;67:633–645. doi: 10.1016/j.molcel.2017.06.035. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh D., Mallon J., Poddar A., Wang Y., Tippana R., Yang O. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a) Proc Natl Acad Sci. 2018;115:5444–5449. doi: 10.1073/pnas.1718686115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murugan K., Seetharam A.S., Severin A.J., Sashital D.G. Pervasive off-target and double-stranded DNA nicking by CRISPR-Cas12a. BioRxiv. 2019:657791. [Google Scholar]
- 42.Jiang F., Taylor D.W., Chen J.S., Kornfeld J.E., Zhou K., Thompson A.J. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang F., Zhou K., Ma L., Gressel S., Doudna J.A. A Cas9–guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- 44.Fonfara I., Richter H., Bratovič M., Le Rhun A., Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 45.Gao P., Yang H., Rajashankar K.R., Huang Z., Patel D.J. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 2016;26:901–913. doi: 10.1038/cr.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders C., Niewoehner O., Duerst A., Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stella S., Montoya G. The genome editing revolution: a CRISPR-Cas TALE off-target story. Bioessays. 2016;38:S4–S13. doi: 10.1002/bies.201670903. [DOI] [PubMed] [Google Scholar]
- 48.Jones S.K., Hawkins J.A., Johnson N.V., Jung C., Hu K., Rybarski J.R. Massively parallel kinetic profiling of natural and engineered CRISPR. nucleases. BioRxiv. 2019:696393. doi: 10.1038/s41587-020-0646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D., Kim J., Hur J.K., Been K.W., Yoon S., Kim J.S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34:863–868. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- 50.Gao L., Cox D.B.T., Yan W.X., Manteiga J.C., Schneider M.W., Yamano T. Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol. 2017;35:789–792. doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim S.K., Kim H., Ahn W.C., Park K.H., Woo E.J., Lee D.H. Efficient transcriptional gene repression by type V-A CRISPR-cpf1 from Eubacterium eligens. ACS Synth Biol. 2017;6:1273–1282. doi: 10.1021/acssynbio.6b00368. [DOI] [PubMed] [Google Scholar]
- 52.Zetsche B., Heidenreich M., Mohanraju P., Fedorova I., Kneppers J., DeGennaro E.M. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35:31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campa C.C., Weisbach N.R., Santinha A.J., Incarnato D., Platt R.J. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat Methods. 2019;67:887–893. doi: 10.1038/s41592-019-0508-6. [DOI] [PubMed] [Google Scholar]
- 54.Li L., Wei K., Zheng G., Liu X., Chen S., Jiang W. CRISPR-Cpf1-Assisted multiplex genome editing and transcriptional repression in Streptomyces. Appl Environ Microbiol. 2018;84:e00827–e00918. doi: 10.1128/AEM.00827-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang M., Mao Y., Lu Y., Tao X. Zhu J kang. Multiplex gene editing in rice using the CRISPR-cpf1 system. Mol Plant. 2017;10:1011–1013. doi: 10.1016/j.molp.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X., Wang J., Cheng Q., Zheng X., Zhao G., Wang J. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov. 2017;3:17018. doi: 10.1038/celldisc.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu R., Qin R., Li H., Li D., Li L., Wei P. Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol J. 2017;15:713–717. doi: 10.1111/pbi.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim H., Kim S.T., Ryu J., Kang B.C., Kim J.S., Kim S.G. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun. 2017;8:14406. doi: 10.1038/ncomms14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y., Long C., Li H., McAnally J.R., Baskin K.K., Shelton J.M. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci Adv. 2017;3:e1602814. doi: 10.1126/sciadv.1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsukamoto T., Sakai E., Iizuka S., Taracena-Gándara M., Sakurai F., Mizuguchi H. Generation of the adenovirus vector-mediated CRISPR/Cpf1 system and the application for primary human hepatocytes prepared from humanized mice with chimeric liver. Biol Pharm Bull. 2018;41:1089–1095. doi: 10.1248/bpb.b18-00222. [DOI] [PubMed] [Google Scholar]
- 61.Lee J.G., Ha C.H., Yoon B., Cheong S.-A., Kim G., Lee D.J. Knockout rat models mimicking human atherosclerosis created by Cpf1-mediated gene targeting. Sci Rep. 2019;9:2628. doi: 10.1038/s41598-019-38732-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.English M.A., Soenksen L.R., Gayet R.V., de Puig H., Angenent-Mari N.M., Mao A.S. Programmable CRISPR-responsive smart materials. Science. 2019;365:780–785. doi: 10.1126/science.aaw5122. [DOI] [PubMed] [Google Scholar]
- 63.Jasin M., Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmid-Burgk J.L., Höning K., Ebert T.S., Hornung V. CRISPaint allows modular base-specific gene tagging using a ligase-4-dependent mechanism. Nat Commun. 2016;7:12338. doi: 10.1038/ncomms12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E.J. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peters J.E., Makarova K.S., Shmakov S., Koonin E.V. Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc Natl Acad Sci. 2017;114:E7358–E7366. doi: 10.1073/pnas.1709035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strecker J., Ladha A., Gardner Z., Schmid-Burgk J.L., Makarova K.S., Koonin E.V. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019;365:48–53. doi: 10.1126/science.aax9181. [DOI] [PMC free article] [PubMed] [Google Scholar]