Abstract
The Candida (C.) albicans complex includes C. albicans, C. dubliniensis, C. stellatoidea, and C. africana, with the last mentioned as an important emerging agent of vulvovaginal candidiasis (VVC). The aim of the study was to identify C. africana and C. dubliniensis and assess their drug susceptibility in vaginitis. One-hundred Candida isolates of the C. albicans complex from women diagnosed with vaginitis and from vaginal samples in the culture collection of a medical mycology laboratory were examined. Species of the C. albicans complex were identified with conventional and molecular methods using polymerase chain reaction (PCR) for amplification and sequencing of the internal transcribed spacer (ITS) region, PCR for partial amplification of hyphal wall protein 1 (HWP1) gene and duplex PCR. The effects of antifungal drugs were evaluated according to standard broth microdilution protocols.
Ninety-seven C. albicans (97%) and three C. africana (3%) isolates were identified. Results of susceptibility testing revealed one isolate of C. africana to be resistant to both clotrimazole and fluconazole, and one showed reduced susceptibility to itraconazole.
Identification of Candida species especially C. africana in vaginitis is crucial, there are varying levels of resistance to antifungal drugs.
Keywords: Microbiology, Mycology, Microbial genomics, DNA sequencing, Antimicrobial, Women's health, Candida albicans complex, Candida dubliniensis, Candida africana, Drug susceptibility, Vulvovaginal candidiasis, HWP1
Microbiology; Mycology; Microbial genomics; DNA sequencing; Antimicrobial; Women's health; Candida albicans complex; Candida dubliniensis; Candida africana; Drug susceptibility; Vulvovaginal candidiasis; HWP1.
1. Introduction
Species in the C. albicans complex can colonize in the vaginal tract. Through changes in the relationship between the host and this commensal yeast, an invasive form may occur (Goncalves et al., 2016) causing mucosal infection and vulvovaginal candidiasis (VVC) (Hedayati et al., 2015). In 2001, C. albicans isolates of African and German patients with atypical phenotypes were described as a novel species, C. africana (Tietz et al., 2001). C. africana produces a germ tube but not chlamydospores. C. albicans remains the most common pathogenic yeast isolated from vaginal specimens of VVC patients, but the closely-related C. africana and C. dubliniensis have often been mistaken for C. albicans. C. africana has been isolated from candidal balanoposthitis in China (Hu et al., 2015), and clinical isolates have been reported as the cause of VVC in Germany, China, Italy, Iran, Nigeria, Spain, the United Kingdom, Argentina, Colombia, among other countries (Tietz et al., 2001; Alonso-Vargas et al., 2008; Romeo and Criseo, 2009; Dieng et al., 2012; Nnadi et al., 2012; Borman et al., 2013; Shan et al., 2014; Rodriguez-Leguizamon et al., 2015; Yazdanparast et al., 2015; Theill et al., 2016). The vaginal candidiasis studies in Iran have been identified susceptible (Yazdanparast et al., 2015) and resistant (Majdabadi et al., 2018) antifungal drugs C. africana isolates.
Molecular methods are suitable for identification of Candida species. The use of the ribosomal DNA (rDNA) ITS region for sequence-analysis appears to be the most sensitive and specific method for accurate and reliable molecular identification of Candida species (Merseguel et al., 2015; Zarrinfar et al., 2016). The sequencing of a short fragment of the ITS2 region has been shown effective in detecting the presence of C. africana in clinical Candida infections (Borman et al., 2013). Duplex PCR is a suitable method for identification of isolates such as C. albicans and C. dubliniensis (Ahmad et al., 2012). Amplification of the HWP1 gene was the first molecular method for discriminating C. albicans, C. dubliniensis, and C. africana (Romeo and Criseo, 2008). Many antifungal agents are effective treating VVC, but some vaginal Candida isolates have developed resistance to antifungal drugs. Assessment of antifungal susceptibility of pathogenic yeasts is essential to proper therapy and will allow evaluation of their efficacy and treatment results as well as prevention of drug resistance (Mukasa et al., 2015; Fornari et al., 2016). However, there is limited information about the prevalence and antifungal susceptibility of C. africana in VVC in Iran. The aim of this study was to identify C. africana and C. dubliniensis in vaginal samples from VVC patients and to evaluate their antifungal susceptibility pattern.
2. Materials and methods
2.1. Patients, sample collection and definitions
The present study analyzed clinical isolates of vaginal discharge from 300 non-pregnant patients aged 18–57 years with suspected VVC admitted to the Shahid Akbar Abadi hospital and Shahriar health centers from June through December 2016, along with culture samples from our laboratory. Patients had not used antifungal drugs within the preceding four weeks. Samples were collected with sterile cotton swabs, and microscopic examination was done to identify yeast forms or pseudohyphae. In addition, 30 Candida spp. of the C. albicans complex obtained from VVC patients were selected from the culture collection of the medical mycology laboratory in our department. All specimens were cultured on CHROMagar Candida medium (CHROMagar, France) at 37 °C for 24–48 hrs for detection of mixed Candida spp. infections. A single colony of each Candida isolate on CHROMagar Candida medium was selected and cultured on Sabouraud dextrose agar (SDA) medium with chloramphenicol at 30 °C for 48–72 hrs for further use. The germ tube test, chlamydospore-forming assay on corn meal agar (CMA) with 1% Tween 80 and growth on SDA medium with chloramphenicol at 42 °C and 45 °C was conducted.
2.2. DNA extraction
A single colony of each clinical isolate from CHROMagar Candida medium was cultured on yeast extract peptone dextrose (YEPD) agar and incubated at 37 °C for 24–48 hrs. Genomic DNA was extracted from yeast cultures using the Qiagen DNA tissue kit (Germany). The extracted DNA was stored at -20 °C until use.
2.3. PCR amplification and sequencing of ITS region
The universal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCC GCTTATTGATATGC-3′) were used to amplify the ITS1-5.8S-ITS2 region (Fujita et al., 2001; Ciardo et al., 2006) under the following conditions: 98 °C at 5 min; 35 cycles of 30 s at 98 °C, annealing for 30 s at 56 °C, 30 s at 72 °C, and a final extension of 5 min at 72 °C. The PCR products were sequenced by Macrogen (Korea). Sequences were compared with reference data available from the GenBank database using the BLAST sequence search tool (http://www.ncbi.nlm.nih.gov/BLAST).
2.4. Duplex PCR
C. albicans and C. dubliniensis strains were analyzed by duplex PCR with paired primers CALF (5′-TGGTAAGGCGGGATCGCTT-3′) and CALR (5′-GGTCAAAGTTTGAAGATATAC-3′) for C. albicans and CDUF (5′-AAACTTGTCACGAGATTATTTTT-3′) and CDUR (5′-AAAGTTTGAAGAATAAAATGGC-3′) for C. dubliniensis (Ahmad et al., 2012). Specific primers used the following PCR protocol: 95 °C for 5 min; 30 cycles of 1 min at 95 °C, 30 s at annealing temperature 55 °C, and 1 min at 72 °C; and a final extension of 10 min at 72 °C.
2.5. PCR amplification of HWP1 gene
The PCR primers (CR-f 5′-GCTACCACTTCAGAATCATCATC-3′ and CR-r 5′-GCACCTTCAGTCGTAGAGACG-3′) used to amplify the HWP1 gene for detection C. albicans, C. africana and C. dubliniensis, as previously described by Romeo and Criseo (2008).
Reference strains used as control were C. albicans (ATCC10231) and C. dubliniensis from the archives of department of Medical Parasitology and Mycology, School of Medicine, Iran University of Medical Sciences. C. dubliniensis was 100% confirmed by sequencing to species level.
2.6. In vitro antifungal susceptibility testing
The in vitro susceptibility was determined by the Clinical and Laboratory Standards Institute (CLSI) (CLSI M27-S3 2008; CLSI M27-S4 2012). Briefly, final inocula of 0.5×103 to 2.5×103 colony-forming units/ml were distributed in 96 well microtiter plates in RPMI 1640 medium buffered to pH 7.0 with 0.165M morpholinepropanesulfonic acid with diluted antifungal drugs and incubated at 35 °C. The antifungals used were amphotericin B, itraconazole, fluconazole, and clotrimazole (Sigma, Germany), and all tests were duplicated. C. glabrata CBS 138 was used as quality control strain. According to CLSI M27-S3 criteria for fluconazole, the sensitivity profile is classified as sensitive (≤8 μg/ml), dose-dependent sensitive (16–32 μg/ml), and resistant (≥64 μg/ml), and breakpoints for itraconazole is sensitive (≤0.125 μg/ml), dose-dependent sensitive (0.25–0.5 μg/ml), and resistant (≥1 μg/ml).
2.7. Ethical approval
This research was approved by Ethics Committee of Iran University of Medical Sciences, under Ethics Committee number 95-01-30-27842.
2.8. Informed consent
Written informed consent was obtained from all patients.
3. Results
3.1. Patients
All patients diagnosed with VVC (23%) were in the 18–50 years age range. None exhibited diabetes, immunodeficiency, or other chronic disease. The archival information on VVC patients indicated a very similar age range, 18–50 years, and without disease.
3.2. Yeast isolates
One hundred isolates belonging to the C. albicans complex were identified, 97 as C. albicans (data not shown) 97% and three (3%) as C. africana. C. africana produced small turquoise-green colonies on CHROMagar Candida and did not produce chlamydospores on CMA medium. C. albicans isolates proliferated at 42 °C and 45 °C, while the C. africana isolates did not.
3.3. Duplex PCR assay
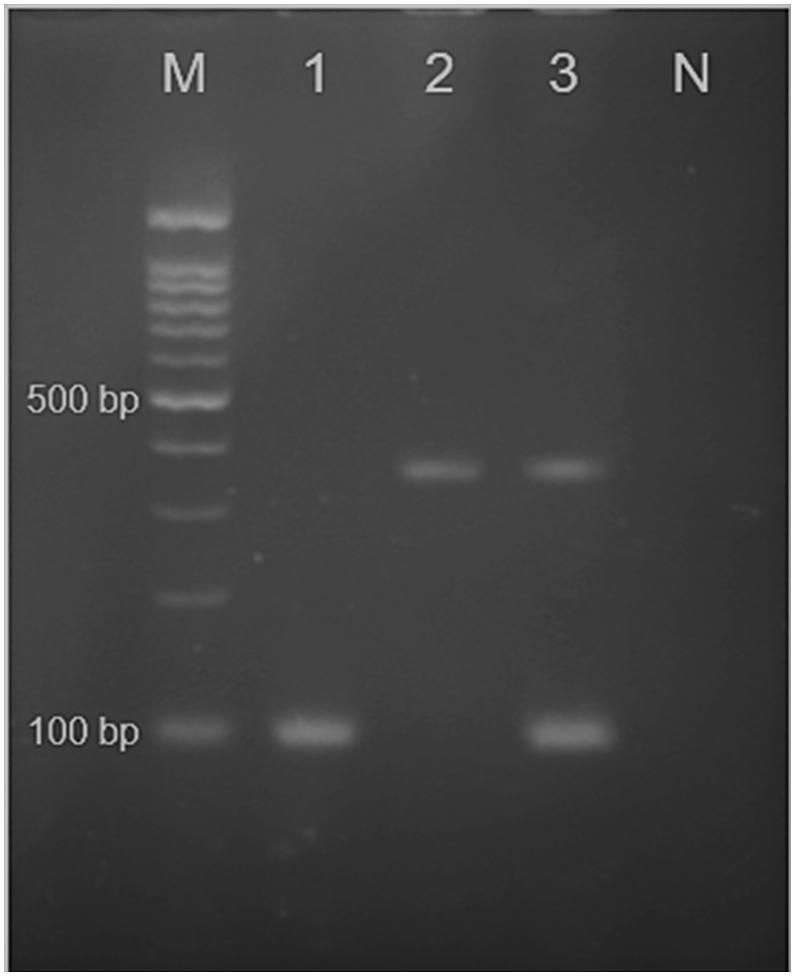
The results of amplification of isolates with CALF/CALR and CDUF/CDUR specific primers for C. albicans and C. dubliniensis, respectively, for duplex PCR assay, were determined for a ~100 bp fragment of C. albicans and a ~325 bp fragment of C. dubliniensis (Figure 1). No C. dubliniensis isolates were found in this study.
Figure 1.
Agarose gel of duplex PCR using primers CALF, CALR, CDUF andCDUR. Lane 1: C. albicans clinical isolate (~100 bp); Lane 2: positive control C. dubliniensis (~325 bp) from the archives of the Department of Medical Parasitology and Mycology; Lane 3: positive controls C. albicans (ATCC10231) and C. dubliniensis. N: negative control. M: marker 100 bp.
3.4. PCR amplification and sequencing of ITS region
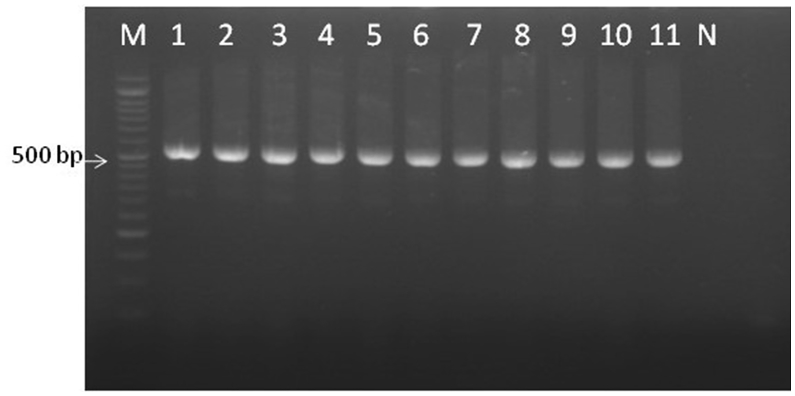
Amplification of all clinical isolates of C. albicans complex with ITS1 and ITS4 primers yielded fragments of 530 bp for both C. albicans and C. africana (Figure 2). The ITS region sequences of Candida spp. clinical isolates were compared in the GenBank database using BLAST. All clinical isolates showed 100% and 99% similarity to C. albicans or C. africana. The ITS sequences of C. africana isolates were deposited in GenBank under accession numbers MG757669, MG757670, and MG757671.
Figure 2.
Clinical isolates of C. albicans complex analyzed with ITS1 and ITS4 universal primers; 530 bp fragment produced. Lanes 1–8: C. albicans; Lanes 9–11: C. africana; N: negative control. M: marker 50 bp.
3.5. PCR amplification with specific primers of HWP1 gene
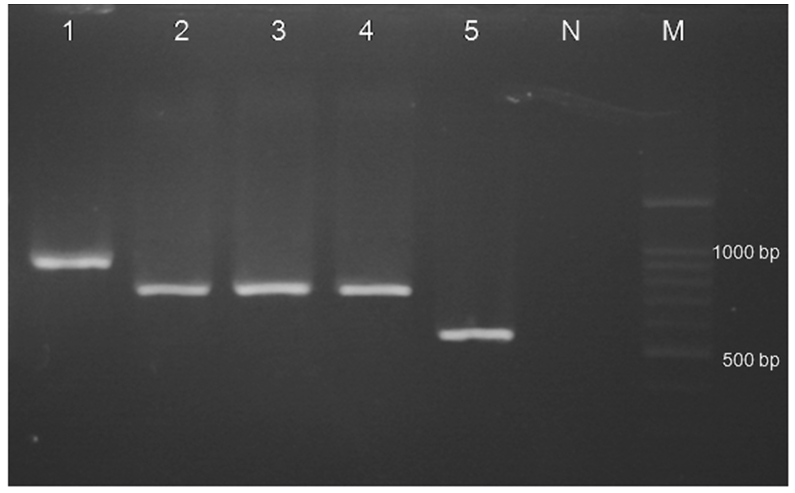
The results of partial amplification of isolates with specific primers of HWP1 gene yielded fragments of ~900 bp, ~700 bp and ~560 bp for C. albicans, C. africana and C. dubliniensis, respectively (Figure 3).
Figure 3.
Agarose gel of PCR amplification with specific primers of HWP1 gene; Lane 1: C. albicans (ATCC10231) ~900 bp; Lanes 2–4: C. africana (~700 bp); Lane 5: positive control of C. dubliniensis (~560 bp) from the archives of the Department of Medical Parasitology and Mycology; N: negative control. M: marker 100 bp.
3.6. Antifungal susceptibility testing
Candida africana isolates were classified as sensitive, dose-dependent sensitive, or resistant to antifungal agents based on their minimum inhibitory concentration (MIC) according to the M27-S3 protocol (Table 1). One isolate of C. africana isolates showed resistance to both clotrimazole (MIC ≥ 1 μg/ml) (Pelletier et al., 2000) and fluconazole (MIC ≥ 64 μg/ml). One isolate showed susceptibity in a dose-dependent manner to itraconazole (0.25–0.5 μg/ml). Clinical C. africana isolates were susceptible to amphotericin B.
Table 1.
In vitro susceptibility profile of three C. africana vaginal isolates using the breakpoints of CLSI M27- S3a.
| Isolates | Antifungal agents | Dosage Range μg/ml |
Number of isolates |
||
|---|---|---|---|---|---|
| Sb | S-DDc | Rd | |||
| C. africana | Fluconazole | 0.25–64 | 2 | - | 1 |
| Clotrimazole | 0.06–16 | 2 | - | 1 | |
| Itraconazole | 0.06–16 | 2 | 1 | - | |
| Amphotericin B | 0.06–2.0 | 3 | - | - | |
CLSI document M27- S3 (2008).
S: sensitive.
S-DD: susceptible dose-dependent.
R: resistant.
4. Discussion
The female genital tract is a common site of pathogenic growth of Candida spp., and VVC is a common infection. C. albicans is reported to be the most common yeast species involved in VVC (Alizadeh et al., 2017; Kord et al., 2017; Khorsand et al., 2015). Species of C. albicans complex include C. albicans, C. dubliniensis, C. africana and C. stellatoidea that are important in vaginitis, with C. albicans the primary agent of Candida infections. C. dubliniensis was identified as the most common cause of oral candidiasis in HIV-infected individuals (Sullivan et al., 2005). Accurate identification of these closely related yeasts is facilitated by the use of both conventional and molecular methods. C. africana (6%) has been reported to be more prevalent than C. dubliniensis (Borman et al., 2013), with worldwide distribution identified from vaginal discharge, candidal balanoposthitis, and blood culture (Odds et al., 2007; Romeo and Criseo, 2011; Romeo et al., 2013; Hu et al., 2015). The present study confirmed C. africana (3%), in the C. albicans complex, as an important agent of VVC. In order to administer effective treatment, the species must be identified, since some show resistance to antifungal drugs. The HWP1 gene amplification is adequate to identify species of this complex (Hu et al., 2015), and sequencing of the ITS region has proven to be a feasible method for the reliable identification of clinically important yeasts. C. dubliniensis and C. stellatoidea were not detected in our study. Other studies of VVC have also not reported C. dubliniensis isolate (Nnadi et al., 2012; Shan et al., 2014; Hu et al., 2015; Ngouana et al., 2015) while some have found it to have higher prevalence than C. africana (Borman et al., 2013; Theill et al., 2016). Romeo et al. (Romeo and Criseo, 2009) studied isolates of C. albicans (338), C. africana (27), and C. dubliniensis (11) from several anatomical sites and found C. albicans (89.9%) to be the most common species, followed by C. africana (7.2%) and C. dubliniensis (2.9%), with C. africana isolated only from vaginal secretions. An investigation from China (Hu et al., 2015) found five C. africana isolates (6.3%) involved in candidal balanoposthitis to be susceptible to fluconazole, itraconazole, voriconazole, posaconazole, caspofungin, flucytosine, micafungin, and amphotericin B. Shan et al. (2014) reported that C. africana isolates (1.5%) from vaginal specimens from China were susceptible to nystatin, fluconazole, itraconazole, miconazole, and clotrimazole, while another study reported five (4.38 %) C. africana VVC isolates to be susceptible to tested antifungal agents (Yazdanparast et al., 2015). Theill et al. (2016) showed low MIC values of nystatin, fluconazole, itraconazole, voriconazole, clotrimazole, and terbinafine to C. africana and C. dubliniensis vaginal isolates. Borman et al. (2013) showed C. africana isolates to be susceptible to tested antifungal agents suitable for VVC treatment. Ngouana et al. (2015) reported a C. africana isolate in vaginal samples from HIV infected patients to be resistant to ketoconazole and exhibit reduced susceptibility to amphotericin B. Majdabadi et al., isolated two C. africana of vaginal candidiasis in which one species was resistant to fluconazole and itraconazole, the other one to itraconazole (Majdabadi et al., 2018). We found one C. africana vaginal isolate resistant to both clotrimazole and fluconazole, and another one showed dose-dependent susceptibility to itraconazole.
5. Conclusion
Our finding of C. africana as a rare agent of vaginitis resistant to both clotrimazole and fluconazole with reduced susceptibility to itraconazole is important in VVC treatment. Identification of C. africana as an uncommon agent of VVC is critical, since this yeast showed a range of susceptibility to antifungal agents.
Declarations
Author contribution statement
S. Farahyar: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
S. Izadi: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
M. Falahati: Conceived and designed the experiments.
E. Razmjou: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Z. Ghahri-Mobaser: Performed the experiments; Contributed reagents, materials, analysis tools or data.
M. Ashrafi-Khozani, M. Roudbary, M. Rahimi: Performed the experiments.
S. Ansari: Contributed reagents, materials, analysis tools or data.
A. Fattahi: Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by Iran University of Medical Sciences, Tehran, Iran (grant no. 27842).
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at GenBank under the accession numbers MG757669, MG757670, and MG757671.
References
- Ahmad S., Khan Z., Asadzadeh M., Theyyathel A., Chandy R. Performance comparison of phenotypic and molecular methods for detection and differentiation of Candida albicans and Candida dubliniensis. BMC Infect. Dis. 2012;12:230. doi: 10.1186/1471-2334-12-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh M., Kolecka A., Boekhout T., Zarrinfar H., Ghanbari Nahzag M.A., Badiee P., Rezaei-Matehkolaei A., Fata A., Dolatabadi S., Najafzadeh M.J. Identification of Candida species isolated from vulvovaginitis using matrix assisted laser desorption ionization-time of flight mass spectrometry. Curr. Med. Mycol. 2017;3(4):21–25. doi: 10.29252/cmm.3.4.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Vargas R., Elorduy L., Eraso E., Francisco Cano J., Guarro J., Pontón J., Quindós G. Isolation of Candida africana, probable atypical strains of Candida albicans, from a patient with vaginitis. Med. Mycol. 2008;46(2):167–170. doi: 10.1080/13693780701633101. [DOI] [PubMed] [Google Scholar]
- Borman A.M., Szekely A., Linton C.J., Palmer M.D., Brown P., Johnson E.M. Epidemiology, antifungal susceptibility, and pathogenicity of Candida africana isolates from the United Kingdom. J. Clin. Microbiol. 2013;51(3):967–972. doi: 10.1128/JCM.02816-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciardo D., Schär G., Böttger E., Altwegg M., Bosshard P. Internal transcribed spacer sequencing versus biochemical profiling for identification of medically important yeasts. J. Clin. Microbiol. 2006;44(1):77–84. doi: 10.1128/JCM.44.1.77-84.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) Clinical and Laboratory Standards Institute; Wayne, Pennsylvania: 2008. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Third Informational Supplement. CLSI Document M27-S3. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) Clinical and Laboratory Standards Institute; Wayne, Pennsylvania: 2012. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Fourth Informational Supplement. CLSI Document M27-S4. [Google Scholar]
- Dieng Y., Sow D., Ndiaye M., Guichet E., Faye B., Tine R., Lo A., Sylla K., Ndiaye M., Abiola A., Dieng T., Ndiaye J.L., Le Pape P., Gaye O. Identification of three Candida africana strains in Senegal. J. Mycol. Med. 2012;22(4):335–340. doi: 10.1016/j.mycmed.2012.07.052. [DOI] [PubMed] [Google Scholar]
- Fornari G., Vicente V.A., Gomes R.R., Muro M.D., Pinheiro R.L., Ferrari C., Herkert P.F., Takimura M., Carvalho N.S., Queiroz-Telles F. Susceptibility and molecular characterization of Candida species from patients with vulvovaginitis. Braz. J. Microbiol. 2016;47(2):373–380. doi: 10.1016/j.bjm.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S.I., Senda Y., Nakaguchi S., Hashimoto T. Multiplex PCR using internal transcribed spacer 1 and 2 regions for rapid detection and identification of yeast strains. J. Clin. Microbiol. 2001;39(10):3617–3622. doi: 10.1128/JCM.39.10.3617-3622.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves B., Ferreira C., Alves C.T., Henriques M., Azeredo J., Silva S. Vulvovaginal candidiasis: epidemiology, microbiology and risk factors. Crit. Rev. Microbiol. 2016;42(6):905–927. doi: 10.3109/1040841X.2015.1091805. [DOI] [PubMed] [Google Scholar]
- Hedayati M.T., Taheri Z., Galinimoghadam T., Aghili S.R., Yazdani Cherati J., Mosayebi E. Isolation of different species of Candida in patients with vulvovaginal candidiasis from Sari, Iran. Jundishapur J. Microbiol. 2015;8(4) doi: 10.5812/jjm.8(4)2015.15992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Yu A., Chen X., Wang G., Feng X. Molecular characterization of Candida africana in genital specimens in Shanghai, China. BioMed Res. Int. 2015;2015:185387. doi: 10.1155/2015/185387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorsand I., Ghanbari Nehzag M.A., Zarrinfar H., Fata A., Naseri A., Badiee P., Najafzadeh M.J. Frequency of variety of Candida species in women with Candida vaginitis referred to clinical centers of Mashhad, Iran. Iran. J. Obstet. Gynaecol. Infertil. 2015;18(168):15–22. [Google Scholar]
- Kord Z., Fata A., Zarrinfar H. Molecular Identification of Candida species isolated from patients with vulvovaginitis for the first time in Mashhad. Iran. J. Obstet. Gynaecol. Infertil. 2017;20(4):50–57. [Google Scholar]
- Majdabadi N., Falahati M., Heidarie-Kohan F., Farahyar S., Rahimi-Moghaddam P., Ashrafi-Khozani M., Razavi T., Mohammadnejad S. Effect of 2- phenylethanol as antifungal agent and common antifungals (amphotericin B, fluconazole, and itraconazole) on Candida species isolated from chronic and recurrent cases of candidal vulvovaginitis. Assay Drug Dev. Technol. 2018;16(3):141–149. doi: 10.1089/adt.2017.837. [DOI] [PubMed] [Google Scholar]
- Merseguel K.B., Nishikaku A.S., Rodrigues A.M., Padovan A.C., e Ferreira R.C., de Azevedo Melo A.S., Briones M.R., Colombo A.L. Genetic diversity of medically important and emerging Candida species causing invasive infection. BMC Infect. Dis. 2015;15:57. doi: 10.1186/s12879-015-0793-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukasa K.J., Herbert I., Daniel A., Sserunkuma K.L., Joel B., Frederick B. Antifungal susceptibility patterns of vulvovaginal Candida species among women attending Antenatal clinic at Mbarara Regional Referral Hospital. South West. Uganda. 2015;5(4):322–331. doi: 10.9734/BMRJ/2015/13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngouana T.K., Krasteva D., Drakulovski P., Toghueo R.K., Kouanfack C., Ambe A., Reynes J., Delaporte E., Boyom F.F., Mallie M. Investigation of minor species Candida africana, Candida stellatoidea and Candida dubliniensis in the Candida albicans complex among Yaoundé (Cameroon) HIV-infected patients. Mycoses. 2015;58(1):33–39. doi: 10.1111/myc.12266. [DOI] [PubMed] [Google Scholar]
- Nnadi N.E., Ayanbimpe G.M., Scordino F., Okolo M.O., Enweani I.B., Criseo G., Romeo O. Isolation and molecular characterization of Candida africana from Jos, Nigeria. Med. Mycol. 2012;50(7):765–767. doi: 10.3109/13693786.2012.662598. [DOI] [PubMed] [Google Scholar]
- Odds F.C., Bougnoux M.E., Shaw D.J., Bain J.M., Davidson A.D., Diogo D., Jacobsen M.D., Lecomte M., Li S.-Y., Tavanti A. Molecular phylogenetics of Candida albicans. Eukaryot. Cell. 2007;6(6):1041–1052. doi: 10.1128/EC.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier R., Peter J., Antin C., Gonzalez C., Wood L., Walsh T.J. Emergence of resistance of Candida albicans to clotrimazole in human immunodeficiency virus- infected children: in vitro and clinical correlations. J. Clin. Microbiol. 2000;38(4):1563–1568. doi: 10.1128/jcm.38.4.1563-1568.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Leguizamon G., Fiori A., Lopez L.F., Gomez B.L., Parra-Giraldo C.M., Gomez-Lopez A., Suarez C.F., Ceballos A., Van Dijck P., Patarroyo M.A. Characterising atypical Candida albicans clinical isolates from six third-level hospitals in Bogota, Colombia. BMC Microbiol. 2015;15:199. doi: 10.1186/s12866-015-0535-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo O., Criseo G. First molecular method for discriminating between Candida africana, Candida albicans, and Candida dubliniensis by using hwp1 gene. Diagn. Microbiol. Infect. Dis. 2008;62(2):230–233. doi: 10.1016/j.diagmicrobio.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Romeo O., Criseo G. Molecular epidemiology of Candida albicans and its closely related yeasts Candida dubliniensis and Candida africana. J. Clin. Microbiol. 2009;47(1):212–214. doi: 10.1128/JCM.01540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo O., Criseo G. Candida africana and its closest relatives. Mycoses. 2011;54(6):475–486. doi: 10.1111/j.1439-0507.2010.01939.x. [DOI] [PubMed] [Google Scholar]
- Romeo O., Tietz H.-J., Criseo G. Candida africana: is it a fungal pathogen? Curr. Fungal Infect. Rep. 2013;7(3):192–197. [Google Scholar]
- Shan Y., Fan S., Liu X., Li J. Prevalence of Candida albicans-closely related yeasts, Candida africana and Candida dubliniensis, in vulvovaginal candidiasis. Med. Mycol. 2014;52(6):636–640. doi: 10.1093/mmy/myu003. [DOI] [PubMed] [Google Scholar]
- Sullivan D.J., Moran G.P., Coleman D.C. Candida dubliniensis: ten years on. FEMS Microbiol. Lett. 2005;253(1):9–17. doi: 10.1016/j.femsle.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Theill L., Dudiuk C., Morano S., Gamarra S., Nardin M.E., Méndez E., Garcia-Effron G. Prevalence and antifungal susceptibility of Candida albicans and its related species Candida dubliniensis and Candida africana isolated from vulvovaginal samples in a hospital of Argentina. Rev. Argent. Microbiol. 2016;48(1):43–49. doi: 10.1016/j.ram.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Tietz H.J., Hopp M., Schmalreck A., Sterry W., Czaika V. Candida africana sp. nov., a new human pathogen or a variant of Candida albicans? Mycoses. 2001;44(11-12):437–445. doi: 10.1046/j.1439-0507.2001.00707.x. [DOI] [PubMed] [Google Scholar]
- Yazdanparast S.A., Khodavaisy S., Fakhim H., Shokohi T., Haghani I., Nabili M., Gholami H., Ahmadi I., Badali H. Molecular characterization of highly susceptible Candida africana from vulvovaginal candidiasis. Mycopathologia. 2015;180(5-6):317–323. doi: 10.1007/s11046-015-9924-z. [DOI] [PubMed] [Google Scholar]
- Zarrinfar H., Kaboli S., Dolatabadi S., Mohammadi R. Rapid detection of Candida species in bronchoalveolar lavage fluid from patients with pulmonary symptoms. Braz. J. Microbiol. 2016;47(1):172–176. doi: 10.1016/j.bjm.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]