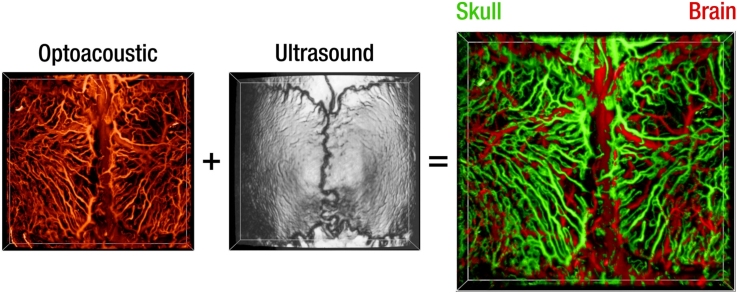
Graphical abstract
Keywords: Optoacoustic microscopy, Photoacoustic microscopy, Ultrasound biomicroscopy, Murine calvaria, Bone vasculature, Vasculature segmentation
Abstract
Bone microvasculature plays a paramount role in bone marrow maintenance, development, and hematopoiesis. Studies of calvarian vascular patterns within living mammalian skull with the available intravital microscopy techniques are limited to small scale observations. We developed an optical-resolution optoacoustic microscopy method combined with ultrasound biomicroscopy in order to reveal and discern the intricate networks of calvarian and cerebral vasculature over large fields of view covering majority of the murine calvaria. The vasculature segmentation method is based on an angle-corrected homogeneous model of the rodent skull, generated using simultaneously acquired three-dimensional pulse-echo ultrasound images. The hybrid microscopy design along with the appropriate skull segmentation method enable high throughput studies of a living bone while facilitating correct anatomical interpretation of the vasculature images acquired with optical resolution optoacoustic microscopy.
1. Introduction
Bone microvasculature links the bone marrow, which is responsible for producing all blood cells and shown to influence tumor growth [1,2], with the rest of the body. The relatively thin skullcap offers a unique window into fully functional bone [3] and is partially accessible for microscopic imaging techniques after scalp removal [4]. Current imaging tools reveal subtleties of the intricate vascular anatomy, bone marrow [[4], [5], [6], [7]], and meningeal lymphatic vessels [8,9]. Yet, intravital imaging with high spatial resolution over large fields of view remains challenging due to the physical limitations imposed by the heterogeneous bone structure, its curvature, optical scattering and absorption [2,[5], [6], [7]].
Skull vessels and bone-marrow compartments can be visualized at high resolution by means of histological techniques applied on thin ex vivo slices [10,11]. Intravital microscopy enables in vivo observation of bone-marrow microvasculature using a minimally-invasive procedure [7]. Also, confocal and two-photon microscopy have been used to examine the hematopoietic stem cell niche [3], flow dynamics [5], and endothelial microdomains for tumor engraftment in the bone marrow microvasculature [4]. Nonetheless, purely optical approaches suffer from small field-of-view and shallow penetration depth, preventing studies across large-scale vascular network over the whole calvaria.
Optoacoustic microscopy (OAM) is a useful tool to image morphology and function of murine cerebral vasculature [12,13] as it does not require exogenous contrast agents, due to the high optical absorption of hemoglobin in the visible spectral range. Previous OAM research focused on studying cerebral vasculature by using juvenile (4 week old) mice where the skull tends to be significantly less vascularized, hence providing a clear view of the underlying cerebral vasculature. In these previous studies, both the skull and its vasculature were therefore largely ignored [12] or viewed as an undesired obstruction [14]. OAM images may often lead to misinterpretation of the vasculature function [15] due to the lack of macroscopic anatomical reference and acoustic distortions introduced as the ultrasound (US) waves traverse the skull bone [16,17,14]. Previous attempts to observe skull vasculature using OAM and discerning it from brain vasculature required the use of multiple wavelengths and contrast agents [18], which imposed significant complexity on the image acquisition. Ning et al. recently presented a method utilizing US guided OAM, where the US image of the skull was used to dynamically focus the OA excitation into cerebral vasculature [19]. However, the study focused solely on cerebral vasculature by using juvenile mice. Virtual craniotomy correction can potentially be applied when the additional information regarding the skull’s geometry and its elastic constants can be extracted from co-registered pulse-echo US images [14]. The latter method yet requires development of an accurate and computationally-extensive wave propagation model to handle thermoelastic sources inside the bone.
Here we present a simple approach to segment the murine skull vasculature in an OAM image enabling high-throughouput studies of the skull vasculature over large field of view with high spatial resolution.
2. Methods
2.1. Experimental procedure
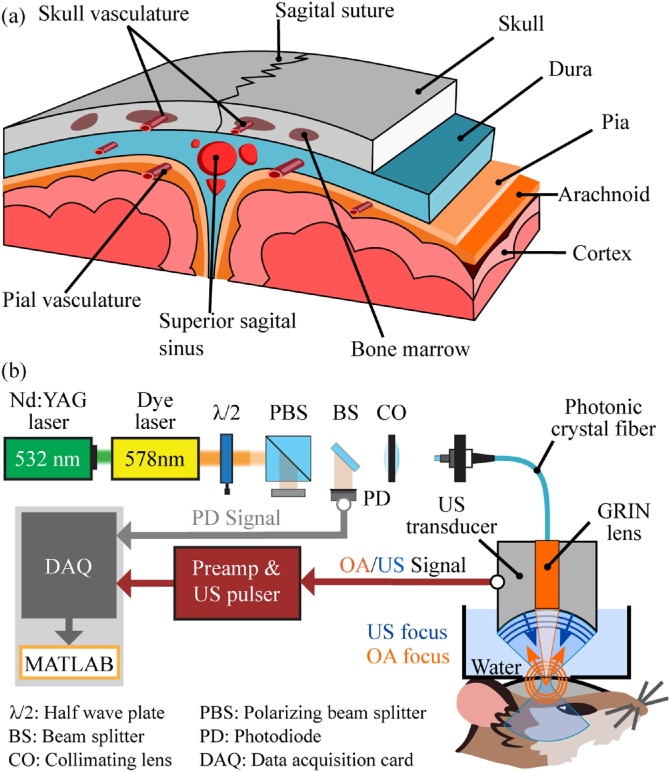
To reveal the intricate anatomy of the murine calvarian and cortical microvasculature, skull and brain (Fig. 1(a)), a hybrid optoacoustic-ultrasound biomicroscope has been employed (Fig. 1(b)) [20,13]. The pulse-echo US modality provides an accurate image of the skull bone, due to the strong mismatch in the elastic properties of water or soft tissue and bone. Skull and brain vasculature are subsequently imaged by focusing nanosecond laser pulses (578 nm) with a custom-designed gradient index (GRIN) lens and detecting the generated OA responses by the same spherically focused PVDF transducer (30 MHz central frequency, 100 % bandwidth, Precision Acoustics, UK) used for the US pulse-echo imaging. Thanks to the low numerical aperture (NA = 0.025) of the GRIN lens, a single two-dimensional scan (5.5 mm × 6 mm) provides three-dimensional imaging of large curved areas covering parietal and parts of the frontal and interparietal bones (see Fig. 2) with a lateral resolution better than 20 μm without the need to refocus the optical beam [13].
Fig. 1.
(a) Illustration of the vascular anatomy of the mouse head. (b) Schematic of the data acquisition with the hybrid optoacoustic-ultrasound biomicroscope.
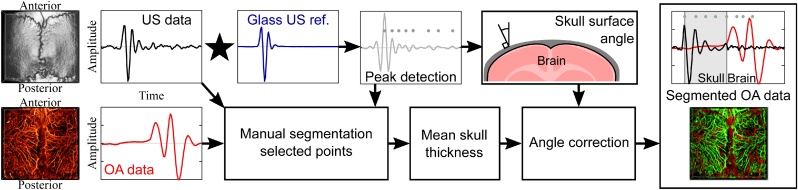
Fig. 2.
Flow chart of the segmentation method. First, the pulse-echo US data is cross-correlated with a water glass interface reference measurement for detecting the positive peaks. A manual segmentation procedure, performed by visual inspection of US and OA waveforms in a few selected locations in the image, yields a mean skull thickness. The effective skull thickness is calculated as a function of skull angle and mean skull thickness for the whole volume and subsequently used to segment the OA data.
In this study, three-dimensional OA and US images were acquired from 14 weeks old female C57Bl/6 mice (Charles River, United States) whose scalp was removed. All animal experiments were performed in full compliance with the institutional guidelines of the Helmholtz Center Munich and with approval from the Government District of Upper Bavaria.
2.2. Segmentation method
To analyze the relatively large number of vessels found in the OA images of the mouse head, we developed a vessel segmentation method using the complementary information of the OA and US datasets (Fig. 2). Due to the strong mismatch in elastic properties between bone and soft tissue, a high amplitude echo first arrives from the skull’s outer surface followed by backscattered signals of smaller amplitude produced by vessels, bone marrow compartments, inner skull surface, and reverberations thereof (Fig. 2, US data). Shape and amplitude of the reflections do not solely depend on the skull structure and its elastic constants, but also on the transducer geometry and its position relative to the skull. Therefore, a reference is needed to disentangle the contribution of the transducer from the skull’s response. For this, we used an echo recorded from a flat water/glass interface measured for different depths in steps of 10 μm (Glass US ref. in Fig. 2).
For a single pixel in the scanning plane, the glass reference with the same (or closest) depth as the skull is selected to perform cross-correlation. The maximum of the cross-correlation function is used to determine the skull's outer surface depth while the remaining positive peaks and their amplitudes are stored. Repeating this process for all the scanned points yields the skull’s outer surface and subsurface reverberations. We further assumed that one of the cross-correlation peaks corresponds to the inner skull surface, which decreases the size of the problem and poses it as a classification problem. However, using solely the US data to find the inner skull surface is not trivial since peaks corresponding to vascular structures can mistakenly be classified as originating from the skull surface. Therefore, the OA waveform completes the puzzle by showing whether a peak exists close to the skull’s outer surface corresponding to a vessel. This information is used to manually segment selected points of the image and calculate the mean skull thickness. Due to the skull’s curvature, the effective thickness seen from the top increases together with the skull’s slope, i.e. towards the side of the parietal bones. Thus, the effective thickness has to be calculated before projecting the mean thickness to the entire imaged volume.
3. Results
The large depth of field of the focused excitation laser beam (< 20 μm beam waist over ∼2 mm) [13,20] spares the depth scanning along the z direction, thus allows for the 3D image acquisition from 6 × 6 × 2 mm3 FOV in less than 90 s.
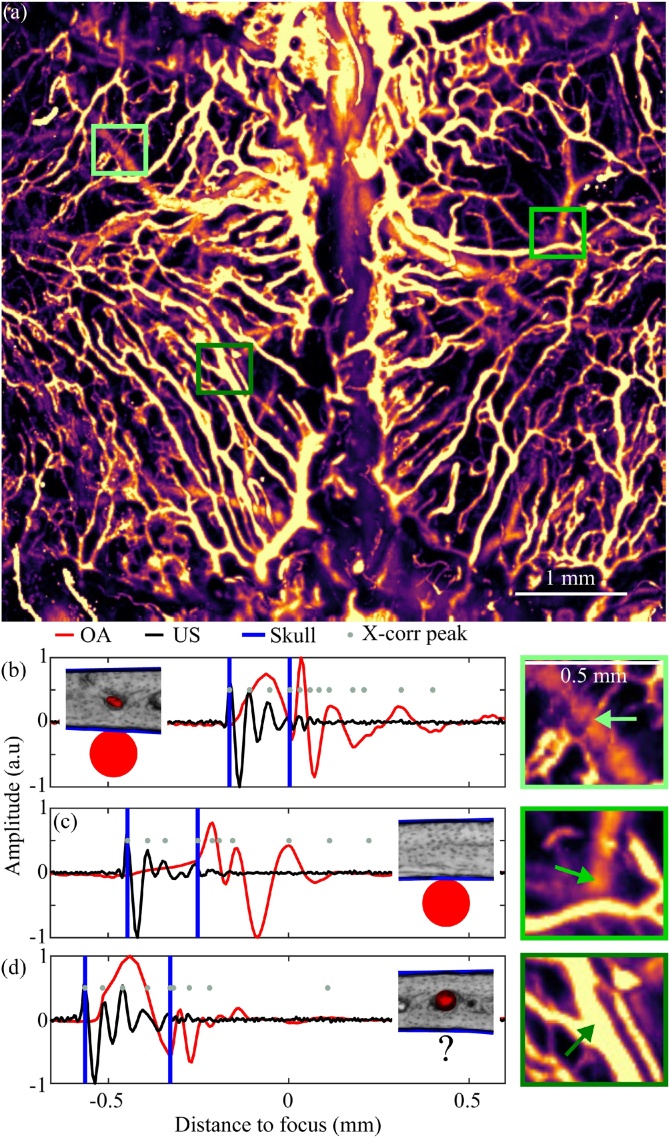
A maximum amplitude projection of the OAM image across left and right parietal bones (Fig. 3(a)) is used for the manual selection of a few points to visually inspect OA and US waveforms. Three distinct cases can be observed in the images: (1) calvarian and cerebral vasculature are detectable (Fig. 3(b)); (2) only cerebral vasculature is detectable (Fig. 3(c)); (3) only skull vasculature is detectable (Fig. 3(d)). Selection of 4 more points to increase the robustness of the mean thickness estimation yields a mean thickness of 207 μm (min. 164 μm, max. 253 μm), which is then fed into the skull vasculature segmentation step. Fig. 4 demonstrates the final result of the proposed segmentation algorithm, where the differences in the segmentation in comparison to the raw OA images from Fig. 3 can be best appreciated in the selected zoomed regions (Fig. 4(b)–(d)).
Fig. 3.
Example of the manual skull segmentation at selected locations. (a) Maximum amplitude projection of the raw OA data. Rectangles indicate the position of the individual points. The individual OA and US waveforms are shown for points where (b) calvarian and cerebral vasculature are detected, (c) the vasculature is underneath the skull bone, and (d) calvarian vasculature is detected.
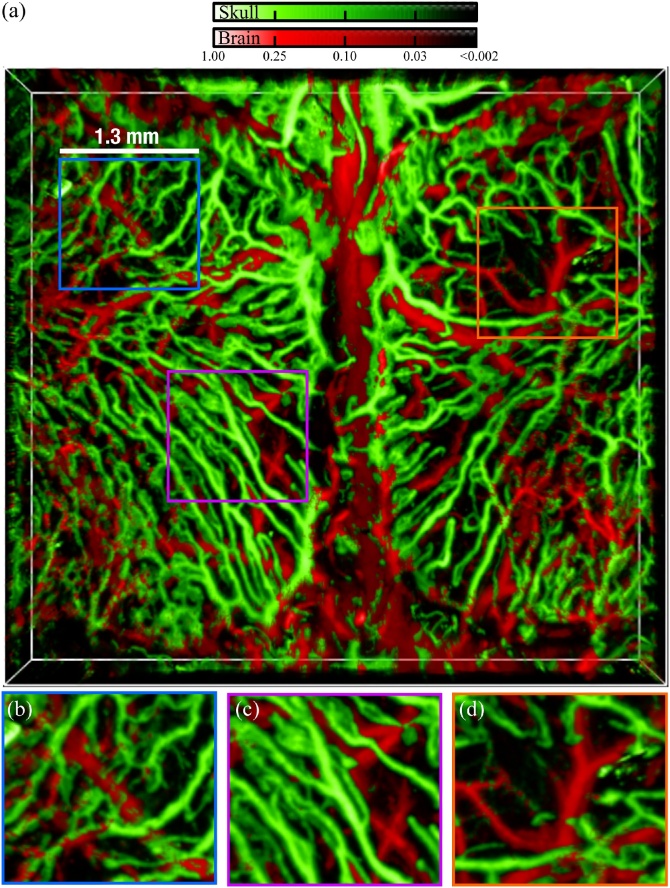
Fig. 4.
(a) Three-dimensional maximum amplitude projection of the segmented vasculature. Augmented portions of the images are shown in (b)–(d).
The fine skull capillaries (Fig. 3, Fig. 4(b), green coloured) appear broken after the segmentation, which is attributed to the stronger signal amplitude generated by the cortical vasculature (waveform in Fig. 3(b)). Two overlapping vessels in Fig. 4(c) are located below the skull, while the skull vasculature exhibits characteristic loops surrounded by what seems to be the bone marrow compartments – greater in size than the typical vessel diameter but generate weaker OA signals as compared to the calvarian vasculature. A region lacking calvarian vasculature has further been identified (Fig. 4(d)) at a point where the right parietal bone is located equidistant to the sagital and right coronal suture. It is very likely that bone angiogenesis [10,11] is taking place and additional calvarian vessels could be observed beyond the 14 weeks of age.
4. Discussion
The proposed method showed an accuracy of 92 % when compared against manual segmentation. Pial vessels were mislabeled in 6 % of cases, whereas skull vessels were confounded only by 2 %.
Some segmentation inaccuracies can be observed in locations where the calvarian vasculature grows under the skull (Fig. 4(a), bottom right, center left). Presumably, the homogeneous skull thickness assumption does not hold everywhere, and some skull regions might present thinner or thicker bone, which calls for development of more sophisticated segmentation algorithms based on locally homogeneous thickness.
Another source of inaccuracies close to the image margins can be attributed to the invalidity of the cross-correlation hypothesis. The angle of the skull surface with respect to the transducer axis may deviate significantly from the reference water/glass measurements that were performed at normal incidence. One possible solution could be the extension of the reference measurements to include angles other than normal incidence. However, departing from the normal incidence approximation would imply a wave propagation regime where glass may not accurately mimic acoustic propagation in the bone. Another option would be extending the wave model developed in [14] to handle sources inside the solid and use a model-based approach to combine multiple sources located at different depths (Fig. 3(b)). However, modelling an independent ultrasound wave propagation problem for each point in the scanning plane may consume unreasonable computational resources.
At certain incidence angles, the outer skull surface might no longer generate the maximum reflection peak, thus its accurate detection in those regions (mainly leftmost parietal) would rely on shallower cross-correlation peaks higher than 50 % of the maximum.
The uniqueness of our approach consists in using a low numerical aperture beam while retrieving spatially resolved information along the axial dimension via the time-resolved optoacoustic waveforms. Note that this option does not exist in other optical microscopy techniques (e.g. confocal microscopy) where an attempt to overcome the skull’s curvature and provide a similarly large field of view by using low numerical aperture optics would readily result in a loss of the axial resolving capacity [22,23]. On the other hand, camera-based planar fluorescence optical imaging can provide comparable or even larger field of view, again at the expense of the lack of depth resolution and additional image blurring due to the photons being scattered from the deep brain structures [24,25]. Most optical methods would require a contrast agent or fluorescent labels, whereas our method is label free. In addition, high resolution intravital optical microscopy methods require significantly longer scanning times in the range of 30−40 min in order to cover the mouse skull e.g. by employing 3D mosaics approaches [26]. It has also been reported that mosaic images are prone to artifacts [4] due to variations in tissue optical properties.
Manual selection of a few points ensures a physics-based choice of the skull thickness instead of a trial and error approach solely based on visual feedback. Since physical modeling of the focused transducer-skull system at millions scanning positions is impractical, we restated the problem as a classification one, amenable for machine learning techniques to be implemented in the future work.
Beyond the vessel segmentation, the US modality can be used to obtain additional information on the skull anatomy and correlate its subsurface porosity to the vessel density. Our segmentation method could also be used in conjunction with multiple illumination wavelengths to determine the oxygen saturation inside the bone [13]. The versatility of our technique could also be aided by development of dedicated optoacoustic labels targeting specific proteins or cell types similar to those in use by fluorescent imaging techniques.
5. Conclusions
Hybrid optoacoustic-ultrasound microscopy is suitable for large-scale imaging of calvarian and cerebral microvasculature in living rodents using a minimally-invasive procedure. Our method allows the quantitative analysis of vascular networks and can be used in high-throughput studies of the calvarian and cerebral vasculature. Although the method relies on manual input, the classification is based on basic physical principles that could potentially be generalised and allow for a fully automated segmentation. Our work thus holds promise for extending the use of optical resolution optoacoustic microscopy beyond very young mouse models and facilitate unambiguous differentiation between the calvarian and cortical vasculature.
Author statement
J.R. conceived the study. H.E. performed the experiments. J.R. preprocessed the experimental data. H.E. and U.H. developed the algorithm. H.E. wrote the algorithm, performed the calculations, and wrote the original draft. D.R. supervised the study. All authors wrote and edited the manuscript.
Funding
This project has received funding from the European Research Council under grant agreement ERC-2015-CoG-682379 (D.R).
CRediT authorship contribution statement
Héctor Estrada: Investigation, Methodology, Software, Visualization, Writing - original draft, Writing - review & editing. Johannes Rebling: Conceptualization, Data curation, Writing - review & editing. Urs Hofmann: Methodology, Writing - review & editing. Daniel Razansky: Supervision, Funding acquisition, Writing - review & editing.
Acknowledgements
We gratefully acknowledge Sven Gottschalk for his work with the animal models and animal protocols. We gratefully acknowledge the valuable help of Wolfgang Sievert, Michael Reiss, Daniela Hladik, and Soile Tapio with the mouse experiments.
Biographies

Héctor Estrada obtained his Ph.D. on physical acoustics from the Universidad Politecnica de Valencia, Spain, in 2011 studying the ultrasound propagation through phononic plates. In 2012 he joined the Institute for Biological and Medical Imaging (IBMI), Helmholtz Zentrum München as a postdoctoral fellow developing hybrid optoacoustic-ultrasound neuroimaging system and studying the acoustic properties of the skull using lasers. He currently works at the University and ETH Zurich developing models and techniques for transcranial ultrasound and optoacoustics. His research interests include optoacoustic and ultrasound imaging as well as guided ultrasonic wave propagation in bone.

Johannes Rebling obtained his master's degree in Optics and Photonics with distinction in 2014, participating in the international Erasmus Mundus program during which he studied at the Karlsruhe Institute of Technology, Karlsruhe, Germany, the Aix Marseille University, Marseille, France, and the Universitat Politecnica de Catalunya, Barcelona, Spain. He joined the Institute for Biological and Medical Imaging (IBMI) at the Helmholtz Zentrum München, Germany for his master's thesis. He finished his Ph.D. under the supervision of Prof. Daniel Razansky early in 2019 and is now focusing his research on the development of high-speed optoacoustic microscopy techniques at the ETH and University Zurich, Switzerland.

Urs A. T. Hofmann received the M.Sc. degree in biomedical engineering from ETH Zurich, Switzerland in 2017. He is currently working towards a Ph.D. degree at ETH Zurich, Switzerland. He worked in different research groups at ETH Zurich and Helmholtz Center Munich both during student projects and as a research assistant. His research focuses on the mechatronic design of imaging systems and their application in the field of biomedical engineering.

Daniel Razansky is Full Professor of Biomedical Imaging at the Faculty of Medicine, University of Zurich and Department of Information Technologies and Electrical Engineering, ETH Zurich. He earned PhD in Biomedical Engineering and MSc in Electrical Engineering from the Technion – Israel Institute of Technology and completed postdoctoral training in bio-optics at the Harvard Medical School. From 2007 until 2018 he was the Director of Multi-Scale Functional and Molecular Imaging Lab and Professor of Molecular Imaging Engineering at the Helmholtz Center and Technical University of Munich. Razansky’s Lab pioneered a number of imaging technologies successfully commercialized worldwide, among them the multi-spectral optoacoustic tomography and hybrid optoacoustic ultrasound imaging. He has authored over 200 peer-review journal articles and holds 15 patented inventions in bio-imaging and sensing. He is a co-founding Editor of the Photoacoustics journal and serves on Editorial Boards of a number of journals published by Nature Publishing Group, Elsevier, IEEE and AAPM. He is also an elected Fellow of the OSA and SPIE.
Contributor Information
Héctor Estrada, Email: hector.estrada@posteo.org.
Daniel Razansky, Email: daniel.razansky@uzh.ch.
References
- 1.Engblom C. Osteoblasts remotely supply lung tumors with cancer-promoting SiglecFhigh neutrophils. Science. 2017;358 doi: 10.1126/science.aal5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao H. A. Leukaemia hijacks a neural mechanism to invade the central nervous system. Nature. 2018;560:55–60. doi: 10.1038/s41586-018-0342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lassailly F., Foster K., Lopez-Onieva L., Currie E., Bonnet D. Multimodal imaging reveals structural and functional heterogeneity in different bone marrow compartments: functional implications on hematopoietic stem cells. Blood. 2013;122:1730–1740. doi: 10.1182/blood-2012-11-467498. [DOI] [PubMed] [Google Scholar]
- 4.Sipkins D.A. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005;435:969–973. doi: 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bixel M.G. Flow dynamics and HSPC homing in bone marrow microvessels. Cell Rep. 2017;18:1804–1816. doi: 10.1016/j.celrep.2017.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuya M. Direct cell-cell contact between mature osteoblasts and osteoclasts dynamically controls their functions in vivo. Nat. Commun. 2018;9:300. doi: 10.1038/s41467-017-02541-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spencer J.A. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature. 2014;508:269. doi: 10.1038/nature13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aspelund A. A dural lymphatic vascular system that drains brain interstitial fluid and macromolecules. J. Exp. Med. 2015;212:991–999. doi: 10.1084/jem.20142290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antila S. Development and plasticity of meningeal lymphatic vessels. J. Exp. Med. 2017;214:3645–3667. doi: 10.1084/jem.20170391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sivaraj K.K., Adams R.H. Blood vessel formation and function in bone. Development. 2016;143:2706–2715. doi: 10.1242/dev.136861. [DOI] [PubMed] [Google Scholar]
- 11.Kusumbe A.P., Ramasamy S.K., Adams R.H. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature. 2014;507:323–328. doi: 10.1038/nature13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao J. High-speed label-free functional photoacoustic microscopy of mouse brain in action. Nat. Methods. 2015;12:407–410. doi: 10.1038/nmeth.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rebling J. Dual‐wavelength hybrid optoacoustic‐ultrasound biomicroscopy for functional imaging of large‐scale cerebral vascular networks. J. Biophotonics. 2018 doi: 10.1002/jbio.201800057. [DOI] [PubMed] [Google Scholar]
- 14.Estrada H. Virtual craniotomy for high-resolution optoacoustic brain microscopy. Sci. Rep. 2018;8:1459. doi: 10.1038/s41598-017-18857-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan B. High-speed widefield photoacoustic microscopy of small-animal hemodynamics. Biomed. Opt. Express. 2018;9:4689–4701. doi: 10.1364/BOE.9.004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kneipp M., Turner J., Estrada H., Rebling J., Shoham S., Razansky D. Effects of the murine skull in optoacoustic brain microscopy. J. Biophotonics. 2016;9:117–123. doi: 10.1002/jbio.201400152. [DOI] [PubMed] [Google Scholar]
- 17.Estrada H., Rebling J., Turner J., Razansky D. Broadband acoustic properties of a murine skull. Phys. Med. Biol. 2016;61:1932. doi: 10.1088/0031-9155/61/5/1932. [DOI] [PubMed] [Google Scholar]
- 18.Hu S., Maslov K., Wang L.V. Second-generation optical-resolution photoacoustic microscopy with improved sensitivity and speed. Opt. Lett. 2011;36:1134–1136. doi: 10.1364/OL.36.001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ning B. Ultrasound-aided multi-parametric photoacoustic microscopy of the mouse brain. Sci. Rep. 2015;5:18775. doi: 10.1038/srep18775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estrada H. Laser Phys. Lett. 2014;11 doi: 10.1088/1612-2011/11/11/115605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White J.G., Amos W.B., Fordham M. An evaluation of confocal versus conventional imaging of biological structures by fluorescence light microscopy. J. Cell Biol. 1987;105:41–48. doi: 10.1083/jcb.105.1.41. Rockefeller University Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng G., Horstmeyer R., Yang C. Wide-field, high-resolution Fourier ptychographic microscopy. Nat. Photonics. 2013;7:739. doi: 10.1038/nphoton.2013.187. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McConnell G., Trägårdh J., Amor R., Dempster J., Reid E., Amos W.B., Bronner M.E. A novel optical microscope for imaging large embryos and tissue volumes with sub-cellular resolution throughout. eLife. 2016;5 doi: 10.7554/eLife.18659. eLife Sciences Publications, Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalchenko V., Israeli D., Kuznetsov Y., Harmelin A. Transcranial optical vascular imaging (TOVI) of cortical hemodynamics in mouse brain. Sci. Rep. 2014;4:5839. doi: 10.1038/srep05839. The Author(s) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahn S., Choe K., Lee S., Kim K., Song E., Seo H., Kim I., Kim P. Intravital longitudinal wide-area imaging of dynamic bone marrow engraftment and multilineage differentiation through nuclear-cytoplasmic labeling. PLoS One. 2017;12(11):1–16. doi: 10.1371/journal.pone.0187660. [DOI] [PMC free article] [PubMed] [Google Scholar]