Abstract
The N-terminal residue influences protein stability through N-end rule pathways. Here, through stability profiling of the human N-terminome, we uncover multiple new features of the N-end rule. In addition to uncovering new specificities of UBR E3 ligases, we characterize two related Cullin-RING E3 ligase complexes, Cul2ZYG11B and CUL2ZER1, that act redundantly to target N-terminal glycine. N-terminal glycine degrons are depleted at native N-termini but strongly enriched at caspase cleavage sites, suggesting roles for the substrate adaptors ZYG11B and ZER1 in protein degradation during apoptosis. Furthermore, ZYG11B and ZER1 participate in the quality control of N-myristoylated proteins, wherein N-terminal glycine degrons are conditionally exposed following a failure of N-myristoylation. Thus, an additional N-end rule pathway specific for glycine regulates the stability of metazoan proteomes.
The ubiquitin-proteasome system (UPS) is the major route through which eukaryotic cells achieve selective protein degradation (1). The specificity of this system is provided by E3 ubiquitin ligases, of which more than 600 are encoded in the human genome. E3 ligases recognize specific sequence elements, known as degrons, that are present in substrate proteins (2). However, whilst a detailed knowledge of the specificity of E3 ligases for degrons will be essential for achieving a systems-level understanding of the UPS, our current knowledge of degron motifs remains remarkably sparse (3).
The first degrons to be discovered were located at the N-terminus of proteins (4). N-terminal degrons are targeted by N-end rule pathways, of which there are two main branches: the Arg/N-end rule pathway, through which UBR-family E3 ligases target N-termini typically generated through endoproteolytic cleavage (5, 6), and the Ac/N-end rule pathway, through which proteins bearing acetylated N-termini are targeted for degradation by the E3 ligase MARCH6 (also known as TEB4) (7, 8). In addition, a Pro/N-end rule pathway was recently described, through which proteins harboring an N-terminal proline residue are degraded by the GID E3 ligase complex (9, 10) (fig. S1). Theoretically these pathways have the capacity to target the majority of cellular proteins, but the extent to which they impact protein stability in a physiological context remains unclear. For example, loss of N-terminal acetyltransferase (NAT) enzymes has minimal effects on protein stability in yeast (11), which is inconsistent with a widespread role for the Ac/N-end rule pathway.
Previously we modified the Global Protein Stability (GPS) system (12, 13) to develop a high-throughput method to characterize degron motifs in human proteins (14). This approach is based on a lentiviral expression vector encoding two fluorescent proteins: DsRed, which serves as an internal reference, and GFP fused to a short peptide of interest, which is translated from an internal ribosome entry site (IRES). As both DsRed and the GFP-peptide fusion protein are expressed from the same transcript, the GFP/DsRed ratio can be used to quantify the effect of the peptide sequence on the stability of GFP (14). Here, by exploiting the ubiquitin-fusion technique (4) to adapt this ‘GPS-peptidome’ approach to search for N-terminal degron motifs, we directly examined the contribution of N-terminal sequences to protein stability in human cells.
Stability profiling of the human N-terminome using GPS-peptidome technology
We synthesized a library of oligonucleotides encoding the first 23 amino acids of the primary isoform(s) of all human proteins, both with and without an initiator methionine (a total of ~50,000 sequences). These were cloned into the ‘Ub-GPS’ expression vector between the ubiquitin gene and GFP (Fig. 1A). Upon expression of the constructs in HEK-293T cells, proteolytic cleavage of the ubiquitin moiety by endogenous deubiquitinating enzymes leads to the exposure of the peptides at the N-terminus of GFP (Fig. 1A). Fluorescence-activated cell sorting (FACS) was used to partition the population into six bins of equal size based on the stability of the peptide-GFP fusion. The abundance of each fusion in each bin was then quantified by Illumina sequencing, with each peptide assigned a protein stability index (PSI) score ranging between 1 (maximally unstable) and 6 (maximally stable) based on the proportion of sequencing reads in each bin (data S1).
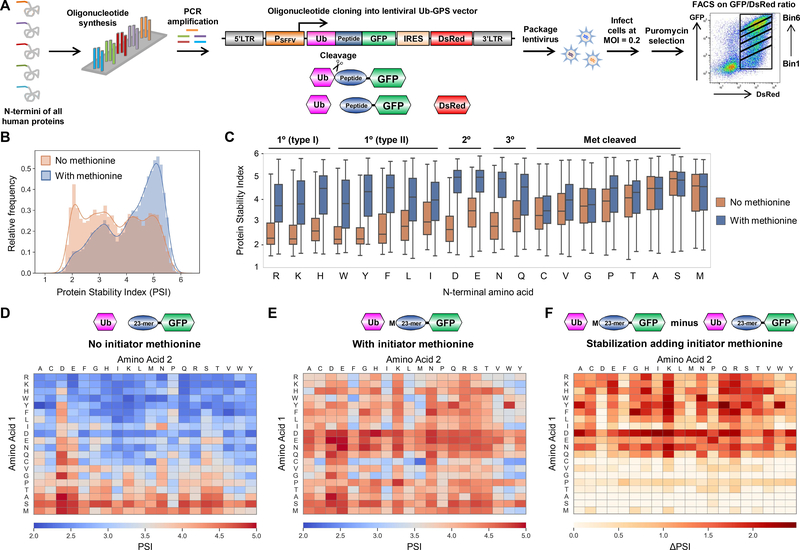
Figure 1. GPS profiling of the human N-terminome.
(A) Schematic representation of the N-terminome GPS screen, in which the first 24 residues of all human proteins were expressed in the Ub-GPS vector as N-terminal fusions to GFP.
(B) Distribution of protein stability scores observed from the screen depicted in (A).
(C) Boxplots showing the distribution of stability scores for all peptides commencing with the indicated amino acid, when encoded either with (blue boxes) or without (orange) an upstream methionine residue.
(D-F) Heatmaps depicting the mean stability score for all peptides commencing with the indicated two amino acids, when encoded either with (E) or without (D) an upstream methionine residue; (F) illustrates the difference between the two.
We began by assessing the effect of the initiator methionine on protein stability. Overall, peptide-GFP fusions lacking an initiator methionine were much less stable than their counterparts with an initiator methionine (Fig. 1B). However, this effect was only observed for certain N-terminal residues (Fig. 1C). Reporters commencing with amino acids bearing small side chains (C, V, G, P, T, A and S) were generally relatively stable, and exhibited little or no difference in overall stability whether or not they were preceded by an upstream methionine residue; this is consistent with the efficient cleavage of the initiator methionine by methionine aminopeptidases when the following amino acid has a sufficiently small radius of gyration (15). In contrast, peptide-GFP fusions commencing with all other residues (except methionine itself) were generally stable only when preceded by an upstream methionine residue, and were greatly destabilized in the absence of an initiator methionine (Fig. 1C–F).
Overall, these data provide strong support for a central role of the Arg/N-end rule pathway in protein quality control. Whereas proteins bearing native N-termini (methionine itself, or C/V/G/P/T/A/S, from which methionine is normally removed (15)) are broadly stable, proteins bearing aberrant N-termini (R/K/H/W/Y/F/L/I/D/E/N/Q, without a preceding methionine) are all highly unstable; the latter residues correspond perfectly to the primary type I (R/K/H), primary type II (W/Y/F/L/I), secondary (D/E) and tertiary (N/Q) N-terminal degrons of the Arg/N-end rule pathway (fig. S1A). Crucially, however, when these residues were preceded by methionine – as they would be in the context of normal protein synthesis – broad stabilization was observed (Fig. 1F).
Computational identification of destabilizing N-terminal motifs
Subsequently we focused on understanding the factors that determined the stability of peptide-GFP fusions synthesized with an initiator methionine. Stability scores for these reporters were distributed bimodally, with approximately one-third of the library exhibiting significant instability (Fig. 1B, blue histogram). One key factor that strongly influenced stability was amino acid composition (Fig. 2, A and B). For example, aspartic acid and glutamic acid were depleted from unstable peptides and enriched among the stable peptides, while hydrophobic residues such as tryptophan, phenylalanine and leucine showed the opposite pattern. This effect is not specific to the N-terminus, however, as previously we found similar rules governed the stability of reporter constructs in which peptides were fused at the C-terminus of GFP (14).
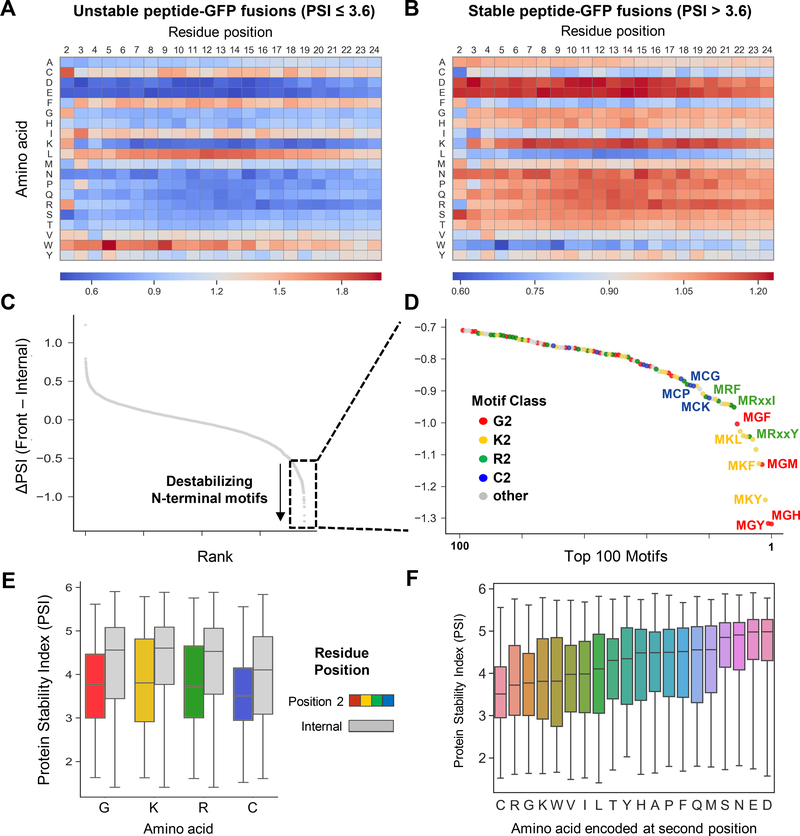
Figure 2. Identification of degron motifs located at protein N-termini.
(A and B) The effect of peptide composition on protein stability. Heatmaps showing the relative depletion (blue) or enrichment (red) of each amino acid across all positions of the 23-mer peptide amongst unstable peptides (A) versus stable peptides (B).
(C-F) Computational prediction of N-terminal degrons. (C) For all possible combinations of di-peptide motifs, the mean difference in stability between peptides containing the motif at the extreme N-terminus (that is, immediately following the initiator methionine) was compared to all peptides containing the motif at any other internal position in the peptide. (D) Classes of N-terminal degrons. The majority of the top 100 predicted destabilizing N-terminal motifs encoded either glycine (G2), lysine (K2), arginine (R2) or cysteine (C2) at the second position; some example motifs are annotated. (E) Boxplots showing the distribution of stability scores for all peptides in which the indicated residues were encoded at the second position (colored boxes) versus any other internal position within the peptide (gray boxes). (F) Boxplots showing the distribution of stability scores for all peptides with the indicated residues encoded at the second position.
Most amino acids exerted a similar effect on stability regardless of their position across the 24-mer peptide, but we noticed that certain residues exerted differing effects specifically when encoded at the second position compared to all other internal positions (Fig. 2, A and B). We therefore performed a computational analysis of the data to identify motifs that might promote instability specifically when located at or near the N-terminus of the peptide. For all possible combinations of di-peptide motifs, we compared the mean stability of all peptide-GFP fusions harboring the motif within the first seven N-terminal amino acids versus those harboring the motif at an internal position in the 23-mer peptide (Fig. 2C and data S2). Strikingly, over 80% of the top 100 candidate destabilizing N-terminal motifs could be grouped into four categories based solely on the identity of the second residue: lysine was present downstream of the initiator methionine in 26 motifs, arginine in 24 motifs, glycine in 22 motifs and cysteine in 9 motifs (Fig. 2D). Globally, reporters encoding these residues at the second position were significantly less stable than reporters containing these residues at an internal position (Fig. 2E). Furthermore, when considering how the overall stability of the N-terminome library varied with the amino acid at the second position, peptides commencing MC-, MR-, MG- and MK- exhibited the lowest mean stability (Fig. 2F). Thus, taking into account initiator methionine removal upstream of amino acids with small side chains, this analysis identified N-terminal glycine and cysteine in addition to MR- and MK- as candidate destabilizing N-terminal motifs.
Exploring the substrate repertoire of UBR family E3 ligases
Next we sought to identify the cellular machinery responsible for the degradation of each class of putative N-terminal degron. The UBR family of E3 ligases target a variety of N-terminal degrons motifs (16), and so we first examined whether ablation of UBR function could stabilize fusion peptides bearing these N-terminal degron motifs. UBR1, UBR2 and UBR4 have been shown functionally to participate in the recognition of N-end degrons (16), and so, through sequential rounds of CRISPR/Cas9-mediated gene disruption, we attempted to create a single cell clone lacking all three of these UBR proteins. Despite screening ~40 clones we were unable to identify a clone in which simultaneous ablation of UBR1, UBR2 and UBR4 proteins was observed by immunoblot, suggesting that such a triple mutant cell may not be viable. However, we were able to generate clones expressing substantially reduced levels of two or more of the proteins (Fig. 3A). To validate a functional lack of UBR activity in these clones, we examined the stability of optimal Arg/N-end rule substrates. Ub-GPS reporters in which the initiator methionine of GFP was replaced with either arginine (R), lysine (K) or tyrosine (Y) were strongly destabilized in wild-type cells, but this effect was abrogated in UBR KO clone #1 and clone #3 (fig. S2A) and completely abolished in clone #2 (Fig. 3B).
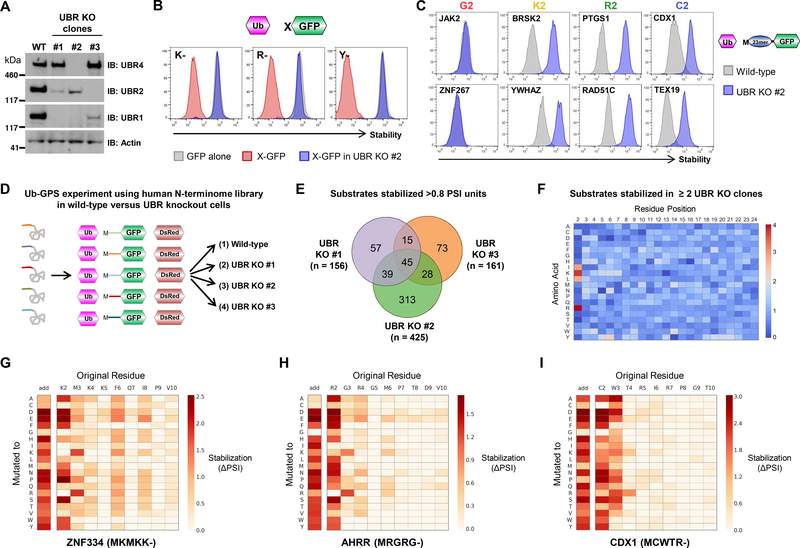
Figure 3. Assessing the repertoire of UBR substrates among the human N-terminome.
(A-C) Assessing UBR-mediated degradation via N-terminal degron motifs. (A) Generation of clones expressing reduced levels of UBR1, UBR2 and UBR4 through CRISPR/Cas9-mediated gene disruption. (B) Functional validation of UBR KO clones. Optimal Arg/N-end rule substrates were highly unstable in wild-type HEK-293T cells, but not in UBR KO clone #2 as measured by flow cytometry. See also fig. S2A. (C) UBR proteins target example peptide-GFP reporters in which lysine, arginine or cysteine, but not glycine, are encoded at the second position. N-terminal peptides derived from the indicated genes were expressed in wild-type or UBR KO clone #2 in the context of the Ub-GPS system and their stability assessed by flow cytometry.
(D-F) Global identification of UBR substrates among the N-terminome library. (D) Schematic representation of the GPS screen designed to identify substrates of UBR family proteins. (E) Venn diagram summarizing the substrates stabilized >0.8 PSI units across the three UBR KO clones. (F) Heatmap showing the relative enrichment (red) or depletion (blue) of each amino acid across all positions of the 23-mer peptide comparing peptides stabilized in two or more of the UBR KO clones relative to the whole N-terminome library. See also fig. S4.
(G-I) Characterization of N-terminal UBR degrons through saturation mutagenesis. Each of the first ten residues of the N-terminal peptides derived from ZNF334 (commencing MK-) (G), AHRR (commencing MR-) (H) and CDX1 (commencing (MC-) (I) were mutated to all other possible residues and their stabilities measured by FACS and Illumina sequencing. The darker the color, the greater the degree of stabilization compared to the wild-type sequence. See also fig. S5.
To assess a possible role for UBR proteins in the targeting of the different classes putative N-terminal degrons, we created a panel of Ub-GPS constructs in which either 23-mer peptides (Fig. 2C) or 3-mer peptides (fig. S2B) harboring example degron motifs downstream of an initiator methionine were fused to the N-terminus of GFP. In both cases, loss of UBR proteins resulted in the stabilization of reporters bearing three of the classes of degrons motifs: MK-, MR- and N-terminal cysteine. However, loss of UBR proteins had little or no effect on the stability of the GFP-fusion proteins bearing N-terminal glycine, suggesting a role for additional E3 ligase(s) in the recognition of this particular N-terminal degron.
It was not surprising that UBR E3 ligases targeted N-terminal cysteine, given that nitric oxide-mediated oxidation and subsequent arginylation of N-terminal cysteine has been shown to render it a substrate for the Arg/N-end rule (17). That said, ATE1 disruption only led to modest stabilization of two peptide-GFP substrates exposing N-terminal cysteine (fig. S2, C and D), suggesting that additional routes to UBR-mediated degradation must also exist.
UBR-mediated degradation of proteins commencing MK- and MR- with an intact initiator methionine was unexpected, however, suggesting that, in addition to targeting truncated proteins bearing abnormal N-termini, UBR ligases might also target certain intact proteins bearing their initiator methionine. To confirm that the initiator methionine of these substrates was indeed intact, and thus rule out the possibility that methionine removal was instead exposing canonical Arg/N-end rule degrons, we examined the N-terminus of two example peptide-GFP UBR substrates expressed in UBR KO clone #2 by mass spectrometry (fig. S3A). In both cases we were readily able to detect the intact N-terminal peptide with the initiator methionine present, while we could not detect any peptides corresponding to a putative processed form without an initiator methionine (fig. S3B).
To further examine this property of UBR ligases, we directly compared the stability of the entire Ub-GPS N-terminome library in wild-type HEK-293T cells versus UBR KO clones #1, #2 and #3 (Fig. 3D and data S3A). Firstly, this revealed that loss of UBR proteins had little effect on the overall stability of reporters synthesized with an N-terminal methionine: only 570 peptide-GFP fusion proteins (<3% of the N-terminome library) exhibited substantial stabilization (>0.8 PSI units) in any of the UBR mutant clones compared to control cells (Fig. 3E). Sequence analysis of the UBR substrates revealed a clear preference for particular N-terminal degron motifs (Fig. 3F and fig. S4A–H). Consistent with our previous data, peptides commencing MC-, MK- and MR- were all enriched. Peptides commencing ML- and MI- were also overrepresented, and for three example peptides in each case we validated that they were indeed stabilized in UBR KO clone #2 (fig. S4I). In S. cerevisiae Ubr1 has been shown to target proteins commencing MΦ- (where Φ is a bulky hydrophobic residue, W/F/Y/L/I) for degradation (18); however, unlike peptides starting ML- and MI-, we did not observe enrichment for peptides starting MF- or MY- among the UBR substrates, and only weak enrichment for peptides starting MW-.
Finally, we noted that only a small proportion of all peptides in the library commencing MK-, MR-, ML- or MI- were UBR substrates, suggesting that additional residues were essential for degron recognition. Indeed, analysis of the composition of all the UBR substrates identified in each category highlighted preferred residues enriched at downstream positions (fig. S4E–H). Furthermore, for some example peptides starting MK-, MR- and MC- we defined the N-terminal UBR degron in detail by performing saturation mutagenesis experiments. We created a Ub-GPS library in which each of the residues from position 2 to position 10 of the 24-mer peptide were mutated to all other possible amino acids, and measured the stability of the resulting peptide-GFP fusions by FACS and Illumina sequencing (data S4A). These experiments confirmed the critical importance of the lysine, arginine or cysteine residue encoded at the second position, but also demonstrated that certain mutations at the third or fourth position along the polypeptide chain could prevent degron recognition (Fig. 3G–I and fig. S5). These data also confirmed the requirement for these degron motifs to be positioned at the extreme N-terminus, as addition of just a single upstream amino acid (that is, immediately after the initiator methionine) resulted in stabilization of the peptide-GFP fusions (Fig. 3G–I and fig. S5, column labeled ‘add’).
N-terminal glycine can act as a potent degron
We next focused on the one class of N-terminal degron motif that was not a substrate for UBR-mediated degradation: N-terminal glycine. To validate that N-terminal glycine did indeed constitute a degron motif, we performed a series of mutagenesis experiments on a panel of unstable Ub-GPS reporters in which 24-mer peptides commencing MG- were fused to the N-terminus of GFP (Fig. 4A and fig. S6A). In each case the glycine residue was indeed critical for instability, as a single substitution converting the glycine residue to serine (G2S) was sufficient to inhibit the degradation of the fusion proteins (Fig. 4A and fig. S6A, left). Moreover, the position of the glycine residue at the extreme N-terminus was also critical, as addition of a single serine residue upstream of the glycine (add S) stabilized the peptide-GFP fusions to a similar extent (Fig. 4A and fig. S6A, center). Finally, consistent with the notion that the initiator methionine is constitutively cleaved when followed by a small residue such as glycine, deletion of the initiator methionine (ΔMet) had no stabilizing effect on any of the peptide-GFP fusions (Fig. 4A and fig. S6A, right).
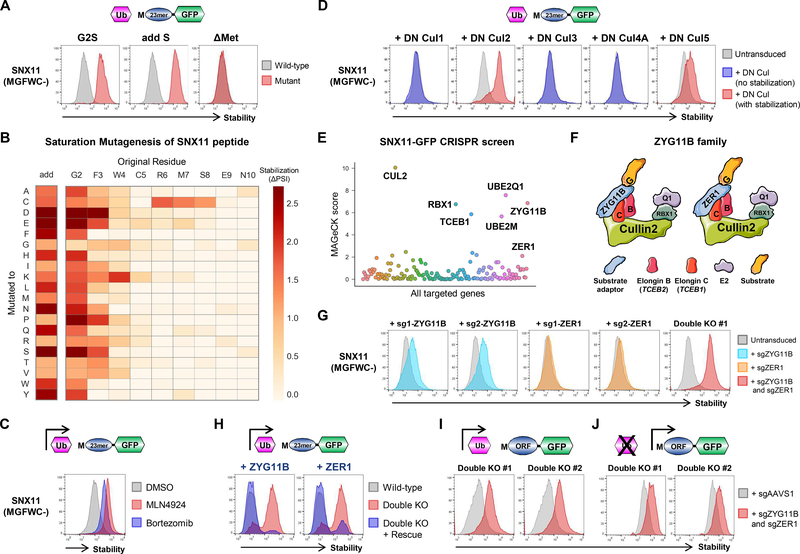
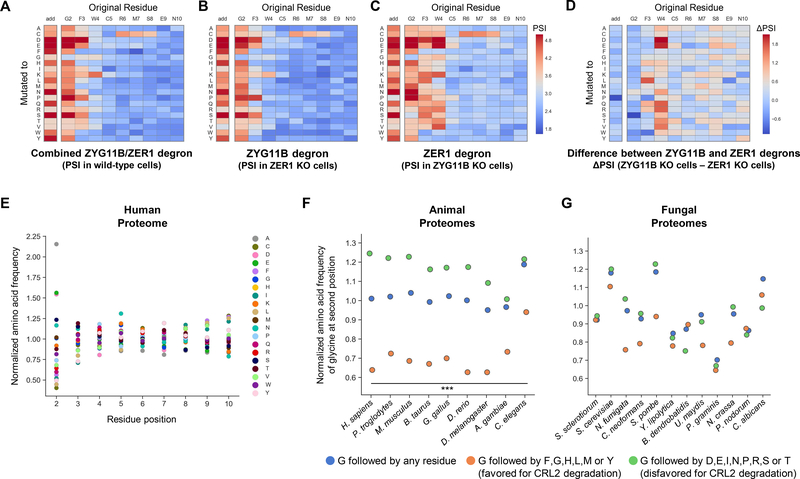
Figure 4. Cul2ZYG11B and Cul2ZER1 target N-terminal glycine.
(A) Glycine at the N-terminus can act as a potent degron. The N-terminal peptide derived from SNX11 was expressed in the context of the Ub-GPS system in HEK-293T cells. The wild-type peptide and a mutant version lacking the initiator methionine (ΔMet) were highly unstable, whilst mutant versions in which the terminal glycine was mutated to serine (G2S) or in which a serine residue was added between the initiator methionine and the glycine residue (add S) were not. See also fig. S6A.
(B) Defining N-terminal glycine degrons through saturation mutagenesis. Each of the first ten residues of the SNX11 peptide were mutated to all other possible residues and their stabilities measured by FACS and Illumina sequencing. The darker the color, the greater the degree of stabilization compared to the wild-type sequence. See also fig. S6B.
(C and D) Cul2 complexes target N-terminal glycine. Stabilization of the SNX11-GFP reporter upon treatment with the CRL inhibitor MLN4924 (C), and following expression of dominant-negative versions of Cullins (D). See also figs. S7 and S8.
(E and F) CRISPR screens identify the Cul2 substrate adaptors responsible for the recognition of N-terminal glycine. (E) Results of the CRISPR screen using the SNX11-GFP reporter, as assessed by MAGeCK. The significantly enriched genes highlight two CRL2 complexes involved in the degradation of proteins bearing N-terminal glycine degrons (F). See also fig. S9.
(G-K) Cul2ZYG11B and Cul2ZER1 cooperate to target N-terminal glycine. (G) Cells stably expressing the SNX11-GFP reporter were transduced with lentiviral vectors expressing Cas9 and sgRNAs targeting either ZYG11B, ZER1 or both and analyzed by flow cytometry; ablation of both ZYG11B and ZER1 was required for full stabilization of the reporter. (H) Exogenous expression of either ZYG11B or ZER1 rescued degradation of the SNX11-GFP reporter in cells lacking endogenous ZYG11B and ZER1. (I and J) Knockout of ZYG11B and ZER1 stabilized full-length SNX11 fused to the N-terminus of GFP, both when expressed in the context of the Ub-GPS system (I) or without upstream ubiquitin fusion (J). See also fig. S10.
For some example peptides we defined the N-terminal glycine degron in detail by performing saturation mutagenesis experiments (data S4A). These confirmed the absolute requirement for the exposure of glycine at the extreme N-terminus, as addition of any single amino acid upstream of the glycine resulted in stabilization of the peptide-GFP fusion (Fig. 4B and fig. S6B–G, column labeled ‘add’). The size of the degron motif appeared to be relatively small, but some substitutions at the residues immediately downstream of the exposed glycine did exert a stabilizing effect (Fig. 4B and fig. S6B–G).
Cul2ZYG11B and Cul2ZER1 target N-terminal glycine
We began the search for the E3 ligase(s) responsible for targeting N-terminal glycine by using the small molecule MLN4924. MLN4924 acts as a broad inhibitor of Cullin-RING ligases (CRLs) by blocking Cullin neddylation (19), thus allowing us to narrow the search to either CRL or non-CRL ligase families. Strikingly, we observed stabilization of all our example Ub-GPS constructs bearing N-terminal glycine upon treatment with MLN4924, implicating CRLs in the recognition of N-terminal glycine (Fig. 4C and fig. S7A).
To obtain an unbiased overview of the potential role of CRLs in the recognition of N-end degrons, we compared the stability of the Ub-GPS N-terminome library in the presence and absence of the CRL inhibitor MLN4924 (fig. S7B and data S5). There was little change in the overall stability of the library in the presence of the drug (fig. S7C), suggesting that recognition of protein N-termini is not a major role of CRLs, but several hundred peptide-GFP fusions did exhibit marked stabilization upon CRL inhibition (fig. S7D). Notably, sequence analysis of the peptide fusions that were stabilized by MLN4924 revealed that a glycine residue at the second position was the most enriched feature (fig. S7, E and F).
Next we sought to identify the specific CRL adaptor(s) responsible for recognition of the N-terminal glycine degron. Employing dominant-negative constructs to inhibit each of the major Cullin proteins expressed in human cells, we determined that either Cul2 or Cul5 was responsible for the degradation of example Ub-GPS reporters harboring glycine at the second position (Fig. 4D and fig. S8). Using these reporter substrates, we performed a series of CRISPR/Cas9-mediated genetic screens using a library of single guide RNAs (sgRNAs) targeting known CRL2/5 BC-box adaptor proteins (fig. S9A). Together these screens identified ZYG11B as the CRL2 substrate adaptor responsible for recognition of the N-terminal glycine degron motif (Fig. 4E, fig. S9B and data S6). Intriguingly, ZER1, which is closely related to ZYG11B (29% amino acid identity) (fig. S9C), was enriched at or approaching the level of statistical significance in several screens, suggesting that these two related adaptors may collaborate in the degradation of proteins exposing N-terminal glycine (Fig. 4F). The third member of the ZYG11 family, ZYG11A, did not score in any of the screens, consistent with RNA-seq data (20) suggesting that it is rarely expressed across human tissues (fig. S9, C and D).
To examine the possibility of cooperation between ZYG11B and ZER1, we performed individual CRISPR/Cas9-mediated gene disruption experiments, ablating the function of ZYG11B or ZER1 either alone or in combination. Loss of ZYG11B alone did indeed stabilize all of the peptide-GFP fusion proteins (Fig. 4G and fig. S10A), but, whilst complete stabilization was observed for two of the reporters (Fig. S10A), only partial stabilization was observed for the others. In contrast, loss of ZER1 alone had little stabilizing effect on any of the reporters; however, simultaneous disruption of both ZER1 and ZYG11B resulted in complete stabilization (Fig. 4G and fig. S10A). Furthermore, ZYG11B and ZER1 both associated with putative substrates bearing N-terminal glycine degrons (fig. S10B), and exogenous expression of either ZYG11B or ZER1 alone in ZYG11B/ZER1 double mutant cells was capable of fully restoring the degradation of a peptide-GFP reporter fusion whose stabilization required ablation of both endogenous ZYG11B and ZER1 (Fig. 4H). Finally, we sought to validate that Cul2ZYG11B and Cul2ZER1 were able to mediate the degradation of full-length proteins bearing exposed glycine residues at their N-termini. We selected a panel of full-length open reading frames (ORFs) whose N-terminal peptides were stabilized by MLN4924 (fig. S7B), and cloned them upstream of GFP. These fusion proteins were all more stable in double mutant cells lacking ZYG11B and ZER1 than in wild-type cells, both when expressed in the context of the ubiquitin fusion system (Fig. 4I and fig. S10C) or without upstream ubiquitin (Fig. 4J and fig. S10D). In addition, for a set of substrates endogenously expressed in HEK-239T cells for which effective commercial antibodies were available, we demonstrated that they were more abundant in ZYG11B/ZER1 double mutant cells than in wild-type cells by immunoblot (fig. S10E).
To obtain a global view of the substrates targeted by these Cul2 complexes, we compared the stability of the Ub-GPS N-terminome library in wild-type cells versus cells lacking either ZYG11B, ZER1 or both ZYG11B and ZER1 (fig. S11A and data S3B). First, this revealed that ZYG11B and ZER1 share the majority of their substrates: there were 115 fusions stabilized in ZYG11B mutant cells and 36 stabilized in ZER1 mutant cells, while 488 were stabilized in the double mutant cells. Sequence analysis of these shared substrates confirmed that N-terminal glycine was the most enriched feature, whilst also highlighting preferred (F, G, H, K and Y) and disfavored (D, E, I, P, S and T) residues at the following position (fig. S11B). Of the substrates that were targeted solely by ZYG11B, over 90% encoded a glycine residue at the second position. Weak enrichment for H, K, L and M was observed at the third position (fig. S11C), which was consistent with our earlier findings that ZYG11B mutant alone prevented the degradation of GFP fusions with N-terminal peptides derived from ZNF267 (commencing MGL-) and JAK2 (commencing MGM-) (fig. S10A). Intriguingly, there was no enrichment of N-terminal glycine among the substrates exclusively targeted by ZER1, with the amino acids H, I, P and Y overrepresented following the initiator methionine (fig. S11D). This finding suggested that (1) any ZER1 substrates bearing an N-terminal glycine were also substrates for ZYG11B, and hence were still targeted for degradation in ZER1 mutant cells, and (2) whilst N-terminal glycine was indispensable for recognition by ZYG11B, in some contexts ZER1 might recognize substrates commencing with residues other than glycine. We characterized one such substrate – the N-terminal peptide derived from KCNT2 commencing MPYL - in detail (fig. S12). In particular, saturation mutagenesis revealed that the hydrophobic residues encoded at the third and fourth position formed a critical part of the ZER1 degron, while some more flexibility was tolerated at the second position (fig. S12G). However, the location of these residues relative to the front of the peptide remained critical, as the addition of a single amino acid upstream of the proline residue prevented degradation (fig. S12G).
Defining the N-terminal glycine degrons recognized by ZYG11B and ZER1
To gain further insight into the specific degron motifs recognized by ZYG11B and ZER1, we examined a larger number of potential peptide-GFP substrates commencing with glycine (fig. S13). These could be divided into three categories: peptides containing degrons fully stabilized upon ZYG11B mutant alone (fig. S13A), peptides containing degrons stabilized partially upon ZYG11B mutant alone, but which required combined ZYG11B and ZER1 mutant for complete stabilization (fig. S13B), and peptides containing degrons for which full redundancy was observed between ZYG11B and ZER1 (fig. S13C). For the vast majority of the peptides in the latter two categories an aromatic residue (H, F or Y) was located downstream of the terminal glycine, supporting the idea that ZER1 might preferentially recognize bulky residues located further along the peptide chain (fig. S13D).
We tested this hypothesis more rigorously by repeating the saturation mutagenesis experiments in the genetic background of either ZYG11B ablation or ZER1 ablation. For peptides targeted by both CRL2 substrate adaptors in wild-type cells, we reasoned that mutagenesis of the peptide in ZYG11B mutant cells would reveal the specific features of the ZER1 degron, and vice versa in ZER1 mutant cells (data S4, B and C). The results for some representative peptides are shown in Fig. 5A–D and fig. S14. These data revealed that mutations conferring instability in wild-type cells were identical to those conferring instability in ZER1 mutant cells (Fig. 5, A and B). Therefore, these residues comprise the minimal N-terminal glycine degron, which is recognized by ZYG11B. On the other hand, the ZER1 degron (as revealed in ZYG11B mutant cells) is more extensive, with mutations two or more residues downstream of the terminal glycine still able to interfere with degradation (Fig. 5, C and D). Overall, these data support a model whereby both ZYG11B and ZER1 target substrates with exposed glycine residues at their N-termini; however, the recognition motif for ZYG11B is relatively small, comprising just the terminal glycine and the following residue, whereas the recognition motif for ZER1 may extend three or more residues along the polypeptide chain and preferentially comprises amino acids with bulky aromatic side chains.
Figure 5. N-terminal glycine degrons are depleted from metazoan proteomes.
(A-D) Defining the degrons recognized by ZYG11B and ZER1 through saturation mutagenesis. A Ub-GPS library was generated in which each of the first ten residues of the SNX11 N-terminal peptide was mutated to all possible amino acids; the library was then expressed in either wild-type (A), ZER1 mutant (B) or ZYG11B mutant (C) HEK-293T cells and the stability of each mutant was measured by FACS followed by Illumina sequencing. The color scale reflects the raw PSI measurement for each peptide-GFP fusion, such that the greater the intensity of the red color, the greater the stabilizing effect of the mutation. The heatmap in (D) illustrates the difference between the PSI in ZYG11B mutant cells versus ZER1 mutant cells; thus, a dark red color indicates mutations which prevent recognition by ZER1 but not by ZYG11B, while a dark blue color indicates mutations which permit recognition by ZER1 but not by ZYG11B. See also fig. S14.
(E) Normalized amino acid frequencies across the first ten residues (following the initiator methionine) of human proteins.
(F and G) Depletion of N-terminal glycine degrons in metazoan proteomes. The normalized amino acid frequency of glycine encoded at the second position in the indicated proteomes is shown by the blue dots, and is further categorized depending on whether the glycine residue is followed by a residue favoring (orange dots) or disfavoring (green dots) CRL2-mediated degradation through ZYG11B and/or ZER1. The relationship observed across animal proteomes (F) is not apparent across fungal proteomes (G), which do not possess a ZYG11 ortholog. (***P < 0.001, Fisher’s exact test).
N-terminal glycine degrons are depleted from metazoan proteomes
Previously we exploited GPS-peptidome technology to identify a suite of degron motifs lying at the C-terminus of human proteins (14). Interestingly, we found that all of these degron motifs were depleted from the human proteome (14), suggesting that our proteomes have evolved to avoid degradation by E3 ligases that target terminal degrons. Therefore, we examined the abundance of N-terminal glycine degrons in eukaryotic proteomes. As is the case for the residue at the extreme C-terminus of eukaryotic proteins (14), the identity of the residue following the initiator methionine at the N-terminus is far more variable than at all neighboring positions, suggesting that its properties are particularly important (Fig. 5E). Nonetheless, glycine is encoded at almost exactly the expected frequency at the second position across a range of metazoan model organisms (Fig. 5F, blue dots). However, classifying glycine residues as those favored (G followed by F, G, H, L, M or Y) or disfavored (G followed by D, E, I, N, P, R, S or T) for CRL2-mediated degradation revealed that, compared to sequences located internally, N-terminal glycine degron motifs are depleted from animal proteomes (Fig. 5F, orange dots), while N-terminal glycine motifs that are not efficiently recognized by ZYG11B and ZER1 are correspondingly enriched (Fig. 5F, green dots). As a control we performed a similar analysis on a panel of reference fungal proteomes, which possess Cul2 but no ZYG11B-family ortholog (21). Consistent with the idea that there should be no selective pressure to avoid N-terminal glycine degrons in the absence of Cul2ZYG11B and Cul2ZER1, no such relationship was observed as in animal proteomes (Fig. 5G). Thus, the avoidance of N-terminal glycine motifs appears to have shaped the composition of metazoan proteomes.
ZYG11B and ZER1 target protein fragments bearing N-terminal glycine following proteolytic cleavage
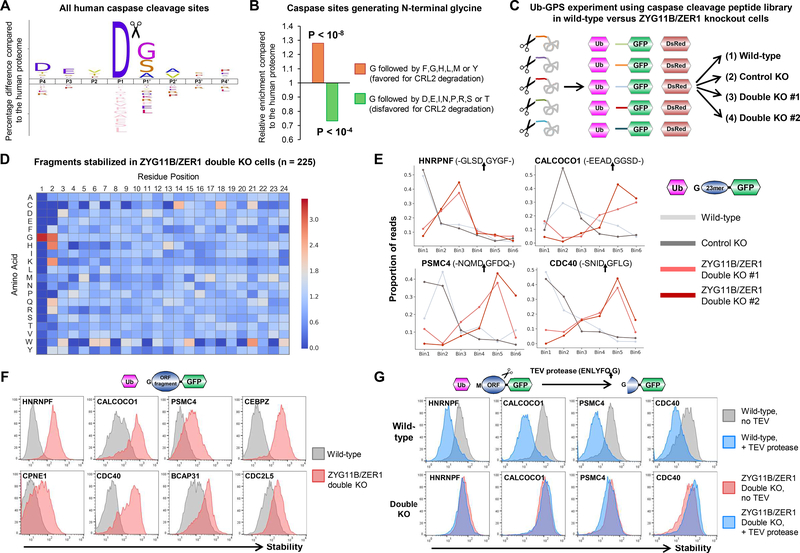
Endoproteolysis generates an additional source of terminal degrons that can be targeted by N-end rule pathways (22–24). Strikingly, caspase cleavage preferentially occurs immediately upstream of glycine residues (Fig. 6A). Indeed, of the ~1800 known human caspase cleavage sites, approximately one-third result in the exposure of glycine at the N-terminus of the downstream fragment (25), suggesting a potential role for ZYG11B and ZER1 in the degradation of cleaved proteins during apoptosis. Moreover, in contrast to the situation at the native N-termini of human proteins (Fig. 5F), we found that degron motifs favoring CRL2-mediated degradation were enriched at the N-termini of caspase cleavage products commencing with glycine (Fig. 6B).
Figure 6. ZYG11B and ZER1 target N-terminal glycine degrons generated through endoproteolytic cleavage.
(A) Caspase cleavage preferentially generates fragments bearing N-terminal glycine. The logoplot depicts the consensus amino acid sequence upstream (P4-P1) and downstream (P1’-P4’) of all annotated caspase cleavage sites annotated in Degrabase (25).
(B) N-terminal glycine degrons are enriched at caspase cleavage sites. Compared to their frequency across the human proteome, preferred glycine degrons (orange bar) are enriched at known caspase cleavage sites, while disfavored glycine degrons (green bar) are depleted.
(C-E) Caspase cleavage events represent a rich source of N-terminal glycine degrons targeted by ZYG11B and ZER1. (C) Schematic representation of the Ub-GPS screen designed to assess the role of ZYG11B and ZER1 in the degradation of caspase cleavage products. (D) Heatmap showing the relative enrichment (red) or depletion (blue) of each amino acid across all positions of the 23-mer peptide comparing peptides stabilized in both ZYG11B/ZER1 double mutant cells compared to the whole caspase cleavage site library. (E) GPS screen profiles of several example substrates bearing N-terminal glycine degrons. The amino acids around the caspase cleavage site (indicated by arrows) are shown. See also fig. S15A.
(F and G) CRL2-mediated degradation of proteolytic cleavage products bearing N-terminal glycine degrons. (F) The full-length fragments lying downstream of the known caspase cleavage sites in the indicated proteins were expressed in the context of the Ub-GPS system, and their stability assessed in wild-type (gray) and dual ZYG11B/ZER1 mutant cells (red) by flow cytometry. (G) Full-length ORFs encoding the indicated genes were modified such that the caspase cleavage site was replaced with a TEV protease cleavage site. Upon TEV expression (blue histograms), destabilization of the downstream cleavage products bearing N-terminal glycine degrons was observed in wild-type cells (top row), but not in dual ZYG11B/ZER1 mutant cells (bottom). See also fig. S15C.
We performed a GPS screen in order to globally assess a potential role for ZYG11B and ZER1 in the removal of protein fragments following proteolytic cleavage. We generated a Ub-GPS peptide library in which the 24 residues downstream of all caspase cleavage events annotated in Degrabase (25) and PROSPER (26) were fused to the N-terminus of GFP, and profiled the stability of these peptide-GFP fusions in wild-type cells versus combined ZYG11B/ZER1 mutant cells (Fig. 6C and data S7). The results confirmed that Cul2ZYG11B and Cul2ZER1 could target many caspase cleavage products for degradation: we identified 225 substrates stabilized >0.5 PSI units in both ZYG11B/ZER1 double mutant lines, of which 219 (97%) harbored an N-terminal glycine residue (Fig. 6D; the GPS profiles of some example substrates are shown in Fig. 6E and fig. S15A).
We validated the findings from the GPS screen in two ways. First, for a panel of example cleavage products exposing N-terminal glycine degrons, we verified that the full-length protein fragments downstream of the cleavage site were stabilized in ZYG11B/ZER1 double mutant cells (Fig. 6F). Second, we verified that these fragments would also be substrates for ZYG11B and ZER1 following endoproteolytic cleavage. We initially attempted to perform these experiments by inducing the dimerization of caspase 9 (27), but in HEK-293T cells this resulted in rapid (<30 min) cell death. Therefore, in order to decouple proteolytic cleavage from cell death, we engineered mutant versions of four example substrates in which the caspase cleavage site was replaced with the Tobacco Etch Virus (TEV) protease cleavage site (Fig. 6G). TEV protease recognizes the amino acid sequence ENLYFQ/G (where / represents the cleavage position), thus exposing an N-terminal glycine on the downstream fragment, and is active when expressed in mammalian cells (28, 29). Upon expression of TEV protease, we observed destabilization of the downstream cleavage products bearing N-terminal glycine degrons in wild-type cells, but this effect was abrogated in ZYG11B/ZER1 double mutant cells (Fig. 6G and fig. S15B). Overall these data support the notion that ZYG11B and ZER1 are likely to be involved in the clearance of proteolytic fragments following caspase cleavage during apoptosis.
ZYG11B and ZER1 function in the quality control of N-myristoylated proteins
Finally we considered whether the recognition of N-terminal glycine degrons might be conditionally regulated through post-translational modifications. Intriguingly, N-myristoylation, the process through which the 14-carbon fatty acid myristate is attached to the N-terminus of a subset of eukaryotic proteins (30), occurs exclusively on N-terminal glycine (Fig. 7A). Given that our mutagenesis experiments showed that addition of just a single amino acid to the N-terminus prevented ZYG11B- and ZER1-mediated recognition, we reasoned that N-myristoylation would prevent CRL2-mediated degradation via N-terminal glycine. Thus we hypothesized that ZYG11B and ZER1 might play an important role in ‘myristoylation quality control’, degrading proteins bearing N-terminal glycine degrons exposed following a failure of N-myristoylation.
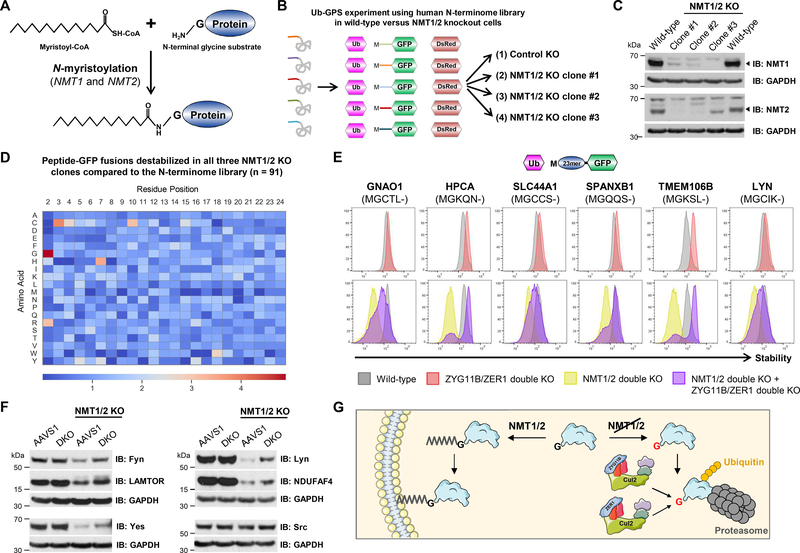
Figure 7. Cul2ZYG11B and Cul2ZER1 target proteins that fail to undergo N-myristoylation for proteasomal degradation.
(A) N-myristoylation of protein substrates occurs on N-terminal glycine.
(B) Schematic representation of the GPS screen using the human N-terminome library designed to assess the effect of loss of N-myristoylation on protein stability.
(C) CRISPR/Cas9-mediated generation of clones with disruptions of NMT1 and NMT2. The protein levels of NMT1 and NMT2 as assessed by immunoblot in the three most optimal clones are shown; arrowheads indicate the bands of the expected molecular weight.
(D) Loss of N-myristoylation destabilizes peptide-GFP fusions commencing with glycine. The heatmap shows the relative enrichment (red) or depletion (blue) of each amino acid across all positions of the 23-mer peptide, comparing the 91 peptides exhibiting significant destabilization in all three NMT1/2 mutant clone relative to the whole N-terminome library.
(E and F) ZYG11B and ZER target N-terminal glycine degrons exposed after a failure of N-myristoylation. (E) The first 24 amino acids from the indicated proteins were expressed as N-terminal fusions to GFP, and their stability in the indicated genetic backgrounds was measured by flow cytometry. The destabilization observed upon loss of NMT1/2 (gold histograms) is rescued in the absence of ZYG11B and ZER1 (purple histogram). (F) The steady-state levels of the indicated substrates of N-myristoylation were assessed by immunoblot in either control (AAVS1) or ZYG11B/ZER1 double mutant cells (DKO), either with or without simultaneous ablation of NMT1/2. Src, which did not exhibit significant destabilization in NMT1/2 mutant cells in the GPS screen, is shown as a negative control.
(G) Model depicting the role of Cul2ZYG11B and Cul2ZER in the quality control of N-myristoylated proteins.
Given that the N-myristoyltransferase enzymes (NMT1 and NMT2 in human cells) require less than the first 20 residues for substrate recognition (30), we reasoned that the peptide-GFP fusion proteins expressed from our N-terminome Ub-GPS library should undergo native N-myristoylation. Therefore, in order to examine the effect of Nmyristoylation on protein stability, we profiled the N-terminome Ub-GPS library in the presence or absence of NMT1/2 (Fig. 7B and data S3C). Although we were not able to generate clones in which both NMT1 and NMT2 were completely ablated following CRISPR/Cas9-mediated gene disruption – a finding consistent with the notion that N-myristoylation is an essential process (31) – we did isolate three clones which retained only residual levels of one NMT enzyme as assessed by immunoblot (Fig. 7C). Strikingly, when we analyzed the composition of all the peptide-GFP fusion proteins whose stability was significantly reduced in all three NMT1/2 mutant clones, we found that N-terminal glycine was the most enriched feature (Fig. 7D). This result strongly supported the idea that a failure to undergo N-myristoylation could lead to instability of the unmodified protein.
To investigate a possible role for ZYG11B and ZER1 in the recognition of proteins exposing N-terminal glycine following a failure of N-myristoylation, we examined the stability of a panel of example substrates (fig. S16A) in which 24 amino acid peptides derived from the N-termini of proteins known to undergo N-myristoylation (32) were expressed in the presence and absence of both NMT1/2 and ZYG11B/ZER1. These peptide-GFP fusion proteins were indeed efficiently myristoylated, as evidenced by membrane localization in wild-type cells but not in NMT1/2 mutant cells (fig. S16B). In support of the data from the GPS screen, in each case we observed destabilization of the peptide-GFP fusion protein upon loss of NMT1/2 (Fig. 7E, gold histograms); moreover, ZYG11B and ZER1 were primarily responsible for this instability, as complete or near-complete re-stabilization was observed upon ablation of both NMT1/2 and ZYG11B/ZER1 (Fig. 7E, purple histograms). The true magnitude of this effect is likely to be even greater, as addition of the small molecule NMT1/2 inhibitor IMP-1088 (33) to the NMT1/2 mutant clones, thereby inhibiting the residual N-myristoyltransferase activity remaining in the cell, further enhanced the destabilization of the peptide-GFP substrates (fig. S16C). Moreover, the small degree of stabilization observed with some of the fusion proteins upon ablation ZYG11B and ZER1 in wild-type (that is, NMT1/2-sufficient) cells (Fig. 7E, top row) suggested that some fraction of protein molecules do normally escape N-myristoylation, emphasizing the necessity for a degradative mechanism to remove these aberrant species.
Lastly, we wanted to validate that endogenous N-myristoylated proteins behave in a similar manner. Indeed, we observed a significant reduction in the steady-state levels of a panel of example substrates in NMT1/2 mutant cells, which was abrogated upon concurrent ablation of ZYG11B and ZER1 (Fig. 7F). In this series, Src serves as a negative control as its N-terminal peptide did not score in the original screen. However, unlike the complete or near-complete stabilization that we observed using the peptide-GFP fusion constructs (Fig. 7E), here dual ZYG11B/ZER1 mutant only resulted in partial re-stabilization. Therefore, in the context of full-length proteins, multiple degrons in addition to N-terminal glycine may be exposed following a failure of N-myristoylation, rendering them substrates for additional E3 ligases. Altogether, these data demonstrate a physiological role for ZYG11B and ZER1 in the surveillance of myristoylated proteins: successful N-myristoylation shields proteins from degradation, but a failure to undergo N-myristoylation results in the exposure of N-terminal glycine degrons and CRL2-mediated degradation (Fig. 7G).
Discussion
Here we exploited GPS technology to directly examine the contribution of N-terminal sequences to protein stability across the human proteome. Unexpectedly we found that, in addition to targeting abnormal proteins lacking an initiator methionine, UBR-family E3 ligases can also target proteins with a native N-terminus in which an arginine or lysine residue follows the initiator methionine. Whilst the initiator methionine is not thought to be removed from proteins commencing MR- or MK-, if some fraction were removed it would reveal basic residues, which are optimal substrates for the canonical Arg/N-end rule pathway. However multiple mass spectrometry approaches designed to catalog N-terminal peptides have found no evidence to support the idea that initiator methionine removal occurs at any appreciable frequency upstream of arginine or lysine (34, 35), and in our own mass spectrometry experiments examining peptide-GFP UBR substrates we were only able to detect N-terminal peptides with their initiator methionine intact (fig. S3). Thus, it seems much more likely that these intact ends are recognized, indicating new degron specificity for UBR proteins.
We also found that cysteine exposed at the N-terminus of GFP conferred instability in a UBR-dependent manner. As noted above, it has previously been shown that nitric oxide-mediated oxidation of N-terminal cysteine renders it a substrate for arginylation by ATE1 and hence UBR-mediated degradation (17). However, in our studies we found that substrates bearing N-terminal cysteine were not stabilized to the same extent in ATE1 mutant cells as in cells lacking UBR proteins; therefore, if UBR proteins do not directly bind N-terminal cysteine, an ATE1-independent pathway must exist that permits this class of degrons to be recognized by UBR E3 ligases.
Most significantly we uncovered an additional branch of the N-end rule centered on N-terminal glycine. There are intriguing mechanistic similarities between the ZYG11B- and ZER1-mediated recognition of N-terminal glycine degrons and the KLHDC2-, KLHDC3 and KLHDC10-mediated recognition of C-terminal glycine degrons (14), with both processes involving multiple related members of CRL2 substrate adaptor families. Like the Kelch repeats found in the KLHDC family proteins, the leucine-rich repeats and the armadillo-like repeats present in the ZYG11 family adaptors also have the propensity to form solenoid structures (36), raising the possibility of a common structural mode through which terminal glycine residues are engaged (37). Furthermore, like their C-terminal counterparts, the ZYG11 family of substrate adaptors have also shaped the proteome, with N-terminal glycine degrons being broadly avoided across metazoa.
Our data suggests two contexts in which the targeting of N-terminal glycine degrons may play an important physiological role. N-myristoylation is a post-translational modification regulating the membrane localization and other properties of several hundred human proteins (30), a group which comprises notable members including Arf family GTPases, G protein alpha subunits and Src family tyrosine kinases (38). We propose a model whereby a failure of N-myristoylation conditionally exposes N-terminal glycine degrons to ZYG11B and ZER1, which are normally occluded upon successful modification. Further work will be required to ascertain whether other classes of terminal degrons function in analogous quality control pathways to ensure the efficient deposition of post-translational modifications.
Furthermore, the strong enrichment for favored ZYG11B/ZER1 glycine degrons at the N-termini of known caspase cleavage products suggested a potential role for these CRL2 E3 ligase complexes during apoptosis. Endoproteolysis has been shown to generate a source of both N-terminal (6) and C-terminal (39) degrons, and we confirmed experimentally that many caspase cleavage events would generate substrates efficiently degraded by Cul2ZYG11B and Cul2ZER1. After glycine, the next most commonly generated N-terminal residue following caspase cleavage is serine, accounting for ~28% of annotated caspase sites (Fig. 6A). Intriguingly, in complete contrast to glycine, serine is the most stabilizing residue when exposed at the N-terminus (Fig. 2A). Indeed, our caspase cleavage product GPS screen data showed that fragments bearing N-terminal serine were generally extremely stable (fig. S15C). This may be useful in circumstances in which caspase may wish to activate a target such as in the case of ATM, whose C-terminal cleavage product acts in a dominant-negative manner to prevent DNA repair during apoptosis (40), or RAD21, whose C-terminal cleavage product acts as a pro-apoptotic factor (41).
The significance of this glycine-specific N-end rule pathway to human biology is underscored by the fact that the frequency of heterozygous loss-of-function mutations in humans for both ZYG11B and ZER1 is far lower than would be predicted. ZYG11B and ZER1 both have a pLi value of 1 in the ExAC database (42), indicating that loss-of-function variants are strongly selected against in the heterozygous state thereby demonstrating potent haploinsufficiency and counter selection in humans. Misregulation of Src-family tyrosine kinases could be deleterious to development. In C. elegans, the ZYG11 ortholog is required for the metaphase to anaphase transition and M phase exit at meiosis II (21, 43, 44). In humans ZYG11B and ZER1 are both expressed in the testes and ovaries, and hence a similar role in the regulation of meiosis could also explain the strong selection against loss-of-function mutations. Altogether, the comprehensive analysis of N-terminal degrons presented here has illuminated multiple new aspects of N-end rule proteolytic pathways and revealed that a family of E3 ligases specific for N-terminal glycine has shaped the human proteome.
Supplementary Material
Acknowledgements
We are grateful to C. Araneo and his team for FACS and we thank A. Varshavsky and J. Wells for advice.
Funding: R.T.T. is a Sir Henry Wellcome Postdoctoral Fellow (201387/Z/16/Z). This work was supported by an NIH grant (AG11085) to S.J.E. and J.W.H. S.J.E. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Competing interests: the authors declare no competing interests.
Data and materials availability: all data is available in the main text or the supplementary materials.
References and Notes
- 1.Kleiger G, Mayor T, Perilous journey: a tour of the ubiquitin-proteasome system. Trends Cell Biol. 24, 352–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ravid T, Hochstrasser M, Diversity of degradation signals in the ubiquitin– proteasome system. Nat. Rev. Mol. Cell Biol 9, 679–689 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mészáros B, Kumar M, Gibson TJ, Uyar B, Dosztányi Z, Degrons in cancer. Sci. Signal 10, eaak9982 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Bachmair A, Finley D, Varshavsky A, In vivo half-life of a protein is a function of its amino-terminal residue. Science. 234, 179–86 (1986). [DOI] [PubMed] [Google Scholar]
- 5.Bartel B, Wünning I, Varshavsky A, The recognition component of the N-end rule pathway. EMBO J. 9, 3179–89 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varshavsky A, The N-end rule pathway and regulation by proteolysis. Protein Sci. 20, 1298–1345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang C-S, Shemorry A, Varshavsky A, N-Terminal Acetylation of Cellular Proteins Creates Specific Degradation Signals. Science 327, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shemorry A, Hwang C-S, Varshavsky A, Control of Protein Quality and Stoichiometries by N-Terminal Acetylation and the N-End Rule Pathway. Mol. Cell. 50, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S-J, Wu X, Wadas B, Oh J-H, Varshavsky A, An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science 355, eaal3655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong C et al. , Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway. Nat. Chem. Biol. 14, 466–473 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Kats I et al. , Mapping Degradation Signals and Pathways in a Eukaryotic N-terminome. Mol. Cell 70, 488–501.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Yen H-CS, Xu Q, Chou DM, Zhao Z, Elledge SJ, Global Protein Stability Profiling in Mammalian Cells. Science 322, 918–923 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Yen H-CS, Elledge SJ, Identification of SCF Ubiquitin Ligase Substrates by Global Protein Stability Profiling. Science 322, 923–929 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Koren I et al. , The Eukaryotic Proteome Is Shaped by E3 Ubiquitin Ligases Targeting C-Terminal Degrons. Cell. 173, 1622–1635 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman F, Stewart JW, Tsunasawa S, Methionine or not methionine at the beginning of a protein. BioEssays. 3, 27–31 (1985). [DOI] [PubMed] [Google Scholar]
- 16.Tasaki T et al. , A Family of Mammalian E3 Ubiquitin Ligases That Contain the UBR Box Motif and Recognize N-Degrons. Mol. Cell. Biol 25, 7120–7136 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu R-G et al. , The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature. 437, 981–6 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Kim H-K et al. , The N-Terminal Methionine of Cellular Proteins as a Degradation Signal. Cell. 156, 158–169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soucy TA et al. , An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 458, 732–736 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Ardlie KG et al. , The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasudevan S, Starostina NG, Kipreos ET, The Caenorhabditis elegans cell-cycle regulator ZYG-11 defines a conserved family of CUL-2 complex components. EMBO Rep. 8, 279–286 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piatkov KI, Brower CS, Varshavsky A, The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc. Natl. Acad. Sci 109, E1839–E1847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piatkov KI, Colnaghi L, Békés M, Varshavsky A, Huang TT, The AutoGenerated Fragment of the Usp1 Deubiquitylase Is a Physiological Substrate of the N-End Rule Pathway. Mol. Cell 48, 926–933 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piatkov KI, Oh J-H, Liu Y, Varshavsky A, Calpain-generated natural protein fragments as short-lived substrates of the N-end rule pathway. Proc. Natl. Acad. Sci 111, E817–E826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crawford ED et al. , The DegraBase: a database of proteolysis in healthy and apoptotic human cells. Mol. Cell. Proteomics 12, 813–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song J et al. , PROSPER: an integrated feature-based tool for predicting protease substrate cleavage sites. PLoS One. 7, e50300 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Straathof KC et al. , An inducible caspase 9 safety switch for T-cell therapy. Blood. 105, 4247–4254 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.To T-L et al. , Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proc. Natl. Acad. Sci. U. S. A 112, 3338–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.To T-L et al. , Rational Design of a GFP-Based Fluorogenic Caspase Reporter for Imaging Apoptosis In Vivo. Cell Chem. Biol 23, 875–882 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright MH, Heal WP, Mann DJ, Tate EW, Protein myristoylation in health and disease. J. Chem. Biol 3, 19–35 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang SH et al. , N -Myristoyltransferase 1 Is Essential in Early Mouse Development. J. Biol. Chem 280, 18990–18995 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Thinon E et al. , Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat. Commun 5, 4919 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mousnier A et al. , Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus. Nat. Chem 10, 599–606 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeom J, Ju S, Choi Y, Paek E, Lee C, Comprehensive analysis of human protein N-termini enables assessment of various protein forms. Sci. Rep 7, 6599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gawron D, Ndah E, Gevaert K, Van Damme P, Positional proteomics reveals differences in N-terminal proteoform stability. Mol. Syst. Biol 12, 858 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gul IS, Hulpiau P, Saeys Y, van Roy F, Metazoan evolution of the armadillo repeat superfamily. Cell. Mol. Life Sci 74, 525–541 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rusnac D-V et al. , Recognition of the Diglycine C-End Degron by CRL2KLHDC2 Ubiquitin Ligase. Mol. Cell 72, 813–822.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thinon E et al. , Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat. Commun 5, 4919 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin H-C et al. , C-Terminal End-Directed Protein Elimination by CRL2 Ubiquitin Ligases. Mol. Cell 70, 602–613.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith GC, d’Adda di Fagagna F, Lakin ND, Jackson SP, Cleavage and inactivation of ATM during apoptosis. Mol. Cell. Biol 19, 6076–84 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen F et al. , Caspase proteolysis of the cohesin component RAD21 promotes apoptosis. J. Biol. Chem 277, 16775–81 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Lek M et al. , Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonneville R, Gönczy P, zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development. 131, 3527–3543 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Vasudevan S, Kipreos ET, CUL-2 and ZYG-11 promote meiotic anaphase II and the proper placement of the anterior-posterior axis in C. elegans. Development. 131, 3513–3525 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Hwang C-S, Shemorry A, Varshavsky A, N-Terminal Acetylation of Cellular Proteins Creates Specific Degradation Signals. Science 327, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aksnes H, Drazic A, Marie M, Arnesen T, First Things First: Vital Protein Marks by N-Terminal Acetyltransferases. Trends Biochem. Sci 41, 746–760 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Li W et al. , MAGeCK enables robust identification of essential genes from genomescale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balachandran RS et al. , The ubiquitin ligase CRL2 ZYG11 targets cyclin B1 for degradation in a conserved pathway that facilitates mitotic slippage. J. Cell Biol 215, 151–166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanjana NE, Shalem O, Zhang F, Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 17, 10 (2011). [Google Scholar]
- 51.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K, Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Thomsen MCF, Nielsen M, Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 40, W281–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.