Abstract
While host proteins incorporated into virions during viral budding from infected cell are known to play essential roles in multiple process of the life cycle of progeny virus, these characteristics have been largely neglected in studies on rabies virus (RABV). Here, we purified the RABV virions with good purity and integrity, and analyzed their proteome by nano LC–MS/MS, followed by the confirmation with immunoblot and immuno-electronic microscopy. In addition to the 5 viral proteins, 49 cellular proteins were reproducibly identified to be incorporated into matured RABV virions. Function annotation suggested that 24 of them were likely involved in virus replication. Furthermore, cryo-EM was employed to observe the purified RABV virions, generating high-resolution pictures of the bullet-shaped virion structure of RABV. This study has provided new insights into the host proteins composition in RABV virion and shed the light for further investigation on molecular mechanisms of RABV infection, as well as the discovery of new anti-RABV therapeutics.
Electronic supplementary material
The online version of this article (10.1007/s12250-019-00157-6) contains supplementary material, which is available to authorized users.
Keywords: Rabies virus (RABV), Purification, Virion structure, Virion proteome
Introduction
Rabies virus (RABV) is a prototypical neurotropic virus causing acute, fatal encephalitis in humans and other animals worldwide. Although rabies can be prevented by appropriate post-exposure prophylaxis (PEP) administration, more than 59,000 people die of this incurable disease annually in the world (Brunker and Mollentze 2018), emerging the significant demand to further investigate the mechanism of RABV life cycle and develop efficient therapeutics. RABV has a non-segmented, single stranded negative RNA genome of about 12 kb, encoding five viral proteins including nucleoprotein (N), matrix protein (M), phosphoprotein (P), RNA-dependent RNA polymerase (L) and glycoprotein (G) (Davis et al.2015). To facilitate the efficient infection and replication, RABV has developed multiple strategies including interaction with host proteins to gain essential functions in viral replicative cycle. An example is that the heat shock proteins 70 (HSP70) was shown to interact with the N protein of RABV and to be present in both the nucleocapsid and purified virions, while down regulation of HSP70, using an inhibitor or RNA interference, results in a significant decrease in the production of viral mRNAs, viral proteins, and virions (Lahaye et al.2012). However, the knowledge on the host protein assembled into RABV virions is limited.
Many viruses hijack the cellular machineries by interaction with a considerable number of host factors for efficient replication. For example, retroviruses, rhabdoviruses, filoviruses, arenaviruses, and paramyxoviruses bud via host endosomal sorting complex required for transport (ESCRT) machinery (Chen and Lamb 2008; Votteler and Sundquist 2013). Human immunodeficiency virus (HIV) is one of the best understood viruses in this respect. ESCRT subunits are recruited at HIV assembly sites and have been detected in the released HIV particles by three dimensional super resolution microscopy (Van Engelenburg et al.2014). ESCRT machinery were also found to be associated with RABV life cycle (Okumura and Harty 2011), but what kind of ESCRT subunits participate in RABV budding has been not investigated.
Sensitive mass spectrometry enables the identification of host proteins incorporated into virions with high accuracy and sensitivity. So far, proteomic studies have been performed on a number of purified virus preparations, including DNA viruses (Stegen et al.2013; Alejo et al.2018) and a broad range of RNA viruses (Shaw et al.2008; Radhakrishnan et al.2010; Nuss et al.2014; Lussignol et al.2016; McKnight et al.2017; Ziegler et al.2018), as well as virus-like particles (Vera-Velasco et al.2018). These studies have revealed substantial host proteins incorporated into viral particles, with several highlighting the utility of proteomics in understanding the mechanisms of virus-host interaction and identification of the essential role of cellular proteins in viral assembly or morphogenesis. For an instance, the proteomic analysis of hepatitis C virions (HCV) has identified the participation of host proteins in viral infection and revealed the interaction of HCV capsid protein with nucleoporin Nup98, a host protein important for viral propagation and morphogenesis (Lussignol et al.2016). Proteomic analysis has also shown the engagement of host proteins in the assembly of respiratory syncytial virus (RSV), with one, heat shock protein 90, playing an important role in the assembly and maturation of the virus particles (Radhakrishnan et al.2010). However, until the present work, the host protein constituents of rabies virions had remained largely unknown, although their identification could significantly enhance our understanding of the pathways of viral budding and entry, and the mechanisms of virus-host interactions required for RABV propagation.
Here, the protein contents of purified RABV were identified by the nano-scale liquid chromatography tandem mass spectrometry (nano LC–MS/MS) approach, revealing 49 virions-associated host proteins with some validated by Western blotting or immunogold labeling. In addition, the potential functional implications of these cellular proteins in the RABV replicative cycle were extrapolated.
Materials and Methods
Viruses and Cells
RABV, CVS-11 strain was propagated in mouse Neuro-2A (N2a) cells in DMEM/F12 (Corning, New York, USA) and Opti-MEM (Gibco, Carlsbad, California, USA) at a 1:1 ratio, supplemented with 2% (V/V) fetal bovine serum (FBS; Biological Industries, Kibbutz Beit Haemek, Israel), penicillin G (100 U/mL) and streptomycin (100 µg/mL). The virus titer was determined as TCID50 according to the Spearman–Kärber method (Dean and Abelseth 1973).
Purification of RABV Particles
N2a cells at 70% confluence were infected with CVS-11 virus at a multiplicity of infection (MOI) of 0.01. The virus-containing supernatant were harvested at 72 h post-infection and clarified by differential centrifugation, first at 3000 ×g for 15 min, then at 10,000 ×g for 30 min, and the supernatants were used in the next step. All clarification and purification processes were carried out at 4 °C to avoid inactivation and maintain stability of virion structure. A 10% iodixanol cushion was prepared by diluting a stock solution of OptiPrep® (60% w/v aqueous iodixanol, Sigma, St Louis, USA) with TNE buffer (50 mmol/L Tris–HCl, 100 mmol/L NaCl, 1 mmol/L EDTA, pH 7.4) and used for pelleting RABV virions by ultracentrifugation at 70,000 ×g for 1 h in a type 45Ti rotor (Beckman Coulter, California, USA) at 4 °C. The pellets were dispersed in 0.5 mL TNE buffer and then overlaid on the top of a discontinuous iodixanol density gradient (10%, 15%, 20%, 25% and 30%). After ultracentrifugation at 100,000 ×g for 1 h in an SW41 rotor (Beckman Coulter), visible virus bands were removed by suction and checked by electron microscopy. In order to obtain higher purity, virus preparations containing bullet shaped particles were subjected to centrifugation at 100,000 ×g in a discontinuous iodixanol density gradient (15%, 20% and 25%) for further purification.
Electron Microscopy of Purified RABV Particles
The resulting RABV preparations were examined by transmission electron microscopy (TEM) and then by cryo-electron microscopy (cryo-EM) to observe their purity and morphology as follows. The purified preparations were adsorbed onto fresh glow-discharged 300 mesh carbon-coated copper grids for 5 min at 20 °C to 25 °C, then negatively stained with 2% phosphotungstic acid (pH 7.0), and imaged using a H-7650 electron microscope (Hitachi, Japan) operating at 80 kV. For cryo-EM, purified virions (3 µL) were frozen onto 200 mesh C-Flat cooper grids (Protochips, USA) by plunging into liquid ethane, using a Vitrobot mark IV specimen preparation unit (FEI, Eindhoven, Netherlands) with settings at 4 °C, 100% humidity and 1.5 s blot time. Cryo-EM images were recorded at a magnification of 96,000 × in an FEI Tecnai Arctica electron microscope (FEI, Hillsboro, USA) at 200 kV, equipped with an FEI 4096 × 4096 CCD camera.
Deglycosylation, SDS-PAGE and Western Blot Analysis
Prior to proteomic analysis, purified virion preps were deglycosylated to increase the sensitivity of detection of peptides containing glycosylation sites, as described previously (Shaw et al.2008; Vera-Velasco et al.2018). Aliquots of purified virus containing 100 µg protein were deglycosylated with PNGase F (New England Biolabs, Ipswich, Mass., USA) as recommended by the manufacturer. Proteins (8 µg) from either purified or deglycosylated virions were separated by 10% SDS-PAGE. For Western blot analysis the proteins were transferred to a nitrocellulose membrane which was then probed with specific RABV glycoprotein polyclonal antibody (made in our laboratory) and Alexa Fluor 680-labeled secondary antibody (Invitrogen, Carlsbad, Cal., USA). Protein bands were observed using an Odyssey infrared imaging system (LI-COR).
Proteomics Analysis
Deglycosylated protein samples were digested in solution with sequencing grade trypsin (Promega, USA) at 37 °C for 15 h with an enzyme: protein ratio of 1:50 (w/w) followed by protein alkylation in 0.1 mol/L iodoacetamide. The resulting peptide mixtures were extracted, desalted with C18 cartridges (Empore™ SPE Cartridges C18, 7 mm bed inner diameter, 3 ml volume, Sigma, USA), dried by vacuum centrifugation, re-solubilized in 0.1% (v/v) formic acid. Proteomic analysis was conducted by nano LC–MS/MS as described previously (Huang et al.2016). Briefly, 2 µg digested peptides from three different preparations were individually separated using C18-reversed phase analytical EASY columns (Thermo Fisher Scientific, USA) at a flow rate of 300 nL/min. Peptides were gradient-eluted into a Q Exactive mass spectrometer (Thermo Fisher Scientific, USA) acquiring fragmentation spectra generated by collision-induced dissociation (CID). Resolution was 70,000 and 17,500 respectively for MS and MS/MS scans at m/z 200. Dynamic exclusion was set to 60 s.
All MS/MS spectral data were searched using the Andromeda search engine and MaxQuant software version 1.3.0.5 (Cox and Mann 2008) against the UniProtKB mouse sequence database (uniprot_Mouse_83482_20170627.) coupled with RABV (CVS-11) protein sequences downloaded from GenBank (https://www.ncbi.nlm.nih.gov/nuccore/GQ918139.1). The resulting data was accepted by controlling the false discovery rate (FDR) to < 0.01% at peptide-level or protein-level. In MaxQuant, the abundance of protein was expressed by intensity based absolute quantification (iBAQ), calculated as the sum of all peptide peak intensities divided by the number of theoretically observable tryptic peptides. Compared with the Top3 approach this provided a more reliable way to obtain absolute protein abundances at a proteome-wide scale, especially in detecting short proteins of MW < 25 kDa (Ahrne et al.2013). A protein was identified to have high-confidence when its iBAQ > 106 and unique peptides ≥ 2. To increase the confidence, proteins identified in three separate assays were included in the candidate list.
Protease K Treatment and Western Blot Analysis
In order to justify the incorporation of host proteins, the purified virions were first subjected to protease K (ProK; Thermo, USA) treatment. After stopping the reaction with phenylmethanesulfonyl fluoride (PMSF; Sigma, USA) and protease inhibitor (Thermo, USA) the treated virions were pelleted through a 10% iodixanol cushion to remove any cleaved peptide. N2a cells were mock infected or infected with CVS-11 for 72 h at an MOI of 0.1. Supernatants of cell extracts were collected following incubation in cell lysis buffer (Cell Signaling Technology, USA) with added protease inhibitor (Thermo, USA) on ice for 20 min, and centrifugation at 14,000 ×g for 15 min. Both cell extracts (25 µg) and untreated or protease-treated virion preparations (10 µg) were analyzed by Western blotting with antibodies against the indicated proteins. The virion glycoprotein (G), nucleoprotein (N) and matrix protein (M) were probed by their specific mouse polyclonal antibodies (made in our laboratory). Rabbit polyclonal antibody against HSC70 (ab137808), cofilin (ab11062), CHMP4B (ab105767), HSP40 (ab69402), VPS37B (ab122450) and mouse polyclonal antibody against CHMP2A (ab67064), mouse monoclonal antibody against β-actin (ab 6276) were obtained from Abcam (Cambridge, UK). Monoclonal antibodies against CD9 (sc-13118), VPS4B (sc-377162) were obtained from Santa Cruz Biotechnology (Santa Cruz, Cal., USA). Rabbit polyclonal antibody against TSG 101 (T5701) was obtained from Sigma (St. Louis, Missouri, USA). Mouse polyclonal antibody against ALIX (#2171) was obtained from Cell Signaling Technology (Danvers, Mass., USA). Detection was performed using species-specific highly cross-adsorbed secondary antibody, Alexa Fluor 680 (A10038 for mouse and A10043 for rabbit (Invitrogen, Carlsbad, Cal., USA) and imaged by LI-COR.
Immunogold Labeling and Electron Microscopy
To localize incorporated proteins on the viral surface, the purified virions were adsorbed onto 300-mesh formvar/carbon-coated nickel grids (PolySciences, Philadelphia, USA) at 20 °C to 25 °C. After 5 min, excess buffer was removed by blotting and the preparations were blocked with 3% BSA in phosphate-buffered saline (PBS, pH 7.4) for 45 min at 20 °C to 25 °C. The blocked virions were incubated with primary antibodies (20 µg/mL in 1% BSA/PBS) for 1.5 h, followed by 10 nm colloidal gold particle-conjugated secondary antibody (ab27241, Abcam, Cambridge, UK) for 40 min at room temperature. The grids were washed thoroughly but gently with PBS, then washed with water before negative staining. Images of virions labeled by colloidal gold particles were captured by TEM as described above.
Results
Virus Cultivation, Purification and Morphology
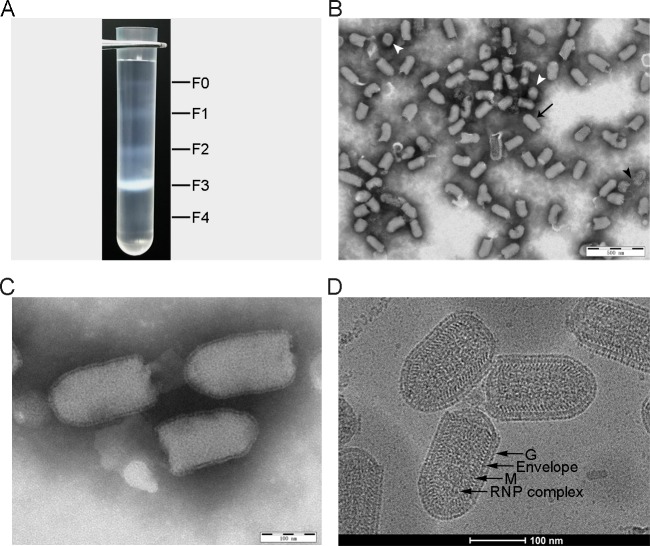
Proteomic analysis of RABV requires purified virions with high purity and an intact structure of typical bullet-shaped morphology. Supernatants of large-scale cultures of CVS-11 with a titer of 108.05 TCID50/mL, were harvested and the virus was purified by ultracentrifugation, resulting in five visible fractions in the iodixanol density gradient (F0-F4) as shown in Fig. 1A. Observation by TEM showed that most particles are in well-defined bullet-shaped with a length of 120–180 nm in the F3 band (20%–25% iodixanol). The truncated or crescent defective interfering (DI) particals with length of 50–70 nm can also be found in the F1 (10%–15% iodixanol) and F2 (15%–20% iodixanol) bands. No virus particles were apparent in the F0 and F4 bands (Supplementary Figure S1). Following further sedimentation through a 15%–25% iodixanol density gradient, the purity and integrity of the virions of three independent purifications were checked by electron microscopy prior to the following proteomic analysis. As shown in Fig. 1B, the purified virus particles were free of cellular debris or vesicles with most having intact, typical bullet shapes of length 120–180 nm. Small numbers of ruptured and DI particles were observed (Fig. 1B). Higher magnification highlighted the characteristic bullet-shaped morphology covered by numerous spikes (Fig. 1C). The purity of the virion preparations was estimated by counting the proportion of virus particles among total particles in images of the three sets of purifications. Of a total 1200 counts, 95.6% were RABV, consisting of 87% intact virions and 8.6% of damaged or DI particles.
Fig. 1.
Purification and electron microscopy (EM) of RABV particles. A Ultracentrifugation of RABV particles in a 10%–30% discontinuous iodixanol density gradient. B The images of purified RABV virions by EM (× 10,000) show multiple intact bullet-shaped virions (black arrow) as well as a few damaged (black arrow head) and DI particles (white arrow head). C High magnification (× 40,000) of intact bullet-shaped RABV virions, highlight the G protein spikes covering the viral surface. D A typical cryo-EM image of intact bullet-shaped RABV virions at × 96,000 clearly showing four distinct layers from the outside to the inside: glycoprotein spike, viral envelope (lipid bilayer), M protein helix and RNP complex.
Cryo-electron microscopy (cryo-EM) has become a robust imaging tool for determining the ultrastructure of viruses at near-atomic resolution (Yuan et al.2018). Images clearly showed four distinct electron-dense layers (Fig. 1D), in which the outermost layer, on the viral surface, consisted of spikes (G protein molecules) over the entire surface apart the planar end. The lipid bilayer of the envelope covering the entire capsid formed the second layer. The third layer consisted of helical structures (M protein molecules), similar to that of vesicular stomatitis virus (VSV) (Ge et al.2010). However, the EM images showed that both G and M layers of the rabies virions were absent from the planar ends, the result is consistent with that of previous publication (Guichard et al.2011). The innermost layer was super-helical structure known to consist of the ribonucleoprotein (RNP) complex VSV (Ge et al.2010) and RABV (Luo et al.2007; Riedel et al.2019).
In order to understand the proteomic characteristics of rabies virions prepared from infected animals, attempts were made to purify the virus from CVS-11 infected mouse brain tissues. Unfortunately, the experiment with various iodixanol density gradients and centrifugation programs failed to remove all brain tissue debris from the virion preparations. Therefore the virus grown on N2a cell lines was eventually used for the proteomic analysis.
SDS-PAGE and Western Blot Analysis of Purified Rabies Virions
SDS-PAGE of purified virions revealed six bands, including two forms of differentially glycosylated viral glycoprotein (GI and GII) (Fig. 2A). Following deglycosylation only one band (G0) was seen. Both glycosylated and deglycosylated G proteins were further identified by Western blotting (Fig. 2B). In addition, analysis also revealed some fainter bands that may represent a low abundance of cellular proteins (Fig. 2A). These results showed that the deglycosylation of proteins in purified virions was complete and that the virion proteins could be used for proteomic analysis.
Fig. 2.
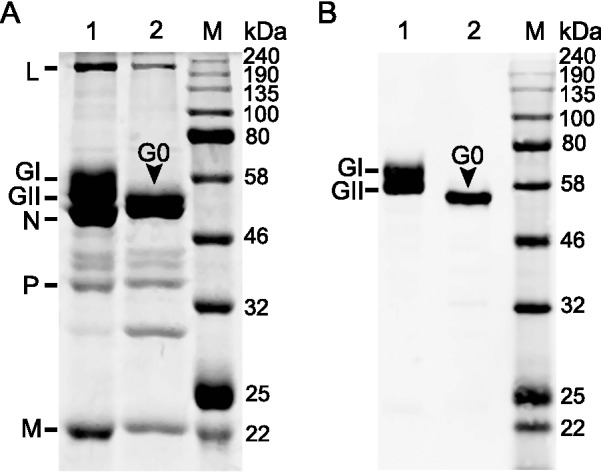
Electrophoresis and Western blot analysis of RABV virion proteins. A SDS-PAGE of the proteins of purified virions (lane 1) and following deglycosylation (lane 2). Eight µg purified virions was loaded in each lane; B Western blot analysis of the viral G protein with its specific polyclonal antibody: two forms of G protein, GI and GII, were detected in purified virions (lane 1), while only one form, G0, was detected following deglycosylation (lane 2).
Proteomic Analysis
Nano LC–MS/MS was utilized to identify the protein composition of purified rabies virions, resulting in identification of 54 high-confidence proteins. These were considered to be incorporated into mature virions based on the following criteria: (1) the proteins were identified in each of three independently purified virion preparations; (2) the abundance of target proteins exceeded 106; (3) each target protein had at least 2 unique peptides. Proteins identified in low abundance or as being unreproducible were likely randomly loaded contaminants or sticky proteins, and were therefore excluded from the viral proteomic composition. Table 1 lists all high-confidence proteins ranked according to their average abundance values. The results are consistent with SDS-PAGE analysis, both showing that the five viral structural proteins were most abundant.
Table 1.
High-confidence proteins identified in purified RABV virions by LC–MS/MS.
| Protein ID | Protein name | Description | Unique peptidesa | Sequence coverage (%)b | Abundancec | Mass (kDa) |
Subcellular locationd | Reported in other virusese |
|---|---|---|---|---|---|---|---|---|
| O92284 | G | Glycoprotein | 37 | 49 | 1.75E+10 | 58.86 | Virion | |
| Q8JXF6 | N | Nucleoprotein | 40 | 80.7 | 1.48E+10 | 50.73 | Virion | |
| P22363 | P | Phosphoprotein | 32 | 84.2 | 1.13E+10 | 33.62 | Virion | |
| P25223 | M | Matrix protein | 8 | 52.5 | 7.75E+09 | 23.13 | Virion | |
| D8VEC2 | L | Large structural protein | 111 | 61.5 | 3.94E+09 | 242 | Virion | |
| P40240 | CD9 | CD9 antigen | 7 | 26.5 | 1.72E+09 | 25.26 | Membrane | IAV1, HAV, MEV, HIV1, ASFV |
| P63168 | DLC8 | Dynein light chain 1 | 2 | 57.3 | 1.09E+09 | 10.37 | Nucleus, mitochondria, cytoskeleton | |
| P63017 | HSC70 | Heat shock cognate 71 kDa protein | 28 | 55.9 | 5.83E+08 | 70.87 | Membrane, nucleus, cytoplasm | RSV, HIV1, HIV2,VSV, RVFV, HSV, ASFV, JUNV |
| P17742 | CyPA | Cyclophilin A | 9 | 68.3 | 5.65E+08 | 17.97 | Cytoplasm | IAV1, MEV, HIV1, HIV2, HIV3, HSV, KSHV |
| P63001 | RAC1 | RAS-related C3 botulinum | 6 | 36.5 | 3.51E+08 | 23.43 | Membrane, cytoplasm | RVFV, HIV2 |
| P18760 | Cofilin-1 | Cofilin-1 | 14 | 58.1 | 3.27E+08 | 24.58 | Cytoskeleton | IAV1, RSV, HIV1, HIV2, HSV |
| P35762 | CD81 | CD81 antigen C | 4 | 30.9 | 2.99E+08 | 25.81 | Membrane | IAV1, HCV, MEV, HIV1, HIV2,VV |
| P61205 | ARF3 | ADP-ribosylation factor 3 | 4 | 57.5 | 2.93E+08 | 20.60 | Golgi | HSV |
| P0CG50 | Ubc | Polyubiquitin-C | 7 | 73.6 | 2.41E+08 | 82.55 | Nucleus, cytoplasm | IAV2, RSV, HIV1, VSV, JUNV |
| O08992 | Syntenin-1 | Syntenin-1 | 8 | 40.5 | 1.77E+08 | 32.38 | Membrane, cytoplasm, cytoskeleton, nucleus, ER, junction | HIV2 |
| Q9WVE8 | PACSIN 2 | Protein kinase C and casein kinase substrate in neurons protein 2 | 13 | 27 | 1.45E+08 | 55.83 | Cytosol, endosome, nucleus, caveola | |
| P10852 | Slc3a2 | 4F2 cell-surface antigen heavy chain | 19 | 39.7 | 1.03E+08 | 58.34 | Membrane | RSV, HIV1, VSV |
| O35566 | CD151 | CD151 antigen | 5 | 17 | 8.78E+07 | 28.25 | Membrane | RVFV |
| P41731 | CD63 | CD63 antigen | 3 | 10 | 8.72E+07 | 26.78 | Membrane | HCV |
| Q9R0P5 | Destrin | Destrin | 6 | 41.2 | 8.62E+07 | 18.52 | Cytoskeleton | IAV1 |
| P60766 | CDC42 | Cell division control protein 42 homolog | 6 | 42.4 | 8.24E+07 | 21.26 | Membrane, cytoskeleton, cytoplasm | IAV2, HIV1, SARS |
| Q9D8B3 | CHMP4B | Charged multivesicular body protein 4b | 7 | 41.1 | 6.07E+07 | 24.94 | Late endosome, cytosol | HAV |
| P63037 | HSP40 | DnaJ homolog subfamily A member 1 | 11 | 36.5 | 5.91E+07 | 44.87 | Membrane, nucleus, ER, mitochondria, cytoplasm | HIV2 |
| P16858 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 9 | 30.6 | 5.73E+07 | 38.65 | Cytoskeleton, cytosol, nucleus | IAV1, RSV, HIV1, HIV2, HIV3, RVFV, IBV, ASFV, KSHV |
| P63242 | EIF5a | Eukaryotic translation initiation factor 5A-1 | 6 | 45.5 | 5.71E+07 | 16.83 | Nucleus, ER | KSHV |
| Q4VAE6 | RhoA | Ras family member A | 5 | 28.5 | 5.67E+07 | 21.80 | Nucleus, ER, cytoplasm | IBV |
| O54946 | HSJ-2 | DnaJ homolog subfamily B member 6 | 6 | 21.9 | 5.16E+07 | 39.81 | Nucleus | |
| P63024 | VAMP3 | Vesicle-associated membrane protein 3 | 2 | 32 | 4.84E+07 | 11.48 | Membrane | |
| Q9WU78 | ALIX | Programmed cell death 6-interacting protein | 30 | 38.9 | 4.84E+07 | 96.31 | Cytoskeleton, cytosol | HAV, HIV1, VSV, HSV, JUNV |
| P63101 | 14–3-3 ζ/θ | 14–3-3 protein zeta/delta | 6 | 37.6 | 4.66E+07 | 27.77 | Cytoskeleton | HIV1, HSV, KSHV, SARS |
| Q62167 | DDX3X | ATP-dependent RNA helicase DDX3X | 5 | 41.2 | 4.13E+07 | 73.10 | Mitochondria, nucleus | HSV, JUNV, SARS |
| P99024 | Tubulin β-5 | Tubulin beta-5 chain | 4 | 35.8 | 4.05E+07 | 49.67 | Cytoplasm, cytoskeleton, microtubules | IAV1, RSV, HIV1, VSV, ASFV |
| P11499 | HSP90β | Heat shock protein HSP 90-beta | 15 | 34.4 | 3.94E+07 | 83.28 | Membrane, cytoplasm, nucleus | RSV, HAV, HIV1, VSV, RVFV, IBV, KSHV, SARS |
| Q91ZR2 | SNX | Sorting nexin-18 | 10 | 20 | 3.76E+07 | 67.79 | Membrane | |
| Q61187 | TSG101 | Tumor susceptibility gene 101 protein | 7 | 19.2 | 3.40E+07 | 44.12 | Endosome, nucleus | HIV1, VSV, JUNV |
| P17182 | ENO1 | Alpha-enolase | 9 | 24 | 3.17E+07 | 47.14 | Membrane, cytoplasm | IAV1, HAV, MEV, HIV1, HIV2, VSV, IBV, ASFV, KSHV |
| Q9DB34 | CHMP2A | Charged multivesicular body protein 2a | 5 | 18 | 3.09E+07 | 25.13 | Cytoplasm, late endosomes | |
| Q9Z127 | SLC7 | Large neutral amino acids transporter small subunit 1 | 5 | 8.8 | 2.94E+07 | 55.87 | Membrane, cytosol | |
| P35278 | Rab5C | Ras-related protein Rab-5C | 4 | 19.7 | 2.91E+07 | 25.35 | Membrane, endosomes | HAV, HIV1, HIV2, ASFV, HSV |
| Q3UFR4 | SLC1 | Amino acid transporter | 8 | 18.9 | 2.60E+07 | 58.36 | Membrane | |
| Q99J93 | IFITM | Interferon-induced transmembrane protein 2 | 2 | 23.6 | 2.30E+07 | 15.74 | Membrane | |
| Q9D1C8 | VPS28 | Vacuolar protein sorting-associated protein 28 homolog | 7 | 36.7 | 2.19E+07 | 25.45 | Endosomes | HIV2 |
| P51150 | RAB7A | Ras-related protein Rab-7a | 6 | 32.9 | 1.88E+07 | 23.49 | Late endosomes | HAV, HIV1, RVFV, HSV, KSHV, JUNV |
| P06837 | GAP43 | Neuromodulin | 7 | 49.8 | 1.79E+07 | 23.63 | Membrane, synapses | |
| P26040 | Ezrin | Ezrin | 8 | 18.3 | 1.77E+07 | 69.41 | Cytoskeleton, cytosol | IAV2, HIV1 |
| P60335 | PCBP1 | Poly(rC)-binding protein 1 | 4 | 18.5 | 1.64E+07 | 37.5 | Nucleus, cytoplasm | HIV1, HIV2, SARS |
| P16045 | Galectin-1 | Galectin-1 | 3 | 28.1 | 1.61E+07 | 14.87 | Cell surface, extracellular matrix | HIV2 |
| P62331 | ARF6 | ADP-ribosylation factor 6 | 3 | 21.1 | 1.11E+07 | 20.08 | Cytoplasm, cytosol, early endosomes | |
| Q91YD9 | nWASP | Neural Wiskott-Aldrich syndrome protein | 5 | 12.6 | 9.01E+06 | 54.27 | Cytoplasm, cytoskeleton, nucleus | |
| P61089 | Ube2 | Ubiquitin-conjugating enzyme E2 N | 5 | 33.6 | 6.31E+06 | 17.14 | Cytoplasm, nucleus | |
| B2RRX1 | β-actin | Beta-actin | 3 | 54.1 | 4.96E+06 | 41.74 | Membrane, cytoskeleton, cytosol | IAV1, HIV1, HIV2, VSV, RVFV, IBV, HSV, KSHV |
| Q8R0J7 | VPS37B | Vacuolar protein sorting-associated protein 37B | 4 | 17.9 | 4.30E+06 | 31.06 | Late endosomes, cytoplasm | |
| P46467 | VPS4B | Vacuolar protein sorting-associated protein 4B | 7 | 18.2 | 3.62E+06 | 49.42 | Late endosomes | HAV |
| P68040 | RACK1 | Receptor of activated protein C kinase 1 | 4 | 11.7 | 2.58E+06 | 35.08 | Membrane, cytoplasm, nucleus |
aThe number of unique peptides correspond to the maximal values among the three biological replications.
bThe percentages of sequence coverage based on peptides with unique sequences.
cAverage abundance expressed by iBAQ calculated from three separate determinations.
dSubcellular location was investigated using the Uniprot database. Endoplasmic reticulum, ER.
eVirus names: influenza A virus (Shaw et al.2008 for IAV1, Mindaye et al. 2017 for IAV2); HAV: hepatitis A virus (McKnight et al.2017); MEV: measles virus (Sviben et al.2018); HIV: human immunodeficiency virus (Linde et al.2013 for HIV1, Chertova et al. 2006 for HIV2, Saphire et al. 2006 for HIV3); ASFV: African swine fever virus (Alejo et al.2018); RSV: respiratory syncytial virus (Radhakrishnan et al.2010)); VSV: vesicular stomatitis virus (Moerdyk-Schauwecker et al.2009); RVFV: rift Valley fever virus (Nuss et al.2014); HSV: herpes simplex virus type 1 (Stegen et al.2013); KSHV: Kaposi’s sarcoma-associated herpesvirus (Zhu et al.2005); HCV: hepatitis C virus (Lussignol et al.2016); VV: vaccinia virus (Krauss et al.2002); IBV: infection bronchitis virus (Kong et al.2010); JUNV: Junin virus (Ziegler et al.2018); SARS: severe acute respiratory syndrome (Neuman et al.2008).
The 49 cellular proteins had significant abundance values ranging from 1.72E+09 to 2.58E+06, with sequence coverages of between 73.6% and 8.8%. Table 1 also lists their subcellular locations and incorporation into other virions as reported elsewhere. According to the Uniprot Knowledge database, many of the proteins were localized at the cell membrane, cytoplasm and actin cytoskeleton. It is worth noting that more than half of these host proteins (34/49) were also found in the other viruses within 11 viral families (Table 1). Of them, CD9, CD81, cofilin-1, cyclophilin A, GAPDH, enolase, HSC70, HSP90β and RAB5C were the most frequently identified in the virions of different viruses, indicating that they were widely recruited by these viruses to benefit their replicative cycles (Table 1). For example, the presence of cyclophilin A in the virions aids capsid stabilization in both influenza virus and HIV by interacting with capsid protein in the early stage of the viral replication (Liu et al.2009, 2016). As reviewed in a recent publication, heat shock protein 90 is a crucial host factor required by many viruses for multiple phases of their life cycle (Wang et al.2017). These suggest that many viruses, especially enveloped ones, might utilize cellular proteins to complete their replicative cycle.
Validation of Cellular Proteins Incorporated Into Purified RABV Particles
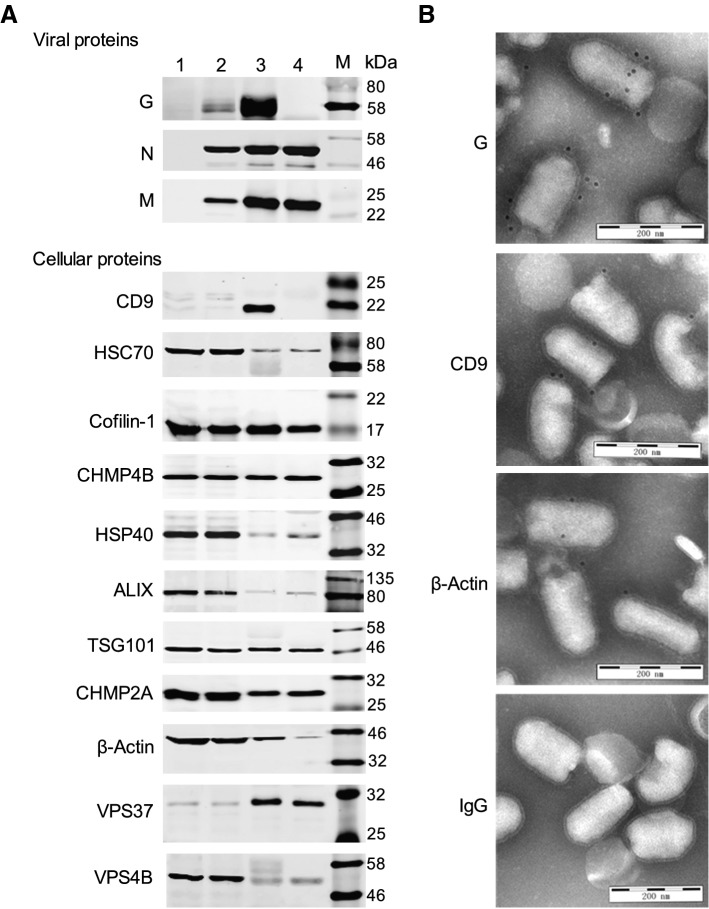
To justify the claim of incorporation, 3 viral proteins and 11 host proteins from Table 1 of different abundances were analyzed by Western blotting and immunogold labeling. A protease protection assay was performed using ProK to degrade any proteins on the surface of the virions, while internal proteins were protected by the lipid envelope. Absence of the viral glycoprotein and the presence of the viral nucleoprotein (N) and matrix protein (M) verified that the ProK digestion was 100% efficient (Fig. 3A). Immunoblotting analysis revealed that HSC70, cofilin, CHMP4B, HSP40, ALIX, TSG101, CHMP2A, VPS37 and VPS4B were all present in the ProK-treated virions, indicating their locations within the virion (Fig. 3A). Extracts from uninfected and infected N2a cells were included as controls to confirm the reactivity of the antibodies and size of the proteins. CD9, however, was completely absent from ProK-treated virions while β-actin was reduced by about 50%, indicating that CD9 was present only on the virion surface, while β-actin was located both inside and outside (Fig. 3A). Immunogold labeling further confirmed the incorporation of these host proteins into the virion surface. As shown in Fig. 3B, gold particle labelling identified CD9 and β-actin as well as the viral G protein on the surface of virus particles. Although it was unrealistic to validate all 49 cellular proteins, confirmation of the incorporation of all 11 proteins demonstrated the high probability of incorporation of all 49 proteins into rabies virions. Clearly, the potential roles of virion-packaged host proteins during RABV infection merit further investigation.
Fig. 3.
Validation of cellular proteins incorporated into the RABV virions. A Detection of 3 viral proteins and 11 cellular proteins by Western blotting from: mock-infected N2a cells (lane 1); CVS-11-infected N2a cells (lane 2); purified virions (lane 3); and ProK-treated virions (lane 4). B Images of immunogold labeling of purified RABV virions targeting the following proteins: RABV G protein, cellular CD9 and β-actin. IgG, included as a control for unrelated immunogold-labeled antibody, did not show colloidal gold particles on the virions.
Analysis of Virion-Packaged Host Proteins Involved in RABV Infection
As is well known, many viruses hijack cellular machinery via host–pathogen interactions to function in their replicative cycles (Robinson et al.2018). To better understand the biological significance of the host proteins ending up in mature rabies virions, functional predictions of the 49 virion-packaged cellular proteins were performed according to the Gene Ontology Database. Results showed that they are grouped into 12 functional categories, including viral transport, protein localization, cytoskeleton organization, and transcription (Fig. 4, Supplementary Table S1). Some proteins had multiple functions, and therefore were classified into multiple categories. Worth noting is that 24 of the 49 proteins were associated with viral processes such as entry (Galectin-1, CD9, CD81, CD151, CD63, IFITM2), genome replication (PCBP1, HSC70, ARF3, CyPA, DDX3X, ENO1, IFITM2, Rack1), assembly (HSP90β), budding (ALIX, CHMP4B, CHMP2A, VPS4B, TSG101, VPS37B, VPS28), release (Rab-7A, cofilin-1) and spread (PACSIN2) (Fig. 4 and Supplementary Table S1).
Fig. 4.
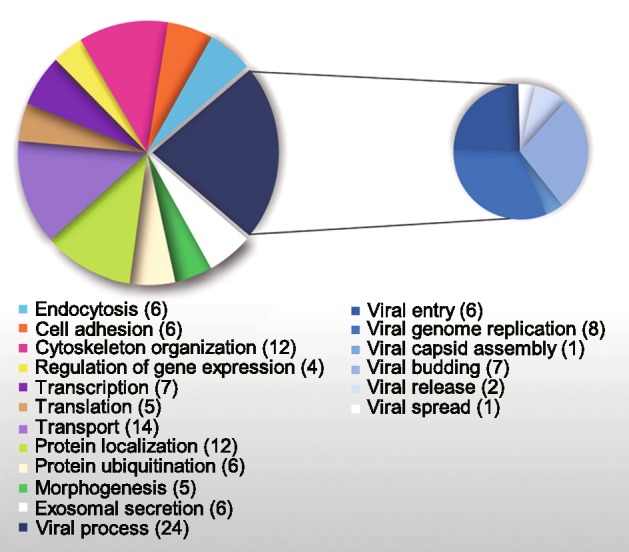
Gene Ontology classification of RABV virions-packaged host proteins.
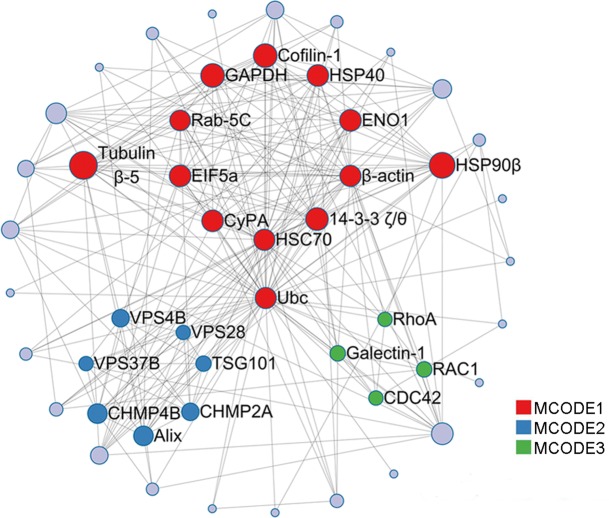
To identify clusters and interactions among the above host proteins, a protein–protein interaction (PPI) network was constructed by Metascape using the BIOGRID6, InWeb_IM7 and OmniPath8 databases (https://metascape.org/). Additionally, the Molecular Complex Detection (MCODE) algorithm was applied to screen densely connected protein groups in the network and to annotate the biological functions of each group. As a result, the 49 cellular proteins were found to form a PPI network with 24 nodes and 25 edges, as shown in Fig. 5, in which three significant modules were identified by the MCODE algorithm to be linked to 9 cell processes (Supplementary Table S2). Enrichment of the MCODE 2-related viral budding process was the most significant with the smallest p-value of 10−21.7, strongly implying the association of the candidate host proteins with viral budding through the endosomal sorting complex required for transport (ESCRT).
Fig. 5.
PPI map of host proteins. Application of the Molecular Complex Detection (MCODE) algorithm to identify densely connected network components. The 49 cellular proteins correspond to 24 nodes (red, blue, and green) and 25 edges (gray) in PPI network, respectively. Three significant MCODEs are displayed on the map by coloring the corresponding nodes.
Discussion
Our previous study identified 50 cellular proteins incorporated into virions of attenuated non-pathogenic RABV vaccine strain SRV9 propagated in epithelium cell line BHK-21 (Tu et al.2015). Although the number of host proteins identified in two studies was almost the same, their proteomic compositions were distinct. To understand the proteomic composition of pathogenic RABV virions infecting nervous cells the present study was conducted, resulting in identification of 49 cellular proteins incorporated into CVS-11 with 11 of them validated by Western blotting or immuno-electron microscopy. Of 49 cellular proteins identified in CVS-11 virions purified from infected N2a cells only 13 were identified in SRV9 virions purified from infected BHK-21 cells, which mainly included 4 cytoskeleton-related proteins (Cofilin-1,GAPDH, Tubulin β-5, β-actin) and 4 virus replication associated proteins (HSC70, ARF3, CyPA, ENO1). The rest 36 proteins were not identified in SRV9 virions. It is interesting to note that ESCRT proteins associated with viral budding identified on CVS-11 virions were not found on SRV9 virions. This suggests that pathogenic and avirulent vaccine RABVs likely incorporate distinct cellular proteins into their virions during budding and release from host cells.
Among the identified cellular proteins in CVS-11 virions, HSC70 and cofilin-1 had previously been reported to be involved in RABV infection. HSC70 protein was reported to interact with RABV leader RNA (leRNA), with its expression level dynamically regulated by RABV infection: down-regulated at an early stage with gradual up-regulation (Zhang et al.2017). Here we demonstrate that HSC70 was packaged inside the virions. Cofilin is known to play an essential role in actin cytoskeleton dynamics, and while its knockdown did not affect the expression of RABV proteins, virion release was inhibited (Zan et al.2016). RABV infection has been reported to up-regulate the expression of phosphorylated cofilin to facilitate actin polymerization for virus budding (Zan et al.2016). Here, we also identified the phosphorylation of cofilin in RABV-infected cells but the protein found on virions was not phosphorylated (data not shown), indicating that cofilin was likely de-phosphorylated when packed into the rabies virion during budding. In addition, PCBP1 was also identified as being incorporated into the rabies virion. PCBP1, an RNA-binding protein, has been reported to interact with the ORF57 gene of Kaposi’s sarcoma-associated herpesvirus (KSHV), being involved in regulation of the expression of both cellular and viral genes through the activated internal ribosome entry site (IRES) (Nishimura et al.2004), however, its function in RABV infection remains unknown. A previous study showed that its highly conserved isoform, PCBP2 (90% aa identity with PCBP1), increased the stability of RABV glycoprotein mRNA through selective interaction with its 3′ UTR (Palusa et al.2012). This suggests that PCBP1 incorporated into virions might act as a functional partner for gene expression and posttranscriptional regulation.
Many enveloped viruses utilize the host ESCRT for budding from the plasma membrane of infected cells (Votteler and Sundquist 2013). In this process viruses employ the ESCRT machinery by selectively using core consensus sequences PPxY, PS/TAP, YxxL and FPIV (in which x denotes any amino acid), of the virus-encoded late domain (L-domain), which results in recruitment of the proteins in ESCRT-I, ESCRT-II, ESCRT-III and VPS4 for viral budding and release (Chen and Lamb 2008). In rabies virus, the M protein contains two L-domains, PPEY and YVPL, which are found in an overlapping fashion (35PPEYVPL41) near its N terminus. Site-directed mutagenesis has confirmed that the PPEY motif is essential for efficient release of rabies virions (Wirblich et al.2008), but the cellular factors required for viral budding have yet to be determined. The PPPY motif has been reported to recruit TSG101 for the budding of Marburg VP40-induced virus-like particles its incorporation within (Urata et al.2007). Likewise, our study has provided evidence for the incorporation of TSG101 into the rabies virion during budding (Fig. 3A), presumably mediated via its binding to the PPEY motif of the M protein. In addition to TSG101, other downstream ESCRT members, VPS28 and VPS37 in ESCRT-I, CHMP2A and CHMP4B in ESCRT-III, and also VPS4B, have been found to be recruited into rabies virions (Fig. 3A and Supplementary Table S1). Elsewhere, ESCRT-III proteins CHMP2A, CHMP4B and VPS4 have been reported associated with the budding of HIV, equine infectious anemia virus (EIAV), and murine leukemia virus (Morita et al.2011; Sandrin and Sundquist 2013; Bartusch and Prange 2016). It is likely from our results, therefore, that these ESCRT proteins are also important for the effective budding and release of RABV. Another L-domain, YxxL, is present as YVPL in RABV M protein, but its function is unknown; however, EIAV uses this motif (as YPDL) within the late assembly domain of the Gag p9 protein to recruit cellular protein ALIX (another rabies virion component) for its budding (Chen et al.2005). Furthermore, ALIX binding to CHMP4 can recruit downstream ESCRT-III subunits (e.g., CHMP2A) and form the CHMP complex by polymerization, compressing the neck of budding viruses to facilitate fission and membrane scission (McCullough et al.2008; Johnson et al.2018). In addition, it is known that the AAA-type ATPase VPS4 A/B recruited by ESCRT-III is essential for disassembly of ESCRT-III spirals and is also likely to provide energy (Adell et al.2014). Here, VPS4B was identified in the RABV virion (Fig. 3A) and its function merits further investigation.
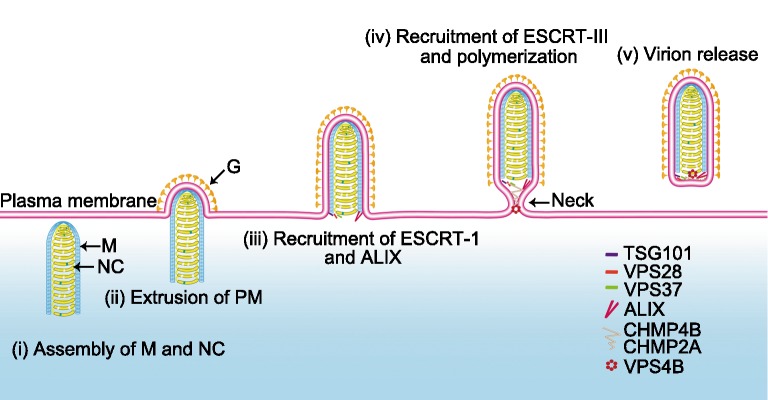
Based on the above analyses a process for RABV egress involving the ESCRT system is proposed in Fig. 6. Since cryo-EM has shown that M protein forms a helical layer it appears that viral morphogenesis takes place in specialized cytoplasmic areas. The genome-containing nucleocapsid (NC) is enfolded within a layer of M helix, which is transported to the plasma membrane (PM) containing the viral G protein, causing extrusion of the membrane to form the bullet-shaped budding site (Schnell et al.1998; Johnson et al.2018). Recruitment of ESCRT at the assembly sites is likely mediated by M protein by two pathways: (1) the PPEY motif recruiting TSG101, VPS28, VPS37 (ESCRTI); and (2) the YVPL motif recruiting ALIX. Downstream CHMP2A and CHMP4B (ESCRT-III) would then be recruited to form the budding neck by polymerization of the ESCRT-III proteins (Shen et al.2014). Since VPS4 can mediate disassembly/remodeling of CHMP2A (Van Engelenburg et al.2014), it may be that VPS4B can remodel CHMP2A and CHMP4B into a filamentous structure to drive membrane scission, ultimately resulting in the release of the mature RABV particles into a new round of infection.
Fig. 6.
Depictive process of rabies virus budding mediated by host ESCRT. (i) The assembled complex of M protein and nucleocapsid (NC) was transport to the plasma membrane (PM). (ii) Recruitment of RABV G protein by M protein to the PM and extrusion of PM. (iii) Recruitment of ESCRT-I proteins TSG101, VPS28, VPS37and ALIX by the two L-domains of M protein. (iv) Recruitment of ESCRT-III proteins CHMP4B and CHMP2A, resulting in polymerization, to form the contractile budding neck. (v) Reorganization of the ESCRT-III complex by VPS4B to release the RABV virions from host cells through membrane scission.
The present study has provided not only new insights into RABV virions but also a candidate protein list to further investigate the molecular mechanisms of RABV infection. They will also benefit screening for target proteins in future development of anti-rabies therapies.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was funded by the National Key Research and Development Plan (Grant No. 2016YFD0500401), National Natural Science Foundation of China (Grant No. 31402214) and China Postdoctoral Science Foundation (Grant No.2014M552638).
Author Contributions
YZ and CT designed the experiments. YZ, YW and YF carried out the experiments. ZT provided specific RABV protein polyclonal antibody. ZL recorded the Cryo-image. YZ wrote the paper. CT and ZL checked and finalized the manuscript. All authors read and approved the final manuscript.
Compliance with Ethics Standards
Conflict of interest
The authors declare no conflict of interest.
Animal and Human Rights Statement
The animal experiments performed in this study were approved by the Administrative Committee on Animal Welfare of the Institute of Military Veterinary, Academy of Military Sciences, China (JSY-DW-2016–02).
Contributor Information
Zhiyong Lou, Email: louzy@mail.tsinghua.edu.cn.
Changchun Tu, Email: changchun_tu@hotmail.com.
References
- Adell MA, Vogel GF, Pakdel M, Muller M, Lindner H, Hess MW, Teis D. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J Cell Biol. 2014;205:33–49. doi: 10.1083/jcb.201310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrne E, Molzahn L, Glatter T, Schmidt A. Critical assessment of proteome-wide label-free absolute abundance estimation strategies. Proteomics. 2013;13:2567–2578. doi: 10.1002/pmic.201300135. [DOI] [PubMed] [Google Scholar]
- Alejo A, Matamoros T, Guerra M, Andres G. A proteomic atlas of the African swine fever virus particle. J Virol. 2018;92:18. doi: 10.1128/JVI.01293-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartusch C, Prange R. ESCRT requirements for murine leukemia virus release. Viruses. 2016;8:103. doi: 10.3390/v8040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunker K, Mollentze N. Rabies virus. Trends Microbiol. 2018;26:886–887. doi: 10.1016/j.tim.2018.07.001. [DOI] [PubMed] [Google Scholar]
- Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: Can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Vincent O, Jin J, Weisz OA, Montelaro RC. Functions of early (AP-2) and late (AIP1/ALIX) endocytic proteins in equine infectious anemia virus budding. J Biol Chem. 2005;280:40474–40480. doi: 10.1074/jbc.M509317200. [DOI] [PubMed] [Google Scholar]
- Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, 2nd, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Davis BM, Rall GF, Schnell MJ. Everything you always wanted to know about rabies virus (but were afraid to ask) Annu Rev Virol. 2015;2:451–471. doi: 10.1146/annurev-virology-100114-055157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean DJ, Abelseth MK. Laboratory techniques in rabies: the fluorescent antibody test. Monogr Ser World Health Organ. 1973;23:73–84. [PubMed] [Google Scholar]
- Ge P, Tsao J, Schein S, Green TJ, Luo M, Zhou ZH. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science. 2010;327:689–693. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P, Krell T, Chevalier M, Vaysse C, Adam O, Ronzon F, Marco S. Three dimensional morphology of rabies virus studied by cryo-electron tomography. J Struct Biol. 2011;176:32–40. doi: 10.1016/j.jsb.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Huang HJ, Liu CW, Huang XH, Zhou X, Zhuo JC, Zhang CX, Bao YY. Screening and functional analyses of nilaparvata lugens salivary proteome. J Proteome Res. 2016;15:1883–1896. doi: 10.1021/acs.jproteome.6b00086. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Bleck M, Simon SM. Timing of ESCRT-III protein recruitment and membrane scission during HIV-1 assembly. Elife. 2018;7:e36221. doi: 10.7554/eLife.36221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Xue C, Ren X, Zhang C, Li L, Shu D, Bi Y, Cao Y. Proteomic analysis of purified coronavirus infectious bronchitis virus particles. Proteome Sci. 2010;8:29. doi: 10.1186/1477-5956-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss O, Hollinshead R, Hollinshead M, Smith GL. An investigation of incorporation of cellular antigens into vaccinia virus particles. J Gen Virol. 2002;83:2347–2359. doi: 10.1099/0022-1317-83-10-2347. [DOI] [PubMed] [Google Scholar]
- Lahaye X, Vidy A, Fouquet B, Blondel D. Hsp70 protein positively regulates rabies virus infection. J Virol. 2012;86:4743–4751. doi: 10.1128/JVI.06501-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde ME, Colquhoun DR, Ubaida Mohien C, Kole T, Aquino V, Cotter R, Edwards N, Hildreth JE, Graham DR. The conserved set of host proteins incorporated into HIV-1 virions suggests a common egress pathway in multiple cell types. J Proteome Res. 2013;12:2045–2054. doi: 10.1021/pr300918r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sun L, Yu M, Wang Z, Xu C, Xue Q, Zhang K, Ye X, Kitamura Y, Liu W. Cyclophilin A interacts with influenza A virus M1 protein and impairs the early stage of the viral replication. Cell Microbiol. 2009;11:730–741. doi: 10.1111/j.1462-5822.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- Liu C, Perilla JR, Ning J, Lu M, Hou G, Ramalho R, Himes BA, Zhao G, Bedwell GJ, Byeon IJ, Ahn J, Gronenborn AM, Prevelige PE, Rousso I, Aiken C, Polenova T, Schulten K, Zhang P. Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat Commun. 2016;7:10714. doi: 10.1038/ncomms10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Green TJ, Zhang X, Tsao J, Qiu S. Conserved characteristics of the rhabdovirus nucleoprotein. Virus Res. 2007;129:246–251. doi: 10.1016/j.virusres.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussignol M, Kopp M, Molloy K, Vizcay-Barrena G, Fleck RA, Dorner M, Bell KL, Chait BT, Rice CM, Catanese MT. Proteomics of HCV virions reveals an essential role for the nucleoporin Nup98 in virus morphogenesis. Proc Natl Acad Sci USA. 2016;113:2484–2489. doi: 10.1073/pnas.1518934113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc Natl Acad Sci USA. 2008;105:7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight KL, Xie L, Gonzalez-Lopez O, Rivera-Serrano EE, Chen X, Lemon SM. Protein composition of the hepatitis A virus quasi-envelope. Proc Natl Acad Sci USA. 2017;114:6587–6592. doi: 10.1073/pnas.1619519114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindaye ST, Ilyushina NA, Fantoni G, Alterman MA, Donnelly RP, Eichelberger MC. Impact of influenza a virus infection on the proteomes of human bronchoepithelial cells from different donors. J Proteome Res. 2017;16:3287–3297. doi: 10.1021/acs.jproteome.7b00286. [DOI] [PubMed] [Google Scholar]
- Moerdyk-Schauwecker M, Hwang SI, Grdzelishvili VZ. Analysis of virion associated host proteins in vesicular stomatitis virus using a proteomics approach. Virol J. 2009;6:166. doi: 10.1186/1743-422X-6-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman BW, Joseph JS, Saikatendu KS, Serrano P, Chatterjee A, Johnson MA, Liao L, Klaus JP, Yates JR, III, Wuthrich K, Stevens RC, Buchmeier MJ, Kuhn P. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Ueda K, Guwanan E, Sakakibara S, Do E, Osaki E, Yada K, Okuno T, Yamanishi K. A posttranscriptional regulator of Kaposi's sarcoma-associated herpesvirus interacts with RNA-binding protein PCBP1 and controls gene expression through the IRES. Virology. 2004;325:364–378. doi: 10.1016/j.virol.2004.04.041. [DOI] [PubMed] [Google Scholar]
- Nuss JE, Kehn-Hall K, Benedict A, Costantino J, Ward M, Peyser BD, Retterer CJ, Tressler LE, Wanner LM, McGovern HF, Zaidi A, Anthony SM, Kota KP, Bavari S, Hakami RM. Multi-faceted proteomic characterization of host protein complement of Rift Valley fever virus virions and identification of specific heat shock proteins, including HSP90, as important viral host factors. PLoS ONE. 2014;9:e93483. doi: 10.1371/journal.pone.0093483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura A, Harty RN. Rabies virus assembly and budding. Adv Virus Res. 2011;79:23–32. doi: 10.1016/B978-0-12-387040-7.00002-0. [DOI] [PubMed] [Google Scholar]
- Palusa S, Ndaluka C, Bowen RA, Wilusz CJ, Wilusz J. The 3' untranslated region of the rabies virus glycoprotein mRNA specifically interacts with cellular PCBP2 protein and promotes transcript stability. PLoS ONE. 2012;7:e33561. doi: 10.1371/journal.pone.0033561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Yeo D, Brown G, Myaing MZ, Iyer LR, Fleck R, Tan BH, Aitken J, Sanmun D, Tang K, Yarwood A, Brink J, Sugrue RJ. Protein analysis of purified respiratory syncytial virus particles reveals an important role for heat shock protein 90 in virus particle assembly. Mol Cell Proteom. 2010;9:1829–1848. doi: 10.1074/mcp.M110.001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C, Vasishtan D, Prazak V, Ghanem A, Conzelmann KK, Rumenapf T. Cryo EM structure of the rabies virus ribonucleoprotein complex. Sci Rep. 2019;9:9639. doi: 10.1038/s41598-019-46126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M, Schor S, Barouch-Bentov R, Einav S. Viral journeys on the intracellular highways. Cell Mol Life Sci. 2018;75:3693–3714. doi: 10.1007/s00018-018-2882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrin V, Sundquist WI. ESCRT requirements for EIAV budding. Retrovirology. 2013;10:104. doi: 10.1186/1742-4690-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire AC, Gallay PA, Bark SJ. Proteomic analysis of human immunodeficiency virus using liquid chromatography/tandem mass spectrometry effectively distinguishes specific incorporated host proteins. J Proteome Res. 2006;5:530–538. doi: 10.1021/pr050276b. [DOI] [PubMed] [Google Scholar]
- Schnell MJ, Buonocore L, Boritz E, Ghosh HP, Chernish R, Rose JK. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 1998;17:1289–1296. doi: 10.1093/emboj/17.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P. Cellular proteins in influenza virus particles. PLoS Pathog. 2008;4:e1000085. doi: 10.1371/journal.ppat.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QT, Schuh AL, Zheng Y, Quinney K, Wang L, Hanna M, Mitchell JC, Otegui MS, Ahlquist P, Cui Q, Audhya A. Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J Cell Biol. 2014;206:763–777. doi: 10.1083/jcb.201403108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegen C, Yakova Y, Henaff D, Nadjar J, Duron J, Lippe R. Analysis of virion-incorporated host proteins required for herpes simplex virus type 1 infection through a RNA interference screen. PLoS ONE. 2013;8:e53276. doi: 10.1371/journal.pone.0053276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sviben D, Forcic D, Halassy B, Allmaier G, Marchetti-Deschmann M, Brgles M. Mass spectrometry-based investigation of measles and mumps virus proteome. Virol J. 2018;15:160. doi: 10.1186/s12985-018-1073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z, Gong W, Zhang Y, Feng Y, Li N, Tu C. Proteomic analyses of purified particles of the rabies virus. Chin J Virol. 2015;31:209–216. [PubMed] [Google Scholar]
- Urata S, Noda T, Kawaoka Y, Morikawa S, Yokosawa H, Yasuda J. Interaction of Tsg101 with Marburg virus VP40 depends on the PPPY motif, but not the PT/SAP motif as in the case of Ebola virus, and Tsg101 plays a critical role in the budding of Marburg virus-like particles induced by VP40, NP, and GP. J Virol. 2007;81:4895–4899. doi: 10.1128/JVI.02829-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Engelenburg SB, Shtengel G, Sengupta P, Waki K, Jarnik M, Ablan SD, Freed EO, Hess HF, Lippincott-Schwartz J. Distribution of ESCRT machinery at HIV assembly sites reveals virus scaffolding of ESCRT subunits. Science. 2014;343:653–656. doi: 10.1126/science.1247786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Velasco NM, Garcia-Murria MJ, Sanchez Del Pino MM, Mingarro I, Martinez-Gil L. Proteomic composition of Nipah virus-like particles. J Proteom. 2018;172:190–200. doi: 10.1016/j.jprot.2017.10.012. [DOI] [PubMed] [Google Scholar]
- Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013;14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jin F, Wang R, Li F, Wu Y, Kitazato K, Wang Y. HSP90: a promising broad-spectrum antiviral drug target. Arch Virol. 2017;162:3269–3282. doi: 10.1007/s00705-017-3511-1. [DOI] [PubMed] [Google Scholar]
- Wirblich C, Tan GS, Papaneri A, Godlewski PJ, Orenstein JM, Harty RN, Schnell MJ. PPEY motif within the rabies virus (RV) matrix protein is essential for efficient virion release and RV pathogenicity. J Virol. 2008;82:9730–9738. doi: 10.1128/JVI.00889-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Wang J, Zhu D, Gao Q, Chen W, Tang H, Wang J, Zhang X, Liu H, Rao Z, Wang X. Cryo-EM structure of a herpesvirus capsid at 3.1 A. Science. 2018;360:3–9738. doi: 10.1126/science.aao7283. [DOI] [PubMed] [Google Scholar]
- Zan J, An ST, Mo KK, Zhou JW, Liu J, Wang HL, Yan Y, Liao M, Zhou JY. Rabies virus inactivates cofilin to facilitate viral budding and release. Biochem Biophys Res Commun. 2016;477:1045–1050. doi: 10.1016/j.bbrc.2016.07.030. [DOI] [PubMed] [Google Scholar]
- Zhang R, Liu C, Cao Y, Jamal M, Chen X, Zheng J, Li L, You J, Zhu Q, Liu S, Dai J, Cui M, Fu ZF, Cao G. Rabies viruses leader RNA interacts with host Hsc70 and inhibits virus replication. Oncotarget. 2017;8:43822–43837. doi: 10.18632/oncotarget.16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FX, Chong JM, Wu L, Yuan Y. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J Virol. 2005;79:800–811. doi: 10.1128/JVI.79.2.800-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler CM, Eisenhauer P, Kelly JA, Dang LN, Beganovic V, Bruce EA, King BR, Shirley DJ, Weir ME, Ballif BA, Botten J. A Proteomics Survey of Junin Virus Interactions with Human Proteins Reveals Host Factors Required for Arenavirus Replication. J Virol. 2018;92:17. doi: 10.1128/JVI.01565-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.