Abstract
The newly emerging human pathogen, Middle East respiratory syndrome coronavirus (MERS-CoV), contains a macro domain in the highly conserved N-terminal region of non-structural protein 3. Intense research has shown that macro domains bind ADP-ribose and other derivatives, but it still remains intangible about their exact function. In this study we report the preliminary structural analysis through solution NMR spectroscopy of the MERS-CoV macro domain. The near complete NMR assignments of MERS-CoV macro domain provide the basis for subsequent structural and biochemical investigation in the context of protein function.
Keywords: Viral macro domain, MERS-CoV, ADP-ribose binding module, NMR spectroscopy
Biological context
Middle East Respiratory Syndrome Coronavirus (MERS-CoV) is a kind of RNA virus belongs to genus Betacoronavirus in Coronavaridae of enveloped, which is a positive-stranded RNA virus and becomes a novel zoonotic virus in recent years (de Groot et al. 2013). MERS-CoV transmits respiratory tract infections from original host, the bat, to human by intermediate host, the camel, although the detail viral life cycle is still unknown (Han et al. 2015). The first human case was reported in 2012 at Saudi Arabia about a man with severe acute pneumonia and secondary renal failure (Zaki et al. 2012). Through close contact to infected patients, the human-to-human transmissions of the MERS-CoV virus spread very quickly. Since the first report, MERS-CoV has caused over 400 people died among 1100 cases in 23 countries. There is no efficient specific drug to use for this newly human pathogen yet, thus drug development is the emergency to conquer for the researchers.
The macro domain, a protein module specifically recognizing ADP ribose (Karras et al. 2005), could be found in many bacteria, archaea, plants and vertebrates, suggesting their evolutionary conservation (Han et al. 2011). Some macro domains possess phosphatase activity towards ADP-ribose-1″-phosphate, which is a by-product of tRNA splicing (Putic et al. 2005). Furthermore, macro domains are found in nonstructural proteins (nsps) of several positive-strand RNA viruses, including alphaviruses, rubella virus, hepatitis E virus (HEV), and coronaviruses [including the severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV)] (Neuvonen and Ahola 2009; Fehr and Perlman 2015; Hurst-Hess et al. 2015; Parvez 2015). The enzymatic activity of viral macro domain is dispensable for coronavirus replication (Putic et al. 2005), however it seems to play a role in the pathogenesis of mouse hepatitis virus infection (Eriksson et al. 2008). In addition, viral macro proteins may act via ADP-ribose binding to influence the cellular macro domains-regulated pathways either to promote virus replication or to inhibit host responses directed against the virus (Neuvonen and Ahola 2009). Since the biochemical, enzymatic and structural analysis of MERS-CoV macro domain is not available, here we present the expression, purification, and chemical shift assignments of MERS-CoV macro domain. The NMR chemical shift assignments serve as the basis for further structural characterization and ligand screening, which provides insight into the conformational properties of this domain in solution and contributes to understanding its function.
Materials and methods
Cloning, overexpression and purification of MERS-CoV macro domain
The DNA sequence containing MERS-CoV macro domain (aa. 1110–1274) was chemical synthesized and cloned into NdeI/XhoI site of pET-28a(+) vector system (Novagen). Bacteria of the E. coli strain BL21(DE3) transformed with the pET-28a (+)-macro domain plasmid were grown in 4 liters of LB medium at 37 °C until the absorbance at 600 nm reach 1.0. Cells were then harvested by centrifugation and re-suspended in one liter of M9 minimal medium supplemented with 1 g/L of 15NH4Cl and 2 g/L of 13C6-glucose (Cambridge Isotope Laboratories). Isopropyl-β-D-thiogalactoside (IPTG, 1 mM) was added to induce His-tagged protein for 20 h at 16 °C. The cell culture was harvested by centrifugation at 6000 rpm.
For purification, cell pellets were re-suspended in 50 mL buffer containing 25 mM sodium phosphate and 100 mM NaCl at pH 7.0 and then disrupted by sonication for 20 min. The cell extract was clarified by centrifugation at 12,500 rpm for 30 min at 4 °C to remove debris. The supernatant was then applied to Ni–NTA column (GE, Healthcare) equilibrated with the same re-suspension buffer, and His-tagged protein was eluted with 200 mM imidazole. The purified His-tagged macro domain was then digested with thrombin for 6 h at 16 °C to remove the His-tag. Finally, macro domain protein (20 kDa, with extra Gly, Ser, His and Met at N-terminus) was purified using size-exclusion Superdex75 XK 16/60 column (GE, Healthcare). The purified protein was concentrated to 0.1–0.5 mM in 20 mM sodium phosphate (pH 6.5) and 100 mM or 150 mM NaCl for NMR structural studies.
NMR spectroscopy
All NMR experiments were carried out at 293 K on Bruker Avance 600 MHz NMR or 800 MHz spectrometers equipped with 5 mm triple resonance cryoprobe and Z-gradient. The data was acquired and processed using the software Topspin2.1 (Bruker, Germany) and further analyzed using Sparky, version 3.114 (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco), following the procedures as described previously (Yang et al. 2010; Chen et al. 2014). 1H chemical shifts were externally referenced to 0 ppm of 2,2-dimethyl-2-silapentane-5-sulfonate, whereas 13C and 15N chemical shifts were indirectly referenced according to IUPAC recommendations (Markley et al. 1998). Protein backbone resonance assignments were based on standard triple resonance experiments (Sattler et al. 1999): HNCACB, CBCA(CO)NH, HNCO and HN(CA)CO. Aliphatic side-chain assignments were primarily done by HCCH-TOCSY and HCCH-COSY with the help of HCC(CO)NH and HBHA(CO)NH experiments.
Assignments extension and data deposition
The backbone resonance assignments were nearly complete. Figure 1 illustrates the 2D (1H–15N) HSQC spectrum and assignments of the amide resonances. Except for the first four amino acids on N-terminal (Gly−2, Ser−1, His0 and Met1) and six Proline residues (Pro1, Pro72, Pro96, Pro118, Pro123, and Pro134), amides of all other residues (158 out of 168) have been assigned under the experimental conditions (pH 6.5 at 293 K). Among these 158 residues, all other backbone resonances (1Hα, 13Cα, 13Cβ and 13C) are 100 % completed. Completeness of 1H resonances assignment, including side-chain, calculated by CYANA3.9 (Güntert 2004) is 86.7 %. Secondary structure elements of MERS-CoV macro domain were identified by calculating the chemical shift deviations of the Cα(ΔδCα) and Cβ(ΔδCβ) from the random coil values and was corroborated by analysis of the chemical shift data using the program TALOS+ (Shen et al. 2009). Positive and negative values of the difference between ΔδCα and ΔδCβ correspond to α-helix and β-sheet secondary structure, respectively and correlated well with TALOS+ index (Fig. 2). Six helices and seven β-strands could be deduced for MERS-CoV moacro domain protein based on the secondary chemical shift analysis, which results in residues 21–27, 47–54. 58–69, 104–115, 134–144 and 156–162 in α-helices and residues 5–10, 14–19, 32–25, 78–81, 89–94, 119–124 and 148–153 forming β-sheets. The resonance assignments have been deposited to the BioMagResBank (http://www.bmrb.wisc.edu/) under the accession number 26657.
Fig. 1.
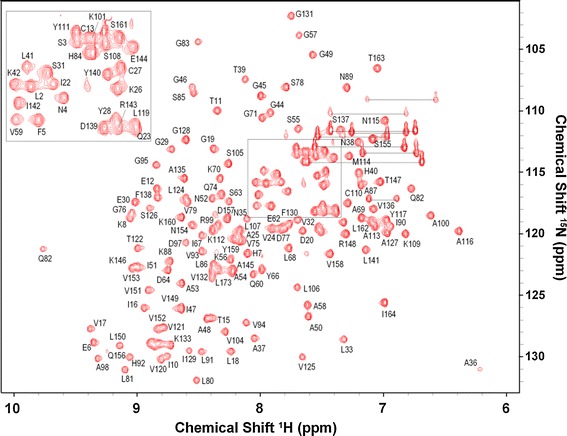
1H–15N HSQC spectrum of MERS-CoV macro domain recorded at 600 MHz cryogenic-probe with pH 6.5 at 293 K. Backbone amide 1H and 15N cross peaks are shown. The horizontal lines connect pairs of the side chain NH2 protons from Asn and Gln residues
Fig. 2.
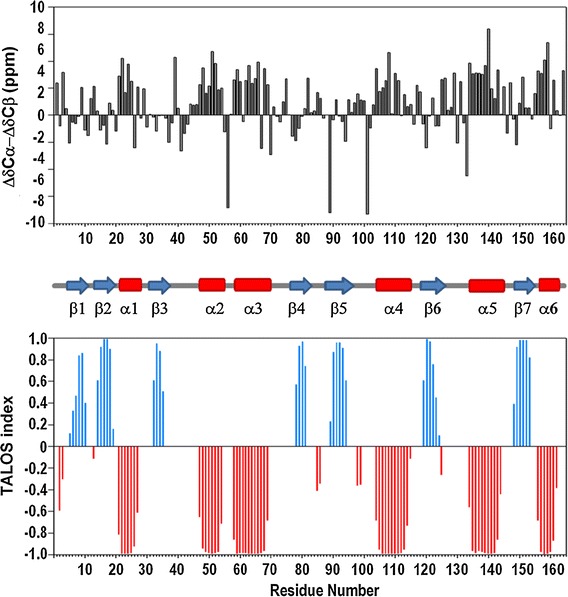
The secondary structure assessment of MERS-CoV macro domain. Upper The parameter ∆δCα − ∆δCβ shows the deviation of Cα and Cβ experimental values from the corresponding random coil values. Positive and negative values suggest α-helix and β-strand structure, respectively. Middle Chemical shift analysis resulting secondary structure elements of macro domain are represented. Lower TALOS+ index shows the prediction of secondary structure distribution based on backbone N, H, Cα, Hα, C and side chain Cβ chemical shift values. Negative and positive values suggest α-helix (in red) and β-strand (in blue) structure, respectively
Acknowledgments
The NMR spectra were obtained at High-Field Nuclear Magnetic Resonance Center (HF-NMRC) and GRC NMR Core Facility in Academia Sinica, Taiwan. This work was supported by the Ministry of Science and Technology, Taiwan (103-2113-M-002-009-MY2), and National Taiwan University (NTU-ERP-104R8600 and NTU-ICRP-104R7560-5) to C.-H. Hsu.
Footnotes
Yi-Ping Huang and Chao-Cheng Cho have contributed equally to this work.
Contributor Information
Chi-Fon Chang, Phone: +886-2-2787-1236, Email: chifon@gate.sinica.edu.tw.
Chun-Hua Hsu, Phone: +886-2-3366-4468, Email: andyhsu@ntu.edu.tw.
References
- Chen YD, Wu SH, Hsu CH. Backbone resonance assignments of the α sub-domain of Brevibacillus thermoruber Lon protease. Biomol NMR Assign. 2014;8:233–236. doi: 10.1007/s12104-013-9490-6. [DOI] [PubMed] [Google Scholar]
- de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RA, Galiano M, Gorbalenya AE, Memish ZA, Perlman S, Poon LL, Snijder EJ, Stephens GM, Woo PC, Zaki AM, Zambon M, Ziebuhr J. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson KK, Cevantes-Barragan L, Ludewig B, Thiel V. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1″-phosphatase, a viral function conserved in the alpha-like supergroup. J Virol. 2008;82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güntert P. Automated NMR protein structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- Han W, Li X, Fu X. The macro domain protein family: structure, functions, and their potential therapeutic implications. Mutat Res. 2011;727:86–103. doi: 10.1016/j.mrrev.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han HJ, Wen HL, Zhou CM, Chen FF, Luo LM, Liu JW, Yu XJ. Bats as reservoirs of severe emerging infectious diseases. Virus Res. 2015;205:1–6. doi: 10.1016/j.virusres.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst-Hess KR, Kuo L, Masters PS. Dissection of amino-terminal functional domains of murine coronavirus nonstructural protein 3. J Virol. 2015;89:6033–6047. doi: 10.1128/JVI.00197-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wuthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the Standardization of Data Bases of Protein and Nucleic Acid Structures Determined by NMR Spectroscopy. J Biomol NMR. 1998;12:1–23. doi: 10.1023/A:1008290618449. [DOI] [PubMed] [Google Scholar]
- Neuvonen M, Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J Mol Biol. 2009;385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez MK. The hepatitis E virus ORF1 ‘X-domain’ residues from a putative macrodomain protein/Appr-1″-pase catalytic-site, critical for viral RNA replication. Gene. 2015;566:47–53. doi: 10.1016/j.gene.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putic A, Filipowicz W, Hall J, Gorvalenva AE, Ziebuhr J. ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J Virol. 2005;79:12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc. 1999;34:93–158. doi: 10.1016/S0079-6565(98)00025-9. [DOI] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angle from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CJ, Chang CF, Huang LY, Chang YY, Shen TL, Hsu CH. Resonance assignments of human C35 (C17orf37) protein, a novel tumor biomarker. Biomol NMR Assign. 2010;4:191–193. doi: 10.1007/s12104-010-9218-9. [DOI] [PubMed] [Google Scholar]
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]


