TO THE EDITOR: Public sharing of research data is being widely promoted. Medical image files contain “metadata” such as the name of the participant, the date of the scan, and the identification number. Such data are typically removed (deidentified) before data sharing, but images of the face in magnetic resonance imaging (MRI) scans remain accessible.
We considered the possibility that a participant in a clinical trial or study may have deidentified MRI data in a publicly shared research database and that someone may seek to identify them by using face-recognition software to compare reconstructed images of the face from cranial MRI with photographs from social media or other sources. This identification would result in an infringement of privacy that could include diagnoses, cognitive scores, genetic data, biomarkers, results of other imaging, and participation in studies or trials (Fig. 1A).
Figure 1. Use of Face-Recognition Software to Identify Study Participants from MRI Scans.
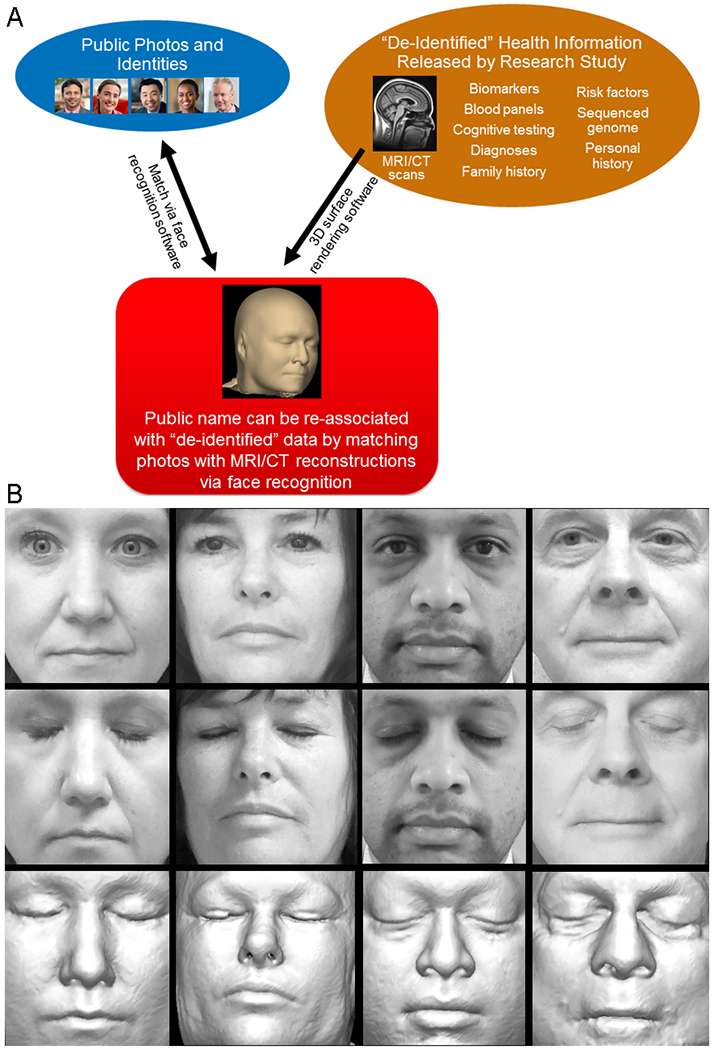
Panel A shows how face recognition hypothetically could be used to identify study participants. Three-dimensional (3D) rendering software can generate realistic facial reconstructions from otherwise deidentified imaging data, and face-recognition software can identify the participants by matching them to publicly available photographs of named persons. (The photos in the upper left are stock photos provided by the Mayo Clinic Division of Media Support Services and do not show participants in our study.) CT denotes computed tomography, and MRI magnetic resonance imaging. Panel B shows examples of the photos of participants in our study (top and middle rows) and corresponding facial reconstructions from structural MRI (bottom row). Photos in which the participants’ eyes were closed (middle) are shown for visual similarity, but we used photos in which the eyes were open (top) for software-based face recognition. These volunteers provided consent to allow publication of their photographs and MRI-based reconstructions. MRI reconstructions largely preserve shapes and relative sizes of facial features, which are used by automated recognition software, but unlike photographs they do not depict hair, lighting, or skin pigmentation and are subject to shape deformations because the participant is lying in a supine position or because of contact with ear padding or the MRI head-coil assembly.
To determine whether face-recognition software could identify individual persons from reconstructed facial images contained in cranial MRI scans, we recruited 84 volunteers between the ages of 34 and 89 years, stratified according to sex and decade of age, and photographed each participant’s face from five slightly varying angles. Each participant had undergone MRI of the head (three-dimensional fluid-attenuated inversion recovery [FLAIR] sequence, conducted with Siemens Prisma scanners) within the previous 3 months in association with their existing participation in the Mayo Clinic Study of Aging or in other studies conducted at the Mayo Clinic Alzheimer’s Disease Research Center.
From each MRI scan, we used an automated system to reconstruct a three-dimensional computer model of the participant’s face and create 10 two-dimensional photograph-like images with random lighting and views of each person (Fig. 1B). We tested publicly available automated face-recognition software (Microsoft Azure), which attempts to match a photograph of a face to a user-defined set of possible faces. We used the MRI-derived images to define a set of 84 possible faces to be recognized by the software, and we used the five actual photographs of each person as the photographs to be matched. For each photograph, the software returned a ranked list of the 50 closest matches from the set of 84 MRI-derived faces, with a confidence score for each. We summed these scores across each participant’s five photos to obtain a ranked list of matches for their set of photographs (a full description of our methods is provided in the Supplementary Appendix, available with the full text of this letter at NEJM.org).
For 70 of the 84 participants (83%), the software chose the correct MRI scan as the most likely match for their photos. The correct MRI scan was among the top five choices for 80 of 84 participants (95%).
In previous studies, 40% of human visual raters could match MRI face reconstructions to photographs with greater-than-chance success rates,1 and automated face-recognition software developed in 2008 could match 27.5% of computed tomography-based face reconstructions to the correct photographs.2 The 83% match rate in our study suggests that face recognition provides a possible means of reidentifying research participants from their cranial MRIs.
The current standard of removing only metadata in medical images may be insufficient to prevent reidentification of participants in research. Existing software for the removal or blurring of faces in medical images is rarely used,3 because these methods can reduce the quality of gray matter volume and cortical thickness measurements4 and may still not fully prevent reidentification.5 Further research is needed to develop improved deidentification methods for medical imaging that contains facial features.
Supplementary Material
Acknowledgments
Supported by grants from the National Institutes of Health (U01 AG006786, P50 AG016574, R01 AG034676, R37 AG011378, R01 AG041851, R01 NS097495, R01 AG056366, and U01 NS100620), the GHR Foundation, the Elsie and Marvin Dekelboum Family Foundation, the Alexander Family Alzheimer’s Disease Research Professorship of the Mayo Clinic, the Liston Award, the Schuler Foundation, and the Mayo Foundation for Medical Education and Research. The NVIDIA Corporation donated the Quadro P6000 GPU used in generating three-dimensional facial reconstructions for this research.
Footnotes
Disclosure forms provided by the authors are available with the full text of this letter at NEJM.org.
References
- 1.Prior FW, Brunsden B, Hildebolt C, et al. Facial recognition from volume-rendered magnetic resonance imaging data. IEEE Trans Inf Technol Biomed 2009;13:5–9. [DOI] [PubMed] [Google Scholar]
- 2.Mazura JC, Juluru K, Chen JJ, Morgan TA, John M, Siegel EL. Facial recognition software success rates for the identification of 3D surface reconstructed facial images: implications for patient privacy and security. J Digit Imaging 2012;25:347–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milchenko M, Marcus D. Obscuring surface anatomy in volumetric imaging data. Neuroinformatics 2013;11:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmes AJ, Hollinshead MO, O’Keefe TM, et al. Brain Genomics Superstruct Project initial data release with structural, functional, and behavioral measures. Sci Data 2015;2:150031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abramian D, Eklund A. Refacing: reconstructing anonymized facial features using GANs. In: Proceedings of the IEEE International Symposium on Biomedical Imaging, Venice, Italy, April 8–11, 2019. (https://ieeexplore.ieee.org/abstract/document/8759515). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


