Abstract
Evidence demonstrating that human rhinovirus (HRV) disease is not exclusively limited to the upper airways and may cause lower respiratory complications, together with the frequency of HRV infections and the increasing number of immunocompromised patients underline the need for rapid and accurate diagnosis of HRV infections. In this study, we developed the first quantitative real-time nucleic acid sequence-based amplification assay with an internal control using molecular beacon probes for selective and sensitive detection of human rhinovirus serotypes. We described a simple method to accurately quantify RNA target by computing the time to positivity (TTP) values for HRV RNA. Quantification capacity was assessed by plotting these TTP values against the starting number of target molecules. By using this simple method, we have significantly increased the diagnostic accuracy, precision, and trueness of real-time NASBA assay. Specificity of the method was verified in both in silico and experimental studies. Moreover, for assessment of clinical reactivity of the assay, NASBA has been validated on bronchoalveolar lavage (BAL) specimens. Our quantitative NASBA assay was found to be very specific, accurate, and precise with high repeatability and reproducibility.
Keywords: Nucleic acid sequence-based amplification, Human rhinovirus, Molecular beacon, Internal control, RNA detection and quantification
Introduction
Human rhinoviruses (HRVs) are the most frequent cause of acute upper respiratory tract infections in humans and are usually responsible for 30–50% of cases of common cold [1–3]. However, they may also be associated with more-severe lower respiratory tract infections. Rhinoviruses have been isolated from cases of cystic fibrosis, otitis media, sinusitis, asthma, exacerbations of chronic obstructive pulmonary disease (COPD), and pneumonia, especially in children, in the elderly, and in immunocompromised patients [4–14]. Evidence demonstrating that HRV disease is not exclusively limited to the upper airways and may cause lower respiratory complications, together with the frequency of HRV infections and the increasing number of immunocompromised patients underline the need for rapid and accurate diagnosis of HRV infections. Two nucleic acid amplification techniques (NAATs) are actually available for the detection of HRV: reverse transcription-PCR (RT-PCR) [15, 16], and nucleic acid sequence-based amplification (NASBA) [17, 18]. NASBA has proven to be highly sensitive, specific, and more rapid than RT-PCR technique [19, 20]. Currently, only qualitative NASBA kits for the detection of HRV are commercially available (registered trademarks owned of bioMérieux, Marcy L’Etoile, France), while there are no quantitative NASBA kits. Some quantitative molecular beacon real-time NASBA assays have been described in the literature, mainly for the identification of human immunodeficiency virus (HIV), respiratory syncytial virus A and B, influenza A virus (H1N1), Trypanosoma brucei, Aspergillus fumigatus, Plasmodium species, and Listeria monocytogenes [21–29]. However, these real-time NASBA assays use mathematical models for the analysis of results that requires employing of specific complex software calibrated to each target [30]. Other quantitative NASBA assays, instead, compute the ratio of the time to positivity (TTP) values for both the target RNA and internal control by using standard curves with a correlation coefficient less than 0.99 (R 2 < 0.99), index of an insensitive assay [19, 24, 26].
Aim of this study was to develop the first quantitative real-time nucleic acid sequence-based amplification assay internally controlled using molecular beacon for selective and sensitive detection of HRV serotypes. Validation and standardization were performed by evaluating diagnostic trueness, precision, and accuracy of real-time NASBA assay.
Materials and Methods
Viral Isolates and RNA Extraction
Prototype human rhinovirus serotype 16 (HRV-16) was obtained from the American Type Culture Collection (ATCC, Manassas, Virginia). Rhinovirus serotype 16 (ATCC VR-283), originally isolated from a human clinical specimen, was extracted by using an automatic extractor NucliSENS easyMAG platform (bioMérieux, France), according to the manufacturer’s recommendations. One-hundred-microliters of HRV-16 were used for the extraction, RNA was eluted in 25 μl of nuclease-free water and stored at −80°C. To evaluate the specificity of the HRV NASBA assay, purified RNA templates from 12 HRV isolates, and 14 selected respiratory viruses other than HRV were used for inclusivity and exclusivity testing (Table 1).
Table 1.
Evaluation of real-time NASBA specificity with HRV isolates and respiratory viruses other than HRV
| Taxon | Provider | Test specificity for: |
|---|---|---|
| HRV | ||
| Inclusivity testing | ||
| Human rhinovirus 1B | ATCC VR-1366 | + |
| Human rhinovirus 2 | ATCC VR-482 | + |
| Human rhinovirus 3 | ATCC VR-483 | + |
| Human rhinovirus 7 | ATCC VR-1601 | + |
| Human rhinovirus 9 | ATCC VR-489 | + |
| Human rhinovirus 16 | ATCC VR-283 | + |
| Human rhinovirus 41 | ATCC VR-339 | + |
| Human rhinovirus 58 | ATCC VR-1168 | + |
| Human rhinovirus 8 | QCMD EQA RV.CV10-02 | + |
| Human rhinovirus 42 | QCMD EQA RV.CV10-01 | + |
| Human rhinovirus 72 | QCMD EQA RV.CV10-03 | + |
| Human rhinovirus 90 | QCMD EQA RV.CV10-05 | + |
| Exclusivity testing | ||
| Influenza A virus (H1N1) | QCMD EQA INFRNA10-08 | − |
| Influenza A virus (H3N2) | QCMD EQA INFRNA10-01 | − |
| Influenza B virus | QCMD EQA INFRNA10-06 | − |
| Human parainfluenza virus 1 | ATCC VR-94 | − |
| Human parainfluenza virus 2 | ATCC VR-92 | − |
| Human parainfluenza virus 3 | ATCC VR-93 | − |
| Human parainfluenza virus 4a | ATCC VR-1378 | − |
| Human parainfluenza virus 4b | ATCC VR-1377 | − |
| Human coronavirus OC43 | ATCC VR-1558 | − |
| Human coronavirus 229E | ATCC VR-740 | − |
| Human coronavirus NL63 | QCMD EQA RV.CV10-10 | − |
| Human coxsackievirus B4 | ATCC VR-184 | − |
| Human respiratory syncytial virus A | ATCC VR-1540 | − |
| Human respiratory syncytial virus B | ATCC VR-1400 | − |
Note: ATCC American type culture collection, QCMD (EQA) quality control for molecular diagnostics (external quality assessment)
In Vitro RNA Transcription
Viral cDNA was generated, first by incubation of random primers (600 ng/μl) and dNTPs (10 mM) (Invitrogen) with 10 μl of HRV-16 RNA for 5 min at 70°C. Subsequently, a mix containing buffer 5× [250 mM Tris–HCl (pH 8.3 at 25°C), 375 mM KCl, and 50 mM DTT], MgCl2 (25 mM), ImpromII RT (1 U/μl), and Recombinant RNasin® Ribonuclease Inhibitor (40 U/μl) (Promega) was added. The total volume (20 μl) of the reaction mixture was incubated for 5 min at 25°C, 60 min at 42°C, and 15 min at 70°C using 9800 Fast Thermal Cycler (Applied Biosystems, Monza, Italy). cRNA production was carried out using T7-RiboMAX Large Scale RNA Production Systems (Promega, USA) at 37°C for 4 h. One-tenth of cRNA product was treated with RQ1 RNase-Free DNase (Promega, USA) at 37°C for 15 min followed by incubation with EDTA for 15 min at 65°C. HRV-16 cRNA was purified using RNAgent kit (Promega, USA) following manufacturer’s instructions. HRV-16 cRNA was quantified using Quant-iT DNA BR assay on Qubit™ fluorometer (Invitrogen, Carlsbad, USA), and the number of molecules per microliter calculated from the molecular weight of HRV-16 amplicon (70,950 MW) and Avogadro number (6.023 × 1023). Ten-fold dilutions of RNA standards were generated in order to amplify from 108 to 1 copy per reaction, and frozen at −80°C until use.
Synthesis of Internal Control (IC) RNA
For the production of internal control (IC) RNA, we used the human U1A housekeeping gene encoding the “A” protein present in the human U1 small nuclear ribonucleoprotein (snRNP) particle. To generate the IC RNA, the U1A molecule was extracted from a clinical specimen, precisely from a human bronchoalveolar lavage sample, and subjected to reverse transcription (RT) reaction by using random primers (600 ng/μl) and ImpromII RT (1 U/μl). IC cDNA product was amplified by using adapted NASBA primers containing the T7 RNA polymerase promoter site. Briefly, 2 μl of IC cDNA was added to 28 μl of PCR solution containing Flexi Buffer 5×, 50 mM MgCl2, 1 unit GoTaq® Hot Start Polymerase (Promega), 200 μM of each dNTP, and 25 μM of each U1A primer. After an initial denaturation step of 2 min at 94°C, the first-round PCR amplification was carried out under the following conditions: 95°C for 0 s, 58°C for 15 s, 72°C for 10 s for 35 cycles, then one cycle at 72°C for 7 min using the 9800 Fast Thermal Cycler (Applied Biosystems). PCR product was transcribed in vitro using T7-RiboMAX, as previously described (see “In vitro transcription” in the “Materials and methods” section). Ten-fold dilutions of IC RNA standards were generated in order to amplify from 108 to 1 copy per reaction.
Real-Time NASBA Assay and Data Analysis
Primers and molecular beacon probes were obtained from literature [31, 32] (Table 2), synthesized by Eurogentec (Seraing, Belgium), and diluted to a final concentration of 100 μmol/l. Moreover, to maximize the oligonucleotides stabilization, we added 60% dimethyl sulfoxide (DMSO) to primers and beacons mixture. The HRV beacon was labeled with FAM at its 5′-end and quencher DABCYL at its 3′-end, while the IC beacon contained a ROX at its 5′-end and a DABCYL quencher at the 3′-end. The stability and predicted structure of the beacons were analyzed by using the European MFOLD server (http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/dna-form1.cgi). Real-time NASBA reaction was performed on a NucliSens EasyQ analyzer (BioMérieux) using the NucliSENS EasyQ Basic Kit Version 2 (bioMérieux, Lyon, France) for the amplification according to the manufacturer’s manual. To obtain the best amplification efficiency, conditions for the real-time NASBA assay were optimized until the best primers, beacons, and KCl concentrations were determined. Titrations of IC RNA (between 10 and 106 copies) and HRV RNA were performed to determine the optimal amount of internal control to generate the greatest dynamic range for the assay without interference with the detection of HRV RNA (data not shown). As a result, each reaction was run with the addition of 105 copies of the IC RNA. Briefly, a total volume of 10 μl of reaction mixture containing 80 mM KCl and 0.3 μM of the HRV- and IC-specific primers was incubated with 2.5 μl HRV RNA and 2.5 μl IC RNA in the presence of 0.05 μM of HRV- and IC-molecular beacons at 65°C for 2 min to denature secondary structure RNA. The reaction was subsequently cooled to 41°C for 2 min to anneal the primers before adding 5 μl of enzyme mixture containing avian myeloblastosis virus retrotranscriptase, RNase H, and T7 RNA polymerase. After a brief centrifugation and gentle mixing by tapping, the tubes were then incubated at 41°C for 90 min. To estimate the dynamic range of the real-time NASBA assay (range of concentrations over which the method performs in a linear manner with an acceptable level of trueness and precision), we used HRV standard dilutions from 108 copies/μl to 1 copy/μl. Sensitivity of NASBA assay was determined by the lowest standard dilution consistently detectable in replicate reactions at frequency of 100%, whereas the limit of detection by the lowest concentration of target quantified. Nuclease-free water was included as the no-template control (NTC) to serve as a control for background fluorescence. The fluorescence signal was measured with an interval time of 45 s for each independent reaction at two wavelengths using the accompanying NucliSENS EasyQ Director software (Version 2.0). Following amplification, a specific fluorescence value was chosen as a positive signal (threshold level = 1.1). Using this value for all reactions in one experiment, the time that the target and IC amplification curves reached the threshold level was recorded as the time to positivity (TTP). HRV TTP values were regressed against the original standard curve to generate a predicted amount of target RNA. In particular, for data analysis, Excel calculation engine was used. NASBA dynamic range was calculated from TTP values regressed against the standard curve by using an Excel spreadsheet that was created “ad hoc” by us. In particular, for each individual HRV standard dilution (from 108 copies/reaction to 1 copy/reaction), fluorescent threshold levels of target close to 1.1 and values of the corresponding minutes were extrapolated by the NucliSENS EasyQ Director software. Therefore, minutes were normalized to a value of threshold level = 1.1, and the natural logarithm of normalized minutes was calculated. Quantification capacity was assessed by plotting the TTP values obtained against the logarithm of the number of target molecules in each dilution and calculating a linear regression.
Table 2.
Primers and molecular beacons for HRV and IC-U1A real-time NASBA assay
| Primer or MBa | Sequence (5′-3′) |
|---|---|
| HRVJ P1 | AATTCTAATACGACTCACTATAGGGAG bACCAMYWTTYTGYSTWGAWAC |
| HRVJ P2 | CTCCGGCCCCTGAATGYGGCT |
| HRVJMB-FAM | CCAAGC cGAYGGGACCRACTACTTTGGGCTTGG |
| U1A P1 | AATTCTAATACGACTCACTATAGGG bAGAGGCCCGGCATGTGGTGCATAA |
| U1A P2 | CAGTATGCCAAGACCGACTCAGA |
| U1AMB-ROX | CGTACG cGATGAAAGGCACCTTCGTGGACGTACG |
Bold sequences corresponding to HRVJ P1 and U1A P1 indicate the stretch of T7 RNA polymerase promoter sequences present in the primer P1 sequences
Bold sequences corresponding to HRVJMB-FAM and U1AMB-ROX indicate the stem sequences present in the molecular beacon sequences
aMB, molecular beacons. 5′-end of the beacon was labeled with FAM or ROX, and 3′-end was labeled with the nonfluorescent quencher DABCYL. b T7 RNA polymerase promoter sequence for NASBA. c Stem sequence for the molecular beacons
Specificity of Real-Time NASBA Assay
The specificity of the developed real-time NASBA assay was verified by in silico studies (analytical specificity) against publicly available sequence databases (BLAST alignment software (http://blast.ncbi.nlm.nih.gov/Blast.cgi)) to evaluate possible cross-reactions with respiratory viruses other than HRV. Experimental specificity was also verified. In particular, sequences of different respiratory viruses were used for exclusivity testing (Table 1).
Validation and Standardization of Real-Time NASBA Assay
To determine the performance of HRV NASBA assay, we assessed the diagnostic trueness, precision, and accuracy of the technique. Specifically, precision was assessed by evaluating repeatability and intermediate reproducibility of NASBA assay. To determine the repeatability, several replicates containing the various amounts of HRV RNA were tested. The repeatability was determined by 10-fold serial dilutions of the HRV RNA standards. In particular, we used four different dilutions (102, 103, 104, and 105 copies/reaction) of quantification standards. Each dilution was analyzed 10 times, with the same method on identical test items in the same laboratory by the same operator using the same equipment. Regarding intermediate reproducibility, each dilution was analyzed with the same method on identical test items in 10 different runs performed by three different operators using different equipment on different days. Moreover, we used the Dixon’s test to examine if one measure from 10 replicate measures that we performed (102, 103, 104, and 105) could be rejected or not, and the Shapiro–Wilk’s test to compare these measures against the normal distribution. Statistical data analyses were performed using the PASW Statistics version 18.0 (SPSS Inc., Chicago, Illinois, USA).
Clinical Specimens
For assessment of clinical reactivity of the assay, respiratory specimens collected from our Virology Unit of the Azienda Ospedaliero-Universitaria San Giovanni Battista, Turin, were tested for HRV by real-time NASBA assay. Clinical specimens included 33 bronchoalveolar lavages (BAL) obtained from 33 transplant patients (M/F, 20/13; mean age, 56.2 years; range 21–88). A number of precautions were undertaken to prevent the occurrence of false-positive results. Each run included control reactions lacking template (no-template controls) to test for the presence of contamination or the generation of nonspecific amplification products under the assay conditions used.
Results
Sensitivity of Real-Time NASBA Assay
Optimal real-time NASBA assay conditions that allowed efficient amplification of the HRV target were established. Sensitivity and limit of detection of NASBA assay were assessed by repeated testing of serial logarithmic dilutions of the HRV RNA standards ranging from 108 to 1 copy/reaction. In particular, HRV NASBA dynamic range was calculated from TTP value (time point at which emitted fluorescence exceeds the baseline emission) regressed against the standard curve by using an Excel spreadsheet created “ad hoc” by us (Table 3). Results from linear regression show that HRV real-time NASBA assay was able to quantify from 108 to 10 copies/reaction. The standard curve of HRV dilutions plotted versus NASBA amplifications (expressed as TPP) is shown in Fig. 1, whereas plots for the amplification of HRV standard dilutions (from 108 to 10 copies/reaction), and the optimal amount of IC RNA (105 copies/reaction), are shown in Fig. 2. The consistency of replicates was measured by the correlation coefficient (R 2), which indicates the linearity of TPP values plotted in the standard curve. The R 2 index for HRV was 0.9948. Sensitivity of real-time NASBA assay was determined by the lowest standard dilution consistently detectable in replicate reactions at frequency of 100%. HRV sensitivity was 10 copies/reaction, whereas the limit of detection for reliable quantification was 1 copy/reaction.
Table 3.
Excel spreadsheet used to calculate HRV NASBA dynamic range
| HRV dilutions | Threshold | Minutes | Normalized minutes threshold 1.1 | Ln normalized minutes (TTP) | HRV ln dilutions |
|---|---|---|---|---|---|
| 108 | 1.10476600 | 15.088 | 15.02290983 | 2.709576358 | 18.4206807 |
| 107 | 1.10571300 | 18.833 | 18.73569362 | 2.930430454 | 16.1180957 |
| 106 | 1.10279600 | 21.82917 | 21.7738249 | 3.080708556 | 13.8155106 |
| 105 | 1.11357000 | 26.3245 | 26.00370879 | 3.258239174 | 11.5129255 |
| 104 | 1.10117700 | 31.57033 | 31.53658585 | 3.451148328 | 9.21034037 |
| 103 | 1.10275300 | 39.81483 | 39.7154331 | 3.681739855 | 6.90775528 |
| 102 | 1.10297100 | 45.81017 | 45.68677418 | 3.821808851 | 4.60517019 |
| 10 | 1.09937600 | 51.05633 | 51.0853093 | 3.933496967 | 2.30258509 |
For each individual HRV standard dilution, fluorescent threshold levels close to 1.1 and values of the corresponding minutes were extrapolated by the NucliSENS EasyQ Director software. Minutes were normalized to a value of threshold level = 1.1, and the natural logarithm (ln) of normalized minutes was calculated. Quantification capacity was assessed by plotting the TTP values obtained against the logarithm of HRV dilutions and calculating a linear regression. TTP time to positivity
Fig. 1.
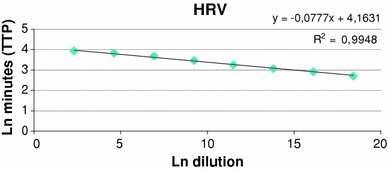
Standard curve of HRV real-time NASBA assay. HRV TTP values were regressed against the standard curve to generate a predicted amount of target RNA. HRV ln dilutions (108 to 10 copies/reaction) on x-axis are plotted against ln minutes (TTP) on y-axis. The coefficient of correlation (R 2) was 0.9948. TTP time to positivity, Ln natural logarithm
Fig. 2.
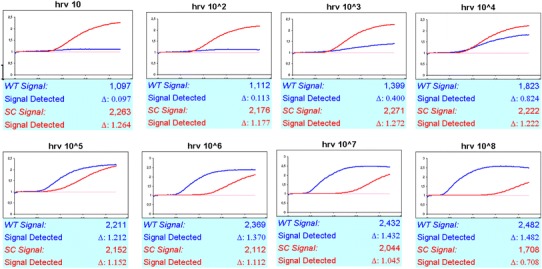
Real-time NASBA amplification plots of HRV (WT) standard dilutions (from 108 to 10 copies/reaction), and the optimal amount of IC (SC) RNA (105 copies/reaction). TPP (in minutes) and fluorescence exceeds background are indicated on x- and y-axis, respectively. WT wild type, IC internal control, SC system control, TPP time to positivity
Specificity of Real-Time NASBA Assay
Based on the data available at the BLAST alignment software, primers were tested in silico for potential cross-reactivity with respiratory viruses other than HRV, and demonstrated no significant homologies to any other sequences. Moreover, HRV primer and probe set, tested on respiratory viruses, was able to detect only HRV isolates, thus being the inclusivity of 100% (Table 1). The assay’s specificity was further demonstrated by its ability to exclude all respiratory viruses other than HRV listed in Table 1. No positive results were demonstrated for the other respiratory viruses, indicating that this molecular assay is highly specific for HRV isolates, thus being the exclusivity of 100% (Table 1).
Validation and Standardization of Real-Time NASBA Assay
Diagnostic trueness of HRV real-time NASBA method, defined as the degree of agreement between the average value obtained from a large series of test results and an accepted reference value, was evaluated. To establish the level of trueness and concordance with the assigned value, data from 10 replicate measures of each dilution that we performed (102, 103, 104, and 105) were analyzed using a Student’s t test to compare the mean concentrations from each dilution with an accepted reference value. The mean concentrations from each dilution for the method are shown in Table 4 with the t test results, which indicate the significance of the differences between each experimental mean and the assigned value. Analysis of the t statistics showed that the method had t-calc values lower than the t-tab value, demonstrating a significant trueness of HRV assay.
Table 4.
Statistical summary of validation and standardisation of HRV real-time NASBA assay
| Ln 102 | Ln 103 | Ln 104 | Ln 105 | |
|---|---|---|---|---|
| Trueness (t test) | ||||
| Experimental mean concentration | 4.18914 | 6.66043 | 9.33646 | 11.44784 |
| Standard deviation | 0.55411 | 0.27162 | 0.11906 | 0.08413 |
| t-calc | 1.67897 | 2.03641 | 2.36935 | 1.72916 |
| t-tab (n = 10) | 2.776 | 2.776 | 2.776 | 2.776 |
| Precision (% coefficient of variation) | ||||
| Repeatability | 13.22732 | 4.07810 | 1.27521 | 0.73493 |
| Intermediate reproducibility | 16.71455 | 3.70736 | 4.861 | 5.53251 |
| Accuracy (% bias inaccuracy) | 9.03457 | 3.58098 | 1.36973 | 0.56511 |
Ln natural logarithm
Precision of method was expressed as the coefficient of variation (CV) in the log10 values of the concentration. Repeatability and intermediate reproducibility of HRV assay were evaluated over different concentrations ranging from 102 to 105 copies/reaction from 10 replicate measures (n = 10) of each reference viral quantification standard within a single run or in 10 different run experiments performed by three different operators. The precision associated with each dilution measurement (102, 103, 104, and 105) was assessed by calculation of the CV for each. The CVs within a single run (repeatability) ranged from 0.73 to 13.23%; whereas, the CVs in different runs (intermediate reproducibility) ranged from 3.71 to 16.71% (Table 4), indicating that the precision of the assay is satisfactory.
Diagnostic accuracy includes both trueness and precision. The measure of accuracy is usually expressed numerically in terms of bias (lack of agreement). Accuracy shall be within ±25% of the accepted reference value over the whole dynamic range, according to document ISO 16140 [33]. Data for the percentage of inaccuracy HRV method are reported in Table 4.
Clinical Performance of Real-Time NASBA Assay
The developed real-time NASBA assay was able to detect HRV RNA in 7/33 (21.2%) BAL specimens. The TTP values of these NASBA-positive samples ranged between 39 and 51 min when plotted against the standard curve in Fig. 1 (data not shown). All the results were validated by the addition of internal control RNA to rule out inhibition of amplification. In all cases, amplification of the control RNA was observed, thus, confirming that all the negative and positive results are valid. Moreover, all negative control reactions were NASBA negative, demonstrating the absence of amplicon contamination.
Discussion
Viral respiratory tract infections have been recognized as a predominant cause of human disease. To improve clinical management of such patients, it is important to obtain an accurate diagnosis and to identify the causative agent early in infection to ensure appropriate treatment. In this study, we developed the first quantitative NASBA assay for the detection of HRV serotypes. By combining NASBA amplification with molecular beacon probes, this assay becomes a real-time analysis tool that offers faster results than conventional RT-PCR technique. Since NASBA amplification involves three separate enzymes each with their own kinetic parameters, variability in every measurement is inevitable [19]. Weusten et al. [30] were the first to describe a mathematical model for RNA amplification of both target and internal calibrator RNA in a molecular beacon-based NASBA reaction to normalize enzyme efficiency differences between reactions. However, the description of this model did not include all of the essential parameters needed to operate the model. Consequently, analysis using this model requires software calibrated to each target and is commercially available for only a few specific targets. Here, we describe an alternative method for normalizing NASBA data by using a simple TPP calculation in the presence of an internal control that reduces the variability between replicates and increases the precision, trueness, and accuracy for predicting unknown concentrations of HRV RNA. It has been shown that tube to tube variation within NASBA can be normalized with the addition of an internal control in each reaction [34, 35]. In particular, the optimal concentration for the internal calibrator should be determined as the concentration providing the greatest dynamic range of amplification of both the target and internal control RNA. In our study, a fixed amount of 105 copies of the internal calibrator RNA was optimal for the assay reported here (data not shown). Furthermore, the addition of an internal control is fundamental to identify false negative results because of reaction failure, and to monitor the effects of unknown sample factors that might interfere with the amplification kinetics. As concerns the importance of primers and KCl for NASBA optimization, their role has not been emphasized to date. NASBA is an isothermal nucleic acid amplification method able to specifically amplify target RNA by using specific primers in a KCl background. Initially, the concentration of the primers relative to the total concentration of amplifiable RNA is very high, is not rate limiting, and relatively small amounts of primers are consumed in depletion of the initially present pool of RNA copies (linear phase of NASBA process). At some time point, obviously, the primers’ concentrations do become rate limiting, and decline to practically zero. At this time point, the DNA intermediate levels have reached their maximum and RNA production proceeds at high speed. From now on, the only reaction that can proceed is T7 RNA polymerase-mediated formation of RNA from the DNA intermediate templates. This time interval represents the second phase of NASBA process characterized by an exponential kinetics. We observed that high concentrations of primers and KCl elongate the linear phase of NASBA process by shorting the exponential amplification; whereas, low concentrations of primers and KCl promote the exponential phase. In particular, in this study we used relatively low concentrations of primers and KCl (0.3 μM and 80 mM, respectively) to elongate the exponential phase of NASBA process, and accordingly, to minimize the reaction-to-reaction variation. By using this simple expedient, we have significantly increased our accuracy, precision, and trueness of prediction over the standard TTP calculations. In summary, we describe a simplified method of calculating unknown concentrations of target RNA using an internal calibrator. This method allowed for greater precision, accuracy, and trueness for predicting HRV RNA over the standard TTP analysis. In conclusion, we described the first real-time NASBA assay capable of accurate quantification of HRV RNA making it a valuable tool in the molecular diagnostics of HRV serotypes.
Footnotes
Francesca Sidoti and Massimiliano Bergallo contributed equally to this work and share first authorship.
References
- 1.Carlsen KH, Orstavik I, Leegaard J, Høeg H. Respiratory virus infections and aeroallergens in acute bronchial asthma. Archives of Disease in Childhood. 1984;59:310–315. doi: 10.1136/adc.59.4.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mertsola J, Ziegler T, Ruuskanen O, Vanto T, Koivikko A, Halonen P. Recurrent wheezy bronchitis and viral respiratory infections. Archives of Disease in Childhood. 1991;66:124–129. doi: 10.1136/adc.66.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mäkelä MJ, Puhakka T, Ruuskanen O, Leinonen M, Saikku P, Kimpimäki M, Blomqvist S, Hyypiä T, Arstila P. Viruses and bacteria in the etiology of the common cold. Journal of Clinical Microbiology. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arola M, Ziegler T, Ruuskanen O, Mertsola J, Näntö-Salonen K, Halonen P. Rhinovirus in acute otitis media. Journal of Pediatrics. 1988;113:693–695. doi: 10.1016/S0022-3476(88)80380-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ireland DC, Kent J, Nicholson KG. Improved detection of rhinoviruses in nasal and throat swabs by seminested RT-PCR. Journal of Medical Virology. 1993;40:96–101. doi: 10.1002/jmv.1890400204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnston SL, Sanderson G, Pattemore PK, Smith S, Bardin PG, Bruce CB, Lambden PR, Tyrrell DA, Holgate ST. Use of polymerase chain reaction for diagnosis of picornavirus infection in subjects with and without respiratory symptoms. Journal of Clinical Microbiology. 1993;31:111–117. doi: 10.1128/jcm.31.1.111-117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraenkel DJ, Bardin PG, Sanderson G, Lampe F, Johnston SL, Holgate ST. Lower airways inflammation during rhinovirus colds in normal and in asthmatic subjects. American Journal of Respiratory and Critical Care Medicine. 1995;151:879–886. doi: 10.1164/ajrccm/151.3_Pt_1.879. [DOI] [PubMed] [Google Scholar]
- 8.Smyth AR, Smyth RL, Tong CY, Hart CA, Heaf DP. Effect of respiratory virus infections including rhinovirus on clinical status in cystic fibrosis. Archives of Disease in Childhood. 1995;73:117–120. doi: 10.1136/adc.73.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson KG, Kent J, Hammersley V, Cancio E. Acute viral infections of upper respiratory tract in elderly people living in the community: Comparative, prospective, population based study of disease burden. British Medical Journal. 1997;315:1060–1064. doi: 10.1136/bmj.315.7115.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitkäranta A, Arruda E, Malmberg H, Hayden FG. Detection of rhinovirus in sinus brushings of patients with acute community-acquired sinusitis by reverse transcription-PCR. Journal of Clinical Microbiology. 1997;35:1791–1793. doi: 10.1128/jcm.35.7.1791-1793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh S, Champlin R, Couch R, Englund J, Raad I, Malik S, Luna M, Whimbey E. Rhinovirus infections in myelosuppressed adult blood and marrow transplant recipients. Clinical Infectious Diseases. 1999;29:528–532. doi: 10.1086/598627. [DOI] [PubMed] [Google Scholar]
- 12.Rabella N, Rodriguez P, Labeaga R, Otegui M, Mercader M, Gurguí M, Prats G. Conventional respiratory viruses recovered from immunocompromised patients: clinical considerations. Clinical Infectious Diseases. 1999;28:1043–1048. doi: 10.1086/514738. [DOI] [PubMed] [Google Scholar]
- 13.Van Elden LJ, van Kraaij MG, Nijhuis M, Hendriksen KA, Dekker AW, Rozenberg-Arska M, van Loon AM. Polymerase chain reaction is more sensitive than viral culture and antigen testing for the detection of respiratory viruses in adults with hematological cancer and pneumonia. Clinical Infectious Diseases. 2002;24:177–183. doi: 10.1086/338238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa, C., Bergallo, M., Astegiano, S., Sidoti, F., Terlizzi, M. E., Gambarino, S., Curtoni, A., Simeone, S., Solidoro, P., & Cavallo, R. (2011). Detection of human rhinoviruses in the lower respiratory tract of lung transplant recipients. Archives of Virology. doi:10.1007/s00705-011-0986-z. [DOI] [PMC free article] [PubMed]
- 15.Gama RE, Horsnell PR, Hughes PJ, North C, Bruce CB, al-Nakib W, Stanway G. Amplification of rhinovirus specific nucleic acids from clinical samples using the polymerase chain reaction. Journal of Medical Virology. 1989;28:73–77. doi: 10.1002/jmv.1890280204. [DOI] [PubMed] [Google Scholar]
- 16.Gambarino S, Costa C, Elia M, Sidoti F, Mantovani S, Gruosso V, Bergallo M, Cavallo R. Development of a RT real-time PCR for the detection and quantification of human rhinoviruses. Molecular Biotechnology. 2009;42:350–357. doi: 10.1007/s12033-009-9164-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuelson A, Westmoreland D, Eccles R, Fox JD. Development and application of a new method for amplification and detection of human rhinovirus RNA. Journal of Virological Methods. 1998;71:197–209. doi: 10.1016/S0166-0934(98)00006-8. [DOI] [PubMed] [Google Scholar]
- 18.Loens K, Ieven M, Ursi D, De Laat C, Sillekens P, Oudshoorn P, Goossens H. Improved detection of rhinoviruses by nucleic acid sequence-based amplification after nucleotide sequence determination of the 5′ noncoding regions of additional rhinovirus strains. Journal of Clinical Microbiology. 2003;41:1971–1976. doi: 10.1128/JCM.41.5.1971-1976.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson SS, Casper ET, Garcia-Rubio L, Smith MC, Paul JH., III Increased precision of microbial RNA quantification using NASBA with an internal control. Journal of Microbiological Methods. 2005;60:343–352. doi: 10.1016/j.mimet.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 20.Loens K, Goossens H, de Laat C, Foolen H, Oudshoorn P, Pattyn S, Sillekens P, Ieven M. Detection of rhinoviruses by tissue culture and two independent amplification techniques, nucleic acid sequence-based amplification and reverse transcription-PCR, in children with acute respiratory infections during a winter season. Journal of Clinical Microbiology. 2006;44:166–171. doi: 10.1128/JCM.44.1.166-171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider P, Wolters L, Schoone G, Schallig H, Sillekens P, Hermsen R, Sauerwein R. Real-time nucleic acid sequence-based amplification is more convenient than real-time PCR for quantification of Plasmodium falciparum. Journal of Clinical Microbiology. 2005;43:402–405. doi: 10.1128/JCM.43.1.402-405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClernon DR, Vavro C, St Clair M. Evaluation of a real-time nucleic acid sequence-based amplification assay using molecular beacons for detection of human immunodeficiency virus type 1. Journal of Clinical Microbiology. 2006;44:2280–2282. doi: 10.1128/JCM.02187-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mens PF, Schoone GJ, Kager PA, Schallig HD. Detection and identification of human Plasmodium species with real-time quantitative nucleic acid sequence-based amplification. Malaria Journal. 2006;5:80. doi: 10.1186/1475-2875-5-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deiman B, Schrover C, Moore C, Westmoreland D, van de Wiel P. Rapid and highly sensitive qualitative real-time assay for detection of respiratory syncytial virus A and B using NASBA and molecular beacon technology. Journal of Virological Methods. 2007;146:29–35. doi: 10.1016/j.jviromet.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 25.Nadal A, Coll A, Cook N, Pla M. A molecular beacon-based real time NASBA assay for detection of Listeria monocytogenes in food products: role of target mRNA secondary structure on NASBA design. Journal of Microbiological Methods. 2007;68:623–632. doi: 10.1016/j.mimet.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Mugasa CM, Schoone GJ, Ekangu RA, Lubega GW, Kager PA, Schallig HD. Detection of Trypanosoma brucei parasites in blood samples using real-time nucleic acid sequence-based amplification. Diagnostic Microbiology and Infectious Disease. 2008;61:440–445. doi: 10.1016/j.diagmicrobio.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 27.Beurskens M, Mens P, Schallig H, Syafruddin D, Asih PB, Hermsen R, Sauerwein R. Quantitative determination of Plasmodium vivax gametocytes by real-time quantitative nucleic acid sequence-based amplification in clinical samples. American Journal of Tropical Medicine and Hygiene. 2009;81:366–369. [PubMed] [Google Scholar]
- 28.Ge Y, Cui L, Qi X, Shan J, Shan Y, Qi Y, Wu B, Wang H, Shi Z. Detection of novel swine origin influenza A virus (H1N1) by real-time nucleic acid sequence-based amplification. Journal of Virological Methods. 2010;163:495–497. doi: 10.1016/j.jviromet.2009.10.025. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Park S, Warn P, Shrief R, Harrison E, Perlin DS. Detection of Aspergillus fumigatus in a rat model of invasive pulmonary aspergillosis by real-time nucleic acid sequence-based amplification. Journal of Clinical Microbiology. 2010;48:1378–1383. doi: 10.1128/JCM.02214-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weusten JJ, Carpay WM, Oosterlaken TA, van Zuijlen MC, van de Wiel PA. Principles of quantitation of viral loads using nucleic acid sequence-based amplification in combination with homogeneous detection using molecular beacons. Nucleic Acids Research. 2002;30:6e26. doi: 10.1093/nar/30.6.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wat D, Gelder C, Hibbitts S, Cafferty F, Bowler I, Pierrepoint M, Evans R, Doull I. The role of respiratory viruses in cystic fibrosis. Journal of Cystic Fibrosis. 2008;7:320–328. doi: 10.1016/j.jcf.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greijer AE, Dekkers CA, Middeldorp JM. Human cytomegalovirus virions differentially incorporate viral and host cell RNA during the assembly process. Journal of Virology. 2000;74:9078–9082. doi: 10.1128/JVI.74.19.9078-9082.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anonymous . Microbiology of food and animal feeding stuffs. Protocol for the validation of alternative method. ISO/DIS 16140. Geneva, Switzerland: International Organization for Standardization; 2002. [Google Scholar]
- 34.Schallig HD, Schoone GJ, Lommerse EJ, Kroon CC, de Vries PJ, van Gool T. Usefulness of quantitative nucleic acid sequence-based amplification for diagnosis of malaria in an academic hospital setting. European Journal of Clinical Microbiology and Infectious Diseases. 2003;22:555–557. doi: 10.1007/s10096-003-0985-4. [DOI] [PubMed] [Google Scholar]
- 35.Peter JB, Sevall JS. Molecular-based methods for quantifying HIV viral load. AIDS Patient Care and STDS. 2004;18:75–79. doi: 10.1089/108729104322802506. [DOI] [PubMed] [Google Scholar]


