Main Text
The revolution in electron cryo-microscopy (cryo-EM) has been amply documented (1,2). The key development leading to the phenomenal improvements in resolution, in which a near-atomic level of detail can now be routinely achieved, was the introduction of direct electron detectors about 7 years ago. But the feasibility of cryo-EM on biological samples was demonstrated 46 years ago when Taylor and Glaeser showed that a frozen protein crystal could be maintained in the hydrated state in the vacuum of an electron microscope (3). The next major advance was when Dubochet and colleagues developed the method of vitrification of samples, in which thin films of water form an amorphous solid (a glass) rather than a crystalline solid when rapidly cooled (4,5). In the almost 40 years that have passed since the vitrification technique was introduced, not a great deal has changed in grid preparation, as most people still rely (whether doing this manually or, more commonly, using commercially available vitrification devices) on applying a small quantity of sample (∼2 μL) to an EM grid, blotting this with filter paper to remove most of the sample, and then plunging this into liquid ethane or propane that is maintained at liquid nitrogen temperatures.
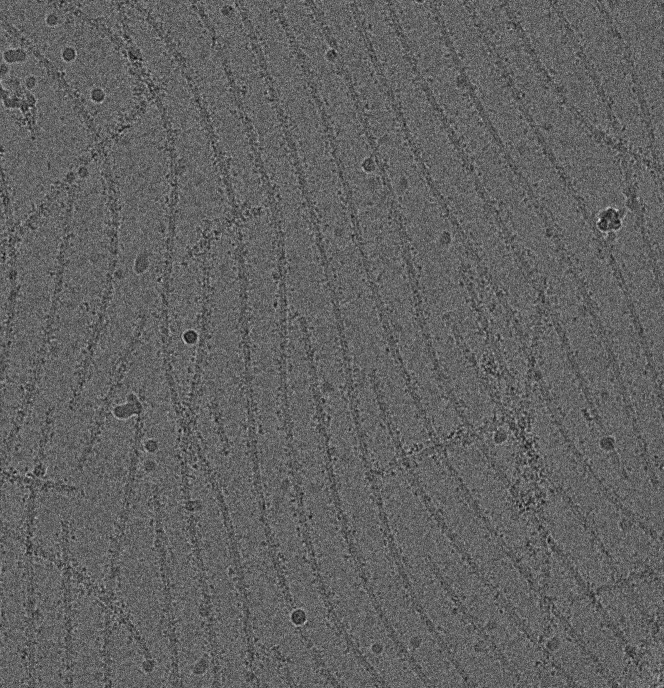
Although this approach to making grids clearly works, as judged by the >3700 structures at better than 4.5 Å resolution submitted to the Electron Microscopy Database (http://www.emdataresource.org) since 2012, it is not necessarily optimal. It has been understood that fluid flow and shear during blotting (6), the exposure of proteins to potential denaturation at the very large air-water interface (7), and the simple loss of most of the sample means that we should be able to do better. Although some using cryo-EM like to think of it as the visualization of molecules in solution, we now know that this is a rather naïve view. In the first place, the thin film in which molecules are embedded when imaged for cryo-EM is a highly anisotropic environment, as opposed to the isotropic environment expected in solution. This is easily seen for filaments (Fig. 1), which are frequently aligned in one direction because of the large fluid flow forces before vitrification. These fluid flow forces can actually place filaments under tension, and we showed that cryo-EM of RecA-DNA filaments oriented in the direction of the flow had a pitch of ∼110 Å, whereas a filament in the same image oriented perpendicular to the flow had a pitch of 96 Å (8). For actin filaments, these forces and thin films can actually make filaments appear anomalously rigid compared to their behavior in solution and shift the distribution of structural states that would otherwise be present (9). In addition to the problems of forces, anisotropy, and potential denaturation, the normal approach to preparing grids is also not entirely reproducible. The ice quality can be quite different between different grids prepared from the same sample, and across any individual grid, one will have a large variation in ice quality. As a result, most grid squares are usually not suitable for high-resolution imaging. This means that time and expertise are typically required for finding the best parts of a grid to image and restrict our ability to more fully automate cryo-EM imaging.
Figure 1.
A cryo-EM image of a field of actin filaments partially decorated with an actin-binding protein. The alignment of the filaments is caused by fluid flow induced by blotting of the sample with filter paper before vitrification. Because one would never see such alignment in solution at equilibrium (in the absence of forces), such images reveal the magnitude of the forces present during the traditional approach to cryo-EM grid preparation.
Glaeser and colleagues have now applied high-speed interference-contrast light microscopy to examine the interface between the fibers in filter paper with water droplets applied to a hydrophilic surface (10). Remarkably, no one appears to have previously tried looking at this during the ∼40 years that people have been preparing cryo-EM grids by blotting with filter paper. As Yogi Berra may or may not have said, “You can see a lot by just looking.” And what Glaeser and colleagues saw was that the nonuniform pattern of fibers in filter paper creates an aqueous film of highly nonuniform thickness. Unexpectedly, when the filter paper is removed, the film does not become uniform in the second or two before vitrification.
What conclusions follow from these surprising observations? The main one is probably that methods that rely upon the classical approach of thinning films using filter paper are doomed to inconsistent and not necessarily reproducible results. Because the actual and potential problems associated with this method of preparing cryo-EM grids have been apparent for some time, attempts are currently being made to use very different approaches (11,12). Although no one expects that such new approaches will have the same impact on cryo-EM as the introduction of direct electron detectors, these new approaches may dramatically improve the throughput in cryo-EM and reduce the amount of expertise currently needed to reach near-atomic resolution for macromolecular complexes.
Editor: Bridget Carragher.
References
- 1.Bai X.C., McMullan G., Scheres S.H. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Kühlbrandt W. Cryo-EM enters a new era. eLife. 2014;3:e03678. doi: 10.7554/eLife.03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor K.A., Glaeser R.M. Electron diffraction of frozen, hydrated protein crystals. Science. 1974;186:1036–1037. doi: 10.1126/science.186.4168.1036. [DOI] [PubMed] [Google Scholar]
- 4.Dubochet J. A reminiscence about early times of vitreous water in electron cryomicroscopy. Biophys. J. 2016;110:756–757. doi: 10.1016/j.bpj.2015.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lepault J., Booy F.P., Dubochet J. Electron microscopy of frozen biological suspensions. J. Microsc. 1983;129:89–102. doi: 10.1111/j.1365-2818.1983.tb04163.x. [DOI] [PubMed] [Google Scholar]
- 6.Galkin V.E., Orlova A., Egelman E.H. Actin filaments as tension sensors. Curr. Biol. 2012;22:R96–R101. doi: 10.1016/j.cub.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glaeser R.M. Proteins, interfaces, and cryo-em grids. Curr. Opin. Colloid Interface Sci. 2018;34:1–8. doi: 10.1016/j.cocis.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu X., Egelman E.H. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J. Mol. Biol. 1992;227:334–346. doi: 10.1016/0022-2836(92)90702-l. [DOI] [PubMed] [Google Scholar]
- 9.Galkin V.E., Orlova A., Egelman E.H. Near-atomic resolution for one state of F-actin. Structure. 2015;23:173–182. doi: 10.1016/j.str.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armstrong M., Han B.-G., Glaeser R.M. Microscale fluid behavior during cryo-EM sample blotting. Biophys. J. 2020;118:708–719. doi: 10.1016/j.bpj.2019.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dandey V.P., Wei H., Carragher B. Spotiton: new features and applications. J. Struct. Biol. 2018;202:161–169. doi: 10.1016/j.jsb.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravelli R.B.G., Nijpels F.J.T., Peters P.J. Automated cryo-EM sample preparation by pin-printing and jet vitrification. bioRxiv. 2019 [Google Scholar]