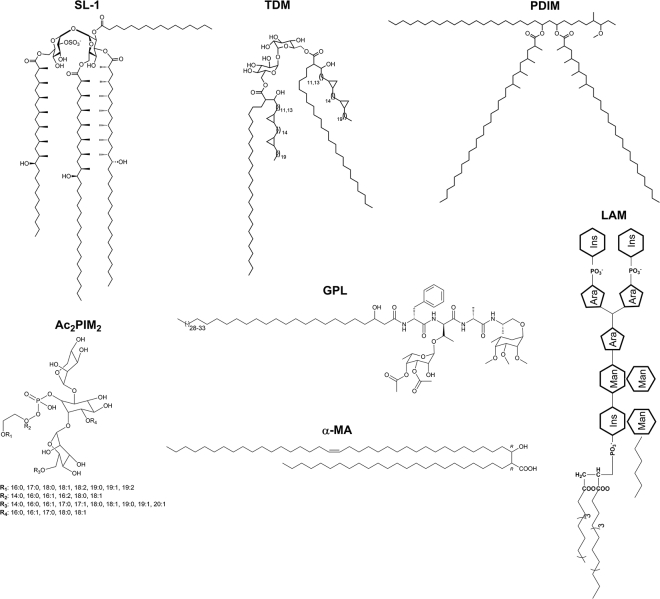
Abstract
Mycobacterium species, including Mycobacterium tuberculosis, employs atypical long (C60–90) and branched lipids to produce a complex cell wall and localizes these toward distinct spatial locations, inner membrane (IM) and outer membrane (OM), thus forming a robust permeability barrier. The properties and functional roles of these spatially orchestrated membrane platforms remain unknown. Herein, we report the distinctive lateral organization, fluidity, and lipid domain architecture of protein-free membranes reconstituted from IM and OM lipids in vitro from M. smegmatis (Msm) underscored by their lipid packing and lipid dynamics. We show that Msm OM, against common notion, is more dynamic and fluid compared with IM and reveal the role of cell wall-associated peptidoglycans and lipoarabinomannan on the Msm OM organization. Overall, these studies indicate that mycobacterial species may regulate their overall membrane functionality by regulating the synthesis of these complex arrays of lipids. Based on the structure-function relationship drawn here, documented alteration in the mycobacterial lipidome during cellular infection and/or drug treatment could reflect a mechanism to fine-tune M. tuberculosis membrane properties to its advantage. These findings are expected to inspire development of lipid-centric therapeutic approaches targeted toward its membrane.
Significance
Limited drug permeability of mycobacterial membranes is a challenging issue to resolve in a quest to develop new antibacterial drugs. One of the factors contributing to this is the lipid composition specificity toward the drug, which underlies both efficacy and toxicity. Mycobacterium synthesizes quite distinct lipid molecules, both chemically and structurally: long (C60–90) chained and branched lipid molecules. These are then localized to different spatial locations (i.e., inner and outer membrane). Investigating the lateral organization, fluidity, and membrane properties of mycobacterial lipids is required to deepen our understanding of these spatially and compositionally distinct membrane platforms and their functional roles in lipid-mediated drug resistance, survival, and pathogenesis that might enable releasing the roadblock in targeting tuberculosis.
Introduction
Mycobacterium tuberculosis (Mtb) is the most successful opportunistic human pathogen, causing tuberculosis—the leading cause of death worldwide from an infectious agent (1). The foremost reason for Mtb’s remarkable defense mechanism and survival from the host inflammatory response is its complex cell wall architecture and associated lipids, common to the Corynebacterium-Mycobacteria group (2, 3, 4, 5). The cell wall comprises inner membrane (IM) consisting of conventional glycerophospholipids and phosphatidyl-dimannosides (5). This layer is followed by a peptidoglycan-arabinogalactan complex, which in turn is covalently linked to the vital long-chain mycolic fatty acids (FAs) (C60–C90) constituting the outer membrane (OM) (6). Intercalated within the outer leaflet of OM are varieties of noncovalently localized, structurally atypical lipids: phthiocerol dimycocerosate (PDIM), trehalose dimycolate (TDM), sulfolipids (SLs), phosphatidylinositol mannosides (PIMs), and lipoarabinomannan (LAM) that harbor extensive methyl branches with exquisite stereochemistry (Fig. 1). Mycobacterial cell membrane provides a robust barrier against many drugs, leading to drug-resistant phenotypes (7), and dictates bacterial physiology by compartmentalizing processes, such as transport of lipid and proteins, protection from hostile environments, and adhesion to host receptors, etc. (8). Given the paramount importance, mycobacterial cell membrane has been the focus of considerable work centered on lipidomic characterization (9,10) and structural investigations afforded by Cryo-EM (11). However, far less is known about the lateral organization of its long-chained lipids within the membrane plane and associated structural and dynamic attributes regulating mycobacterial membrane functions that are implicated in drug interactions and virulence.
Figure 1.
Structures of representative lipids from spatially distinct membrane regions in mycobacterial species. Sulfolipids (SL-1), trehalose dimycolate (TDM), and phthiocerol dimycocerosate (PDIM) reflect the noncovalent lipids in the Mtb outer membrane (OM) along with glycopeptidolipids (GPLs). Lipoarabinomannan (LAM)-lipomannan (LM) and phosphatidylinositol mannosides (PIMs) transverse the peptidoglycan layer. α-Mycolic acids (α-MAs) represent the major component of peptidoglycan-associated lipids (PALs). Tetra-acylated phospho-myo-inositol dimannosides and Ac2PIM2 (and other acylated phosphomannosides) are most abundant in the Mtb inner membrane (IM).
Cellular membrane organization, in general, serves as an orchestrating principle underlying cellular functions by regulating domain dynamics and biomolecular interactions within the membrane plane. Mammalian cells have astounding lipid diversity that generate lateral heterogeneity (from a few nm to μm) to create lipid domains (e.g., rafts) that recruit specific lipids and proteins and fine-tune signaling pathways modulating cell cycle, migration, and immune system (12,13). Though plausible phase separation of mycobacterial membranes forming lateral heterogeneities have been reported (14,15), these studies suffer from low resolution details, thus precluding substantial information on their existence and associated properties. In addition, some bacterial species are known to form functionally relevant lipid rafts enriched in squalenes. However, the presence of such ordered membrane microdomains in mycobacteria has not been shown (15). The indirect presence of the same has been inferred though by monitoring the lateral dynamics of fluorescent lipids probes, showing restricted diffusion in certain regions of the mycobacterial OM (16).
Collectively, our understanding of the mycobacterial membrane architecture with molecular and functional details is still in infancy, largely attributed to their unique lipid repertoire and lack of selective lipid extraction approaches to investigate membrane phase behavior. What makes this even more interesting is that the lipid profiles of drug-resistant and -sensitive strains are different, indicating association of antibiotic susceptibility to lipid compositions and eventually altered membrane organization and functional properties (17,18). For instance, rifampicin-resistant Mtb shows chemical remodeling of its cell membrane, leading to altered drug permeability and virulence (18). This remodeling is synonymous with attuned membrane design and properties with drug resistance phenotype. Furthermore, biosynthesis of mycobacterial lipids and structuring of its cell membrane is actively rewired during host interaction and under various stages of infection, underlining the functional importance of mycobacterial membrane configuration, which remains understudied (3).
Here, we describe the lipid membrane properties of compositionally and spatially distinct mycobacterial lipids and functional significance of this transmembrane bilayer arrangement using M. smegmatis (Msm), a commonly used model specie of mycobacteria, a group including Mtb (5,8). Though Msm has served as an invaluable surrogate for Mtb biology over many decades, studies recapitulating subtle host pathogen interactions need cautious interpretation. However, to our advantage, studies have revealed similarity within the lipidome of both the species, with a recent study showing ∼21–26% altered abundance of certain lipid molecules using comparative lipidomics (10,19). These probably correspond to lipids associated strongly with virulence. Thus, Msm at the moment serves as an appropriate model to investigate lipids and associated membrane properties of Mtb. Needless to say, observations made in Msm would require verification in the homologous system for a complete understanding of the membrane properties within pathogenic species. Keeping this in mind, protein-free lipid extracts from the Msm IM and OM were reconstituted into model membranes in vitro and assessed using atomic force microscopy (AFM), fluorescence, infrared spectroscopy, and two-photon microscopy. The systems designed in this work serve as simplified protein-free membrane mimics of Msm membranes. These experiments demonstrated lipid domains with varying levels of membrane packing, order, hydration, and fluidity. The branched and extremely long lipid structures in OM dictate loose acyl chain packing and higher fluctuations with concomitant high interfacial hydration. The saturated lipid chains in IM govern their tighter packing at the hydrophobic lipid acyl chain region. Specifically, the conformational heterogeneity in the meromycolate chains of mycolic acid (MA) within the peptidoglycan-associated lipids (PALs) play a crucial role in regulating lipid-lipid interactions along the length of the lipid acyl chains. Along with this, the highly branched network of sugar appendages in LAM in OM negates the effect of MA, thus fine-tuning the morphological organization and lipid dynamics in Msm OM systems.
Collectively, a lipid structure-regulated gradient in various membrane properties is spatially obtained within the context of the mycobacterial cell envelope. This indicates that subtle changes in the lipid composition and structure are expected to affect the mycobacterial membrane function profoundly, further underscored by modulated lipidome by the pathogen during infection and therapeutic interventions. Of note, the absence of membrane proteins as well as plausible asymmetry in native mycobacterial cell membrane in contributing to the observed OM and IM membrane properties, and hence the fully functional landscape of mycobacterial membranes, remains unknown at present. This is currently under investigation in our lab using giant plasma membrane vesicles at least for Msm OM and designing layered bilayer structure to capture the native structural details as closely as possible.
Materials and Methods
Extraction of lipids
Msm mc2155 was grown in Middlebrook 7H9 supplemented with 10% (v/v) albumin-dextrose-catalase and 0.2% glycerol under shaking conditions at 37°C. The cells were harvested at log phase (optical density at 600 nm [OD600] of 0.8–3) and washed with distilled water. Lipids were extracted selectively from OM and IM of Msm based on a previously reported method (5). For the recovery of all noncovalently bound OM lipids (OMLs), 10 mg dry weight of cells was extracted with 1 mL of reverse micellar solution (RMS; 10 mM sulfosuccinic acid 1,4-bis (2-ethylhexyl) ester sodium salt (AOT) in heptane). The first extraction was overnight, followed by four re-extractions of 15–30 min each. For the extraction of IM lipids (IMLs), the RMS-extracted cells were thoroughly washed with water and then extracted with chloroform:methanol:water (2:1:0.1) in the same manner as above. All the extractions were carried out in monophasic solutions. For extracting the cell wall-associated lipids, the cells were treated with aqueous lysozyme solutions as reported previously (5). Briefly, 10 mg of cells were treated with 1 mL of sodium phosphate buffer (pH 7.5) containing 10 mg of lysozyme. The suspension was shaken at 150 rpm for 2 h at 37°C.
For extraction of LAM/lipomannan (LMs), cells from chloroform:methanol:water extraction of RMS-treated cells (∼50 mg of dry cell mass) were extracted with 50% ethanol as described in (5). The cells were refluxed in 50% ethanol for 4 h. The cells were pelleted down at 3500 g and reextracted twice in the same manner. The extract was dried and solubilized in 50% phenol (v/v), and the suspension was immersed in 75°C water bath for 20 min. This was followed by incubation on ice for 20 min and left to stand for 20 min at room temperature (RT). The phenol and aqueous layer were separated by centrifugation at RT at 27,000 g. The phenol layer was back extracted twice as mentioned above with LAM/LM partitioning into the aqueous phase. The aqueous phase was then dialyzed against water (molecular weight cutoff 3500).
Recovery of AOT-free fraction of lipids
To remove AOT from the lipid mixture for unbiased studies, the OML mixture was separated and purified using column chromatography using a 100–200 Silica mesh. The mobile phase was a gradient of methanol in chloroform ranging from 1 to 5%. All the lipids were eluted out in 3% MeOH followed by AOT. This was confirmed using thin layer chromatography (TLC) for the separation of each fraction obtained, followed by visualization using anthrone spray.
Liquid chromatography mass spectrometry
Extracted lipids were analyzed in both positive and negative ion mode using a 1290 Infinity Ultra-High-Performance Liquid Chromatography System attached to a 6550 iFunnel Quadrupole Time-of-Flight system (Agilent Technologies, Santa Clara, CA). The lipids were identified using databases available online (9,10). Three independent replicates were performed for all measurements.
Analysis of FAs
Phase transfer catalysis-mediated conversion of FAs to FA methyl esters was done from the lipids in the extracted fractions as described previously (5). Gas chromatography-mass spectrometry (GC-MS) of FA methyl esters was done to obtain quantitative data on the relative content of C16–C20 FAs. An 8°C/min gradient was used, the column used was a Rxi 5-mass spectrometry (30 m), and the detection was done using a time-of-flight mass spectrometer.
Analysis of lipoglycans
The presence of extracted LAM/LM were analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (4% stacking and 16% resolving acrylamide). Gels were stained with silver staining. Western blotting was done using a LAM-specific antibody (NR-13798, BEI Resources, Manassas, VA). The sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels were transferred onto an Immobilon-FL (Merck, Kenilworth, NJ) polyvinylidene difluoride membrane and blocked with 5% bovine serum albumin. The anti-Msm LAM antibody was diluted 1:5000 in 5% bovine serum albumin solution and incubated overnight at 4°C. The membranes were washed and incubated for 1 h with goat anti-mouse (1:5000) secondary antibody conjugated to horseradish peroxidase (Invitrogen, Thermo Fisher Scientific, Waltham, MA). Blots were visualized using a LI-COR Biosciences chemiluminescence imaging system (Lincoln, NE).
Analysis of lipids
Various lipid extracts, including the mycolate methyl esters, which were not quantified in GC-MS, were resolved by TLC using aluminum-backed silica gel plates (Silica gel 60 F254; Merck). Visualization of lipids was done using various spraying agents like anthrone and phosphospray as indicated.
Preparation of liposomes
Liposome suspensions were prepared by the gentle hydration method. The desired lipid ratios for OML+PAL (70.58:29.41 mol%), OML+LAM (97.80:2.20 mol%), and OML+LAM+PAL (OLP) (72.34:1.56:28.80 mol%) were diluted with chloroform in the presence of 1 mol% lipid probes. The lipid solutions were dried under a stream of N2 gas followed by lyophilization overnight. The dried films were hydrated using 20 mM Tris and 5 mM MgCl2 (pH 7.4) buffer and subjected to sonication at 65°C for 15 min, followed by five freeze-thaw cycles to generate large unilamellar vesicles (LUVs).
Fluorescence spectroscopy
The steady-state fluorescence spectroscopic measurements were performed on Varian Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies) with a temperature controller with an accuracy of ±0.1°C. A final lipid concentration of 0.5 mg/mL with probe/lipid ratio of 1:100. Laurdan was excited at 350 nm, (1-(4-trimethylammoniumphenyl)-6-phenyl-1,3,5-hexatriene p-toluenesulfonate) (TMA-DPH) and diphenylhexatriene (DPH) were excited at 352 nm, and the emission was measured from 380 to 570 nm. For calculation of Laurdan generalized polarization (GP) and anisotropy, see Supporting Materials and Methods. Lifetime measurements were performed at RT using Horiba Jobin Yvon FluoroCube IBH (Glasgow, UK) time-correlated single photon counting spectrometer. The samples were excited with a 375-nm Horiba NanoLED diode laser, and the decays were collected at 440 nm at magic angle (54.71) polarization with respect to vertically polarized excitation light. The full width at half maximum of the instrument response function was 250 ps. The decays were fitted to single or biexponential functions by iterative reconvolution using IBH Data Station 6.2 software.
Giant unilamellar vesicle formation and fluorescence microscopy
Lipids in desired ratios were mixed to obtain a concentration of 4 mg/mL. Lauran, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (N-Rh-DPPE), and/or TopFluor-Cholesterol were added to the lipid sample to yield a final probe/lipid molar ratio of 1:100 for Laurdan and 1:1000 for the latter two. Giant unilamellar vesicles (GUVs) were prepared using the electroformation method in a temperature-controlled custom-made chamber using optically transparent and electrically conductive indium tin oxide-coated glass coverslips (SPI Supplies, West Chester, PA). 60 μL of lipid mixture was spin coated (1000 rpm for 20 s) onto the coverslips and subsequently dried under vacuum overnight. The coverslips were placed within the electroformation cell, and the cell was filled with MilliQ water. Lipids were hydrated within the cell at a temperature of 65°C, and low-frequency alternating current field (sinusoidal wave function with a frequency of 10 Hz and an amplitude of 2Vp-p) was applied for 120 min. The cell was gradually cooled to RT before imaging the GUVs. The laser lines used were 488 nm (25 mW Argon laser) and 561 nm (20 mW diode-pumped solid-state laser) for N-Rh-DPPE and TopFluor-Cholesterol, respectively. The GUVs were imaged using a laser-scanning confocal microscope (Carl Zeiss, Oberkochen, Germany) at RT using an Olympus (Tokyo, Japan) 10×/0.45 NA Air objective. The images collected were of 1024 × 1024-pixel resolution. For GP imaging, GUVs were excited at 780 nm using a multiphoton (titanium sapphire) laser (Coherent, Santa Clara, CA). Two images were recorded in the range of 420–450 nm (pink) and 470–520 nm (green) using a λ/4-wave plate located at the input to compensate for any polarization effect and to obtain a circularly polarized light.
| (1) |
GP images were constructed using ImageJ and Eq. 1. GP values were depicted in the range of −1 (highest fluidity) to +1 (lowest fluidity) using a custom color palette. GP distribution obtained from the histogram were fitted to a Gaussian function using Origin Software (Origin Pro 9.1).
Atomic Force Microscopy
AFM imaging was carried out in intermittent contact (alternating current) mode, and force spectroscopy was done in contact mode with MFP-3D atomic force microscope (Asylum Research, Santa Barbara, CA) Silicon nitride cantilevers with spring constant 0.03–0.26 N/m was used. For preparation of supported lipid bilayer, LUVs as prepared above were deposited on Ted Pella mica (9.9 mm in diameter) glued on a 60-mm Petri dish and incubated for 2 h in a water bath at 65°C. Before deposition, mica was cleaved with a scotch tape to form an atomically flat surface. After incubation, the sample was washed thoroughly to remove any unfused liposome. Typical scan rate for images were 0.5 Hz. The images were plane fitted at order zero and flattened at order one. For force spectroscopic analysis, cantilever velocity was maintained at 0.7–1 μm/s and applied force was 30–35 nN. Force maps were recorded in 16 × 16 pixels (for details, see Supporting Materials and Methods). For each of the experiments, at least two independent replicates were performed. All the experiments were carried out at 23 ± 1°C. Super-resolution AFM experiments were performed by a custom-built AFM with an ultralow noise cantilever deflection sensor and a high stability photothermal excitation system. A custom-built phase-locked loop circuit was used for oscillating a cantilever at its resonance frequency with constant amplitude and for detecting frequency shift induced by the interaction force variation. The AFM head was controlled with a custom-built AFM controller. Commercially available stiff Si cantilevers (PPP-NCHAuD; NanoWorld, Neuchâtel, Switzerland) with a spring constant 42 N/m were used to capture the super resolution images.
Fourier transform infrared spectroscopy
LUVs at a concentration of 10 mg/mL in D2O-based buffer containing 20 mM Tris and 5 mM MgCl2 were prepared as mentioned above. The spectra were measured in a temperature-dependent manner using a Bruker Tensor II Fourier transform infrared spectroscopy (FT-IR) spectrometer equipped with liquid nitrogen-cooled Mercury cadmium telluride (HgCdTe) detector and in a liquid mountable cell with CaF2 windows separated by 50-μm Teflon spacers. Each spectrum was obtained by averaging out 128 spectral scans of 2 cm−1 spectral resolution. The sample chamber was purged with dry air throughout the experiment and an external circulating water bath (Huber Ministat 125) was used to control temperature with an accuracy of ±0.1°C. Three independent replicates were performed for all measurements.
Cryo-transmission electron microscopy
Five microliters of lipid were deposited on a carbon-coated holey grid. Excess fluid was blotted away with a Whatmann filter paper followed by plunging into liquid ethane using a Vitrobot (FEI, Hillsboro, OR) with a blot time of 1 s. The sample was imaged using Gatan:Orius charge-coupled device camera under high-resolution Cryo-TEM (JOEL-JEM 2100 electron microscope) at a temperature of −173.45°C. Further analysis of the vesicles was carried out using ImageJ software. For IMLs, each experiment was performed in triplicates, and for other systems, each experiment was performed twice.
Results and Discussion
Mycobacterial complex lipids pack to form stable liposomes: the effect of chain length and branching
Preliminary insights using metabolically labeled lipids in native Msm (and other bacteria) have demonstrated the correlation of MA structure with OM diffusion, specifically linked to drug interaction (16). However, it points toward a huge gap in our understanding of mycobacterial cellular membranes. Studies in cells using limited labeled lipid probes preclude obtaining information about the lipid membrane lateral organization and compartmentalization. Under these circumstances, model membranes composed of complex cellular lipids serve as attractive platforms for investigating membrane structure and dynamics in a controlled fashion, many of which cannot be studied using intact cells. Thus, to accomplish the same for the first time with mycobacterial membranes, we selectively extracted 1) IM, 2) noncovalently bound OMLs in the outer leaflet, 3) PALs within the inner leaflet of OM, and 4) hypermannosylated forms of phosphatidylinositolmannoside such as LAMs (5) from Msm.
RMS has been instrumental to obtain OM components free of IM contaminants across various Corynebacteriaceae species including Msm, but the use of sulfosuccininc acid AOT, the main component of RMS, does pose serious issues. The excess amount of AOT prevents lipidomic characterization of low abundant lipid species and may markedly influence the lipid membrane structure and associated properties within liposomal or solid supported bilayer geometries. To counter this, we developed a chromatography-based method using methanol gradient in chloroform to separate out AOT. We were able to obtain an AOT-free fraction of OMLs confirmed by TLC and mass spectrometric lipidomic characterization (Fig. S1, A, B, and I; Table S1). Next, we confirmed the presence of lipids characteristic to OM such as glycopeptidolipids (GPLs), trehalose dimycolate (TDM), SL, diacyltrehaloses, diacylglycerols (DAGs), triacylglycerols, and some loosely bound phospholipids, such as phosphatidylethanolamines, phosphatidylinositol, etc., after mapping them with the known Mtb LipidDB, MycoMass, and MycoMap databases (Table S1; (9,10)). The constituent FAs such as hydroxyphthioceranic acids (for SL) and mycocerosic/phthioceranic acids (for dimycocerosate/PDIMs) could also be identified using liquid chromatography mass spectrometry (Table S1). We also detected the presence of mycolipenic acids, constituents of long-chained and branched poly (di-, tri-, and penta-) acyl trehalose presumed to be located noncovalently in the outer leaflet of OM.
The fatty esters corresponding to the PALs revealed the presence of α-MAs (Fig. S1 C). In addition, this fraction also demonstrated the presence of other cell wall-associated lipids, such as hydroxyphthioceranic, mycolipenic, and mycocerosic acids, major lipid acyl chain components of SL, dimycocerosate, and poly acyl trehalose lipids, etc., suggesting their peptidoglycan arabinogalactan-associated nature, plausibly attributed to their long chains facilitating interdigitation to the outer leaflet (Table S2). Alternatively, this could also be due to similar lipid components located in both OM leaflets, arguing for membrane symmetry (11). Similarly, signature lipids corresponding to IM, such as diacylated phosphomannosides (Ac2PIM2), PIMs (PIM6), and other acylated phosphatidylinositol mannosides (stained with anthrone, indicating the presence of sugar moiety in the headgroup), cardiolipin, and phospholipid (Fig. S1, D and E; Table S1) could be identified. GC-MS of the constituent FAs of IMLs showed major contribution from C16:0, C22:0, C23:0, C21:0, C35:0, and C21:1 (Fig. S1 H).
Additional TLC investigations were performed to comment on extraction selectivity. In low polarity solvent (i.e., in chloroform/methanol (9.7:0.3)), low polarity lipids were seen only for OML fraction and most likely correspond to GPLs (or yet unidentified nonpolar lipids (5)), which are characteristic of OML fraction as reported previously (5). No lipids were detected under these conditions in IML (Fig. S2). Upon slightly increasing the polarity by using chloroform/methanol/water (100:14:0.8), whereas GPLs in OML were pushed up front, no corresponding bands at the same position in IML were seen. Further, certain bands of lipids in IM were observed with an almost negligible counterpart in OML, suggesting a differential lipid profile of IML and OML under these conditions. Thus, together with mass and TLC investigations, selective extraction and identification of IM and OM characteristic lipids could be accomplished.
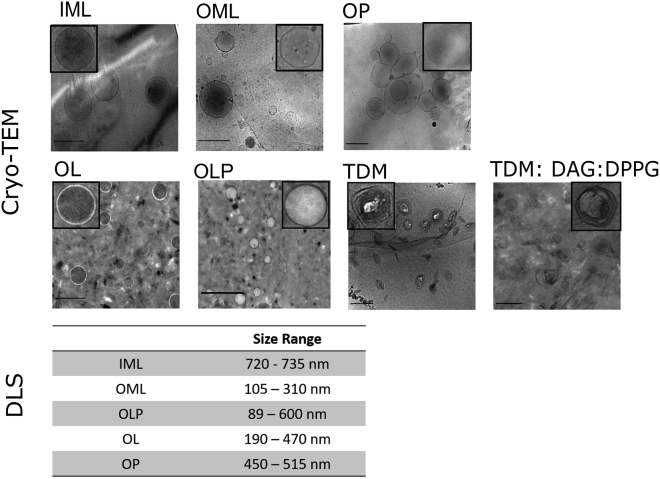
After quantitative identity assessment, we designed membrane models reflective of: 1) loosely bound lipids in the outer leaflet of the OM (denoted as OML), 2) mixed bilayers of OML with covalently attached PALs in the inner leaflet of the OM along with LAM/LM (denoted as OLP or mycomembrane), and 3) inner plasma membrane lipids (IMLs). These model membranes were reconstituted in vitro with lipids extracted from the mentioned regions of Msm cell envelope and were devoid of any proteins. Cryo-transmission electron microscopy (cryo-TEM) micrographs confirmed the formation of unilamellar vesicles by the complex lipid mixtures of Msm IM and OM with sizes of a few hundred nanometers and were either spherical or faceted (Figs. 2 and S2). Vesicle facetation is a distinctive feature observed in gel or the crystalline phase of lipids with high bending rigidity (20). We attribute the vesicle facetation in OML to curvature induced by tighter packing of MAs (including trehalose-based MA lipids). We witnessed a similar behavior previously in a chemically simpler mycomembrane model consisting of TDM, DPPG, and DPPG (Fig. 2; (21)). To precisely understand the structural features of OM, we generated two more systems, OML containing LAM (OL) and OML containing only PALs (OP), to investigate the effects of LAM and MAs in isolation on the conformation of OML. Interestingly, facetation was abolished upon the addition of LAM to OML (in OL), indicating that LAM counteracts the tight packing at the lipid headgroup region by its branched sugar structures. Similarly, it was intensified by the presence of MA in OML containing PALs (OP), further supporting the curvature defects due to MA chain-induced packing with the other lipids. The effect of LAM was dominant because no faceted vesicles were seen in OLP, rather smooth spherical boundary was observed (Fig. 2). We thus suggest that MA-containing lipids and LAM have critical roles in shape reformation of Msm membranes, required for cellular processes such as transport of lipids, proteins, nutrients, and drugs.
Figure 2.
Visualization and size variation of mycobacterial OM and IM lipid vesicles. Shown are cryo-TEM images for IML, OML, OL, OP, and OLP lipid vesicles. cryo-TEM images of TDM and the three-component model membrane of TDM:DAG:DPPG (9.1:18.2:72.7 mol%) are shown as an aid to explain the facetation behavior. The inset images show the enlarged image of a single lipid vesicle. Scale bars, 500 nm. DLS provided the hydrodynamic diameter range observed for IML, OML, OL, OP, and OLP vesicles at 25°C.
The mean (±SEM) bilayer thickness was 7.5 ± 0.2 nm for OML, 7.3 ± 0.2 for OLP, and 6.7 ± 0.1 nm for IML (Fig. S3, A–C) in accordance with the reported thicknesses using cryo-TEM. OL displayed a bilayer thickness of 7.7 ± 0.5 nm, whereas OP (6.6 ± 0.1 nm) had the lowest thickness among OM Msm preparations (Fig. S3, D and E). The presence of large hydrophilic-branched sugar structures protruding from the vesicle surface is expected to increase the bilayer thickness in OL, whereas increased van der Waals interactions by MA reduces the bilayer thickness in OP. The number-weighted average hydrodynamic size of all lipid systems by dynamic light scattering (DLS) was in the range of 120–600 nm (Fig. 2) with negative surface potentials between −16.5 and −25.3 mV (Fig. S3 F).
Distinct hydration pattern along interfacial headgroup and acyl chain dynamics in Msm IM and OM
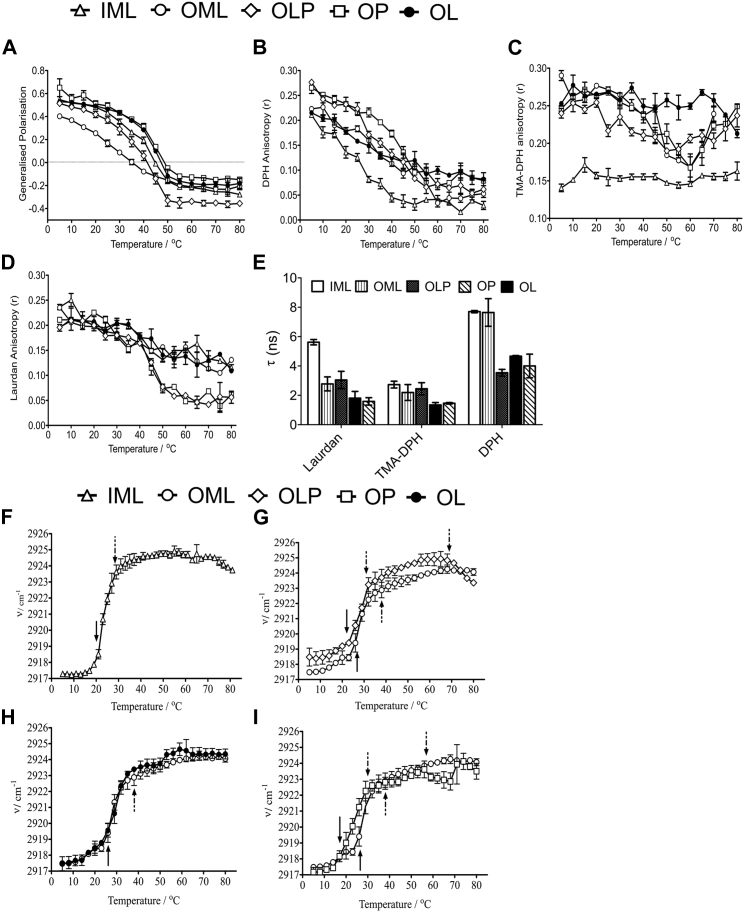
The membrane packing in the Msm model membrane systems in a bilayer depth (using probes with known membrane bilayer depths) and temperature-dependent manner (for studying thermotropic lipid phase transitions) was quantitatively determined. Using Laurdan spectroscopy, GP (Eq. S1) reflective of membrane hydration at the interfacial bilayer region was calculated (22). Lipids constituting the OML demonstrated increased membrane hydration and disorder with a broad lipid phase transition, implying high conformational heterogeneity with varying degrees of hydration (Fig. 3 A). In addition, the lipid headgroup region showed higher dynamics, evident by the low lifetime of Laurdan (Fig. 3 E), indicating a fluid environment favoring faster probe relaxation. On the other hand, IML showed high GP values at physiological temperatures (37°C) compared with OML. This implies a higher inherent packing efficiency in IML, whereas OML with branched chain lipids (as identified) prevent efficient packing; the same is also indicated by the higher lifetime of Laurdan in IML, suggesting slower relaxation (Fig. 3, A and E). Further, most abundant lipid components of IM are acylated PIMs with mannosylated headgroups; however, their acyl chains are mostly saturated as evident by the gas chromatography analysis (Fig. S1 H). This would afford a tighter intermolecular interaction (as seen in other gel or cholesterol-enriched liquid ordered (lo) lipid systems) and thus lead to lower interfacial hydration (high GP). The IML GP decreases above 50°C to the level of OML, probably due to transitions to a more fluid lipid phase, suggesting alike membrane hydration and order at high temperatures between IML and OML.
Figure 3.
Bilayer depth-dependent changes in membrane order and fluidity of mycobacterial lipid membranes. Temperature-dependent changes in (A) Laurdan GP and (B) DPH anisotropy shows microviscosity changes in the deep hydrophobic acyl chain region. (C) TMA-DPH anisotropy is indicative of microviscosity changes in the interfacial region. (D) Laurdan anisotropy depicts microviscosity changes at the lipid headgroup region. (E) Fluorescence lifetimes of the indicated lipid probes are shown. Data presented are mean (±SEM) of three independent experiments. (F–I) Temperature-dependent changes in the acyl chain conformational dynamics of the indicated mycobacterial membranes were measured using FT-IR monitoring νasymCH2. (F) Asymmetric CH2 stretching mode wavenumber in IML is shown. (G–I) Shown are comparisons between the temperature-dependent asymmetric CH2 stretching of (G) OML and OLP, (H) OML and OL, and (I) OML and OP. Black arrows indicate changes in slope, which can be ascribed to the onset of phase transitions. Solid arrows indicate main phase transition, and dashed arrows indicate minor slope change and the appearance of additional phases. Data presented are mean (±SD) of three independent experiments.
Next, the complete Msm OM system (i.e., OLP) displayed a higher GP value compared with its loosely bound lipid counterpart, OML (till 40°C), denoting higher membrane order. This is attributed to strong intermolecular van der Waals interactions rendered by the long, saturated MA chains (as found in the inner leaflet of OM in native mycobacterial cell envelope), facilitating tighter packing of lipid acyl chains and reduced water penetration at the interfacial region. This was further confirmed by studying the liposomes formed using PALs only, which showed a higher membrane order and was also similar to liposomes assembled from pure commercial MAs (Fig. S4 A). The addition of PALs to OML (forming OP) led to a significant increase in the membrane order, thus further supporting the above. This was also concluded by the rescued Laurdan lifetime in OLP compared with OML at 25°C (Figs. 3 E and S5). OLP lifetime was less than IML, indicating that the level of membrane packing and hydration in IML permits slower probe relaxation not clearly evident by GP. At higher temperatures, OLP displayed the lowest GP compared with all other systems, pointing toward comparatively loose packing between headgroups of OML, LAM, and PALs as the major contributing factor. Whereas bulky headgroups of LAM should favor a relatively hydrated membrane interface, the effect is dominated by the PAL fraction in OLP, presumably because of the relatively low amounts of LAM. Furthermore, the gradual increase in the Tm (apparent demixing temperature range for phase separation obtained from GP curve) by the systematic addition of PALs and LAM to OML signifies intricate interactions of these with OML (lipids in the outer leaflet of OM), such as electrostatic headgroup, H-bonding, and van der Waals interactions, leading to a more compact and ordered OM. Collectively, a distinctive membrane hydration and order pattern emerges by various structurally different lipids of the inner and outer layer of the Msm cell envelope. Of note, this may imply that the Msm lipid membranes maintain a membrane order gradient that is temperature sensitive and lipid structure dependent. The role of Msm membrane proteins in tempering with this gradient cannot be ascertained by our studies at the moment. No disintegration or disassembly of lipid systems was obtained at a high temperature as revealed by DLS (Fig. S6). Furthermore, the presence of AOT substantially altered the properties of OML (Fig. S7), implying that separation of AOT fractions during lipid extraction is extremely important.
Membrane fluidity plays an essential role in cellular processes, such as cell division, DNA replication, signal transduction, and biofilm formation. To analyze and quantify the same, steady-state fluorescence anisotropy DPH and TMA-DPH with known membrane position and bilayer depth was used (Fig. 3, B–D; Eq. S2; (21)) (i.e., the probes are located at different depths along the lipid acyl chain in the lipid bilayer). Within the deep hydrophobic region of the membrane bilayer, assessed using DPH, OLP showed the highest anisotropy (∼45–60% in the temperature range investigated compared with IML), arguing for a highly compact conformation of the bilayer interior with increased microviscosity (Fig. 3 B). In addition, no sharp cooperative transition was seen for any OM containing lipid systems, attributed to heterogeneity in the lipid chain composition coupled with coexisting MA chain conformations (23). Upon the addition of PALs to OML (i.e., in OP), a similar behavior to OLP was observed. This indicates that MA chains within the PAL fraction are responsible for the extraordinary compactness of Msm OM, with almost no contribution by LAM. This was supported by higher DPH anisotropy of the PAL system alone and observed a decrease in OML membrane fluidity upon the addition of PALs to OML (Fig. S4 B).
Membrane microviscosity within the interfacial region (TMA-DPH) was lowest for IML (compared with OML and OLP), suggesting substantial differences in the packing of the membrane near the hydrophilic lipid region in all systems, and almost no significant change with temperature was observed (Fig. 3 C). OML-containing systems and IML showed a decreased membrane order (higher fluidity) within bilayer interior and increased order at the headgroup region, especially above 37°C (physiological growth temperature of Msm). This trans-bilayer perturbation effect, reported previously in other systems (24), might be mediated in Msm membranes by the mismatched chain lengths generating free space toward the membrane interior. This free space is then filled by an increasing amplitude of chain fluctuations, thus accounting for higher fluidity. The mismatched chain lengths are most likely rendered by the presence of DAG in OLP/OML/IML and MA in OLP. trans-bilayer perturbation effect was most pronounced at higher temperatures in IML and may be due to C20–C30 carbon-chained IMLs being in the highly disordered conformations, leading to higher hydrophobic mismatch. The temperature-independent membrane fluidity at the interfacial region may be due to tighter contacts induced by the small headgroup of DAG and MA units. Within the lipid headgroup region (near to the interfacial region), OLP displayed a relatively sharp transition with temperature compared with all other investigated systems (Fig. 3 D). Further, IML and OML showed less sensitive membrane fluidity at this region with an increasing temperature. These data indicate that membrane fluidity varies along the length of the lipid acyl chain in various Msm model membranes and may be rationalized to specific structural features, including (but not limited to) chain lengths, level of unsaturation, and branched network of contributing lipids.
For all systems, a higher lifetime of TMA-DPH compared with DPH implies that the deeper, heterogeneous acyl chains are not packed as tightly as near the headgroup region (Fig. 3 E). The presence of branched lipids in OML and OLP could explain this inefficient packing at the headgroup region as the CH3 branches are situated in C1–C8; however, reasons for IML remain unknown.
Orthogonal investigation of the conformational order of acyl chains was performed using infrared spectroscopy (Fig. 3, F–I). The internal vibrational mode of lipid acyl chains (asymmetric CH2; νasymCH2) give rise to vibrational bands that act as reporters of trans-gauche ratio and kinks, thus mapping the conformational dynamics of the lipid chains in a label-free manner. Inner and outer Msm model membranes displayed a broad phase transition over a wide temperature range compared with one-component lipid systems hinting toward coexisting lipid phases with marginal difference in their acyl chain conformations. This was most evident for OM-containing systems harboring MA chains as these exist in a variety of conformational states: the four-chain W-shape with parallel configuration of the chain, folding extended conformation, and fully extended states (23,25). The aforementioned MA phases are separated by small energy differences and thus may account for the shallow νasymCH2-T dependence. The maximum of the νasym CH2 vibrational mode increases gradually with temperature and slope changes (solid and dashed black arrows) suggest onset of lipid phase transitions at ≈ 20°C, ≈ 28°C for IML (Fig. 3F), and at ≈ 28°C, ≈ 38 °C, for OML (Fig. 3G). For both systems, the wavenumber range can be assigned to a solid ordered-like phase. At higher temperatures, the change in the slope indicates the formation of a new phase. This phase region may be attributed to a solid-liquid phase coexistence region. Though the identity and nature of these phases remains currently unknown, based on the microscopy and AFM data at RT, solid-liquid phase coexistence is inferred. Further studies using pressure-dependent FT-IR and molecular simulations would shed light on the structural details of various lipid phases and chain conformations that can exist in these Msm membrane models. For OP and OL (Fig. 3, H and I), additional phase transitions were observed as compared with OML, demonstrating that LAM (decreased νasymCH2) and PALs (increased νasymCH2) induce disordering and ordering within the lipid acyl region, respectively (Fig. S4 C). Interestingly, the coaddition of both LAM and PALs counteracted each other in OLP but displayed relatively more ordered conformation of the lipid acyl chains compared with OML (Fig. 3 G).
Noticeably, the apparent Tm (also from fluorescence studies) for OLP agrees well with the thermodynamic behavior of the mycobacterial cell wall (26), indicating the suitability of the Msm membrane models to mimic the membrane order, conformational dynamics, and physical properties of the natural mycomembrane in a pronounced manner. We show for the first time, to our knowledge, that Msm IM substantially deviates from the OM in the packing of their lipid acyl chains, showing a narrower phase distribution that is temperature independent above 40°C (Fig. 3 F). This suggests that composition and structure of lipids within Msm IM dispose this layer to have compact acyl chain conformation and packing, thus creating a packing gradient from OM to IM, which may have crucial implications for the diffusion of nutrients as well as drugs across the mycobacterial cell envelope.
Phase segregation and lipid microdomains with structure-dependent membrane mechanics
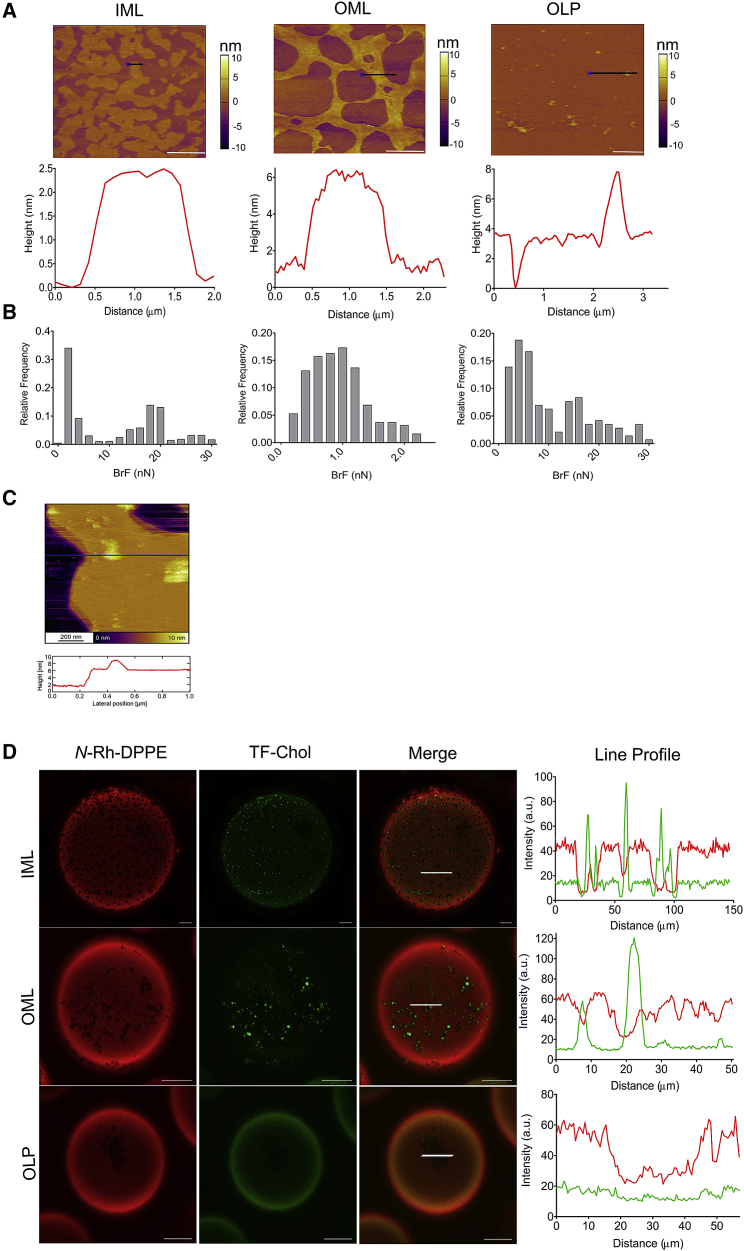
AFM of solid supported bilayers has furnished significant insights into the lateral organization of model and natural membranes, specifically lipid phase segregation and domains/lipid rafts with sub-nm z resolution (27,28). Msm IM and OM displayed lateral lipid phase segregation with at least two discernable regions or lipid phases/domains of varying heights (Fig. 4 A). We refer to them as ordered or disordered lipid regions in mycobacterial model membranes because of the lack of conclusive evidence detailing their identity at present. The height difference between ordered and disordered lipid regions were between 2 and 3.8 nm for all indicated systems, giving a total bilayer height of 7–8 nm, in excellent agreement with intact bacterial cell wall cryoelectron tomography (Fig. 4 A; (11)). The lipid regions had smooth boundaries in IML and OML but were highly irregular in OLP systems, most probably because of LAM and PALs altering the membrane lipid chain packing, leading to attenuated line energy (tension) associated with the rims of the two demixing phases (29). Even though the height mismatch is maximum for OLP, it is not the only contributor to line tension. Membrane defects and curvature rendered by longer and branched lipids in OLP are also some factors to be considered (30). The reported height difference of phase-segregated raft-like model membranes is around 1 nm, implying that the lateral organization and lipid packing in eukaryotic and mycobacterial membrane is markedly different, although the latter shows for the first time, to our knowledge, the ability of Msm lipids to phase segregate, which could be biologically relevant as in eukaryotes.
Figure 4.
Visualization of the domain architecture in mycobacterial model membranes. (A and B) Shown is AFM imaging (A) and force spectroscopy (B) of IML, OML, and OLP. The topographical images show phase separation of these independent lipid systems into ordered and disordered phase at 25°C. Scale bars, 5 μm for IML and 2 μm for OML and OLP. For IML and OLP, the height difference between the two phases is ∼3 nm, whereas for OML, it is 2 nm, suggesting differential lateral packing of these lipids. The BrF distribution shows the mean values centered at 4.24 ± 3.5 nN and 19.9 ± 3.8 nN for IML (n = 500), 0.93 ± 0.46 nN for OML (n = 194), and 5.82 ± 1.1 nN and 19.7 ± 4.61 nN for OLP (n = 144), showing nanomechanical heterogeneity in these samples. Data represented are mean (±SD). (C) Shown is super-resolution AFM image of IML and associated line profile at 25°C. (D) Confocal fluorescence microscopy images of indicated lipid systems are shown. Fluorescence intensity was collected in two channels at 25°C. N-Rh-DPPE was used as a marker labeling preferentially the disordered domains (detected by red channel), TopFluor-Cholesterol partitions preferentially into lo phase of the membrane. Line profile of the GUVs shows differential incorporation of N-Rh-DPPE (red line), and TopFluor-Cholesterol (green line) into GUVs are plotted on the right. Scale bars, 20 μm. To see this figure in color, go online.
Nanomechanical properties measured using AFM force spectroscopy revealed distinct populations of break-through force (BrF), the force required to pierce the membrane bilayer. The distribution of forces in IML and OLP were comparable to those found in membranes, showing liquid disordered (ld) and ordered lipid phase segregation, respectively (31, 32, 33). This partially reinforces the coexistence of ordered and disordered lipid phases in Msm membranes (Fig. 4 B); however, this requires additional experiments. Moreover, the ordered lipid region/phase in Msm membranes may be either lo or solid phases, which cannot be distinguished by our methods at present. Noticeably, the BrF in OML were extremely low, indicating a highly disordered lipid bilayer with heterogeneous and low membrane packing, also supported by GP results. Thus, the highly branched and long chain lipids specific to OM lead to high membrane fluctuations in their acyl chain packing. This implies that lateral structure of OM allows for unrestricted diffusion, whereas compact lateral structure of the second membrane layer (i.e., IM) might act as a checkpoint to regulate diffusion and permeation through mycobacterial membranes for an optimal biological activity. Further, super-resolution AFM afforded significant insights at the nanoscopic level in IML (Fig. 4 C), showing a highly heterogeneous membrane organization within nm regime with a concomitant height profile. Taken collectively, the data point toward a distinctive lateral lipid organization within spatially separated mycobacterial OMs and IMs. Further investigations into their ultrafine structure, domain dynamics, and how the transmembrane domain architecture is biologically relevant are warranted.
Coexistence of ordered and disordered lipid regions in Msm model membranes was also demonstrated in GUVs using confocal microscopy with lipid domain-specific probes that are known to selectively partition into lo and ld phases (Fig. 4 D; (27)). IML GUVs demonstrated a three-phase coexistence region in which three types of domains/regions were distinguishable: dark domains in which no partitioning of any of the lo and ld probes was observed, weakly fluorescent domains harboring lo dyes, and strongly fluorescing domains in which ld dyes partitioned (Figs. 4 D and S8). This aligns with the extended range of AFM BrF in IML, hinting toward minor amounts of gel-like solid ordered domains from which both the fluorescent dyes are excluded. Because lo markers may also partition into the gel phase, thus commenting on dark regions in IML devoid of any fluorophore, we generated GUVs from 1,2-dioleoyl-sn-glycero-3-phosphocholine:1,2-dipalmitoyl-sn-glycero-3-phosphocholine:cholesterol (45:45:5 mol%), which has been previously shown to exhibit a solid-liquid phase coexistence (i.e., gel-ld (34,35)). We could see the presence of dark regions/domains within these GUVs (Fig. S9) in which TopFluor-Cholesterol did not partition, indicating that this lo marker does not partition into the gel phase of phase-segregated lipid membrane, showing gel-liquid phase coexistence. This supports our hypothesis on the presence of ordered gel-like domains within the GUVs made from extracted IMLs (Fig. 4 D). However, as mentioned above, the ordered phase as inferred may be either lo or a solid phase or both and requires additional investigations. OML demonstrated a continuous stretch of connecting intermittent patterns of dark lipid domains devoid of ld probe and strongly fluorescing regions having lo dyes (punctate appearance) (Figs. 4 D and S10). The distinct shape of these fluid domains has its origin in the compositional and the energetic differences between coexisting lipid conformations in OML. On the other hand, OLP demonstrated the presence of disordered regions wherein the ld dye had partitioned, whereas no specific partitioning of lo probes was observed but, instead, a uniform distribution was visualized (Figs. 4 D and S11). Coupled with AFM and Laurdan spectroscopy, this would imply that OLP has a more fluid-like disordered lipid region compared with IML, again implicating the nature and structure of the constituting lipid chains and their packing in regulating membrane bilayer structure and properties.
To delve deeper into the coexistence of distinct lipid regions in mycobacterial model membranes and map the same to the membrane hydration and order, we performed quantitative imaging of lipid order using Laurdan two-photon microscopy. Msm model membranes representative of IM and OM regions from the native cell envelope were used by generating GUVs reconstituted from protein-free lipid extracts from the respective cell envelope regions (Fig. 5; Table 1 and S12).
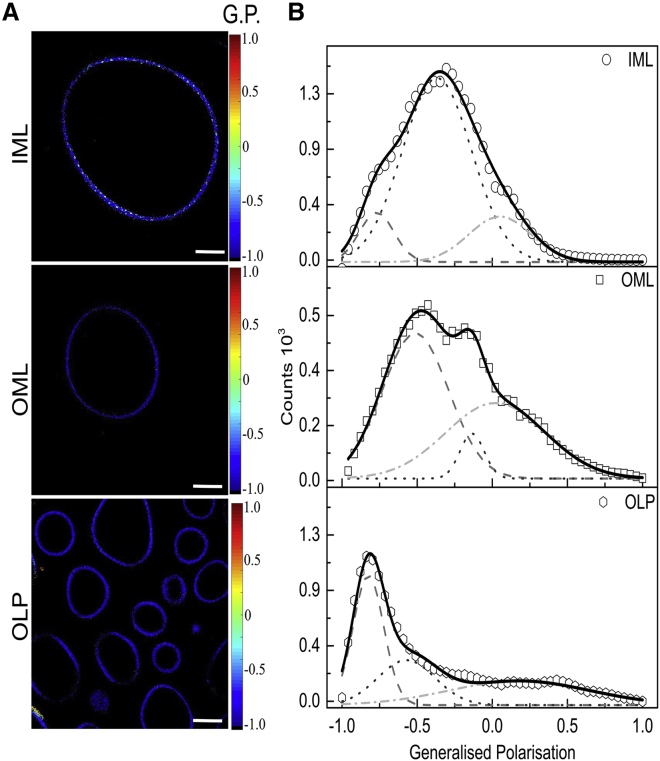
Figure 5.
Membrane lipid order maps of mycobacterial model membrane revealing phase segregation and lipid microdomain formation. (A) Pseudocolored Laurdan GP images of indicated GUVs at 25°C are shown. (B) Shown is the GP distribution from the stack of GP images deconvoluted by fitting Gaussian distribution. Scale bars, 64 μm (n of IML = 39, OML = 85, and OLP = 69). To see this figure in color, go online.
Table 1.
Data from GP Imaging Showing Peak Centroid (C1, C2, and C3) Representing Distinct Lipid Phases and Area Under the Curve of Laurdan-Stained Pixels for the Respective Populations of Different Msm Lipid Systems
| C1 | % Coverage (C1) | C2 | % Coverage (C2) | C3 | % Coverage (C3) | |
|---|---|---|---|---|---|---|
| IML | −0.77 ± 0.02 | 29.03 ± 3.04 | −0.37 ± 0.02 | 45.53 ± 3.37 | 0.15 ± 0.01 | 25.44 ± 1.52 |
| OML | −0.51 ± 0.06 | 60.31 ± 3.38 | −0.13 ± 0.05 | 19.63 ± 3.59 | 0.02 ± 0.07 | 26.08 ± 5.14 |
| OLP | −0.82 ± 0.04 | 55.61 ± 5.08 | −0.57 ± 0.06 | 36.41 ± 3.49 | 0.17 ± 0.05 | 15.52 ± 4.06 |
Error are SD. χ2 for fitting 0.99–0.98.
The ratiometric measurement of fluorescence intensity affords information on the hydration and ordering levels in lipid domains by the parameter of GP. Both IML and OML showed a broad distribution of GP values, indicating a wide distribution of lipid domain population with varying levels of order (indicative by their GP values). On average, three populations of lipid domains may be assigned within IML and OML systems, supporting AFM and microscopy data (based on GP histograms). However, all the three showed higher hydration and lower order within OML (low GP values centered at −0.51, −0.13, and 0.02 in OML), supporting AFM force distribution data. The addition of PALs and LAM to OML (OLP) caused a substantial shift in the lipid population, which was skewed toward fluid lipid population (centered at GP = −0.82). OLP GP distribution indicates that all contributing coexisting lipid phases only marginally differ in hydration and ordering and are devoid of highly ordered gel-like phases, in accordance with other results. This could be rationalized by the opposing effects of LAM and PALs in fine-tuning the overall properties in the OLP system. The ordered and disordered lipid phases in mycobacterial IM and OM appear to be highly distinct owing to their compositional and structural heterogeneity.
Conclusions
Deciphering the spatial organization and structure of mycobacterial lipid membranes greatly facilitates our understating of how the bacterium employs the lipid arcade to form a robust cell envelope to guard the bacterial biology. Long-standing notions majorly supported the mycobacterial cell membrane to form a massive permeability barrier against drugs; however, the role of specific lipids in shaping the membrane is poorly understood. This is even more intriguing when the chemical structure of mycobacterial lipids is taken into account: serpentine-like long carbon chains of 80–90 atoms, excessive methyl branches, and network of sugar appendages. Furthermore, mycobacterial mutants defective in the synthesis of specific cell wall components have been demonstrated with compromised permeability and surprisingly attenuated virulence. These findings highlight the importance of the integrity of the mycobacterial cell membrane for its survival and pathogenesis. Elegant studies have provided indispensable clues toward the structure of the mycobacterial IM and OM, complemented with precise lipidomic compositional analysis.
In this study, we investigate various biophysical membrane properties of the OM and IM lipids of Msm (used as a surrogate for Mtb and reflective of mycobacterial membranes) and correlate the same to the lipid chemical makeup. We designed protein-free membrane systems reflective of Msm IM and OM by in vitro reconstitution of extracted lipids. We show using these model systems that the lipids from the outer mycobacterial membrane have a high conformational heterogeneity attributed to the branched nature of lipids, including a contiguous stretch of methyl branches that prevent efficient chain packing. This accounts for their high interfacial hydration and low membrane order. On the other hand, lipids from the IM demonstrate lower interfacial headgroup hydration, leading to higher membrane order. This is assigned to the saturated nature of lipid acyl chains constituting the IMLs. Within the mycomembrane (i.e., OMLs along with peptidoglycan-associated lipids and LAM (OLP)), PALs and LAM counteract each other’s effect, leading to an overall compact membrane and relatively high order.
Within the deep acyl chain region, OMLs possess low rotational freedom compared with IMLs. This is mostly because of the saturated nature of the IM acyl chains, leading to higher conformational freedom and acyl chain fluctuations; the same would be limited by the methyl branches, cyclopropane rings, and double bonds. Interestingly, mycobacterial lipids, even with their distinct structural attributes, were capable of forming phase-segregated membranes, the biological role of which remains understudied. Whereas IMLs form continuous boundaries of ordered and disordered regions, extractable OMLs formed relatively circular domains, probably because of higher line tension. Mycomembrane displayed intermittent ordered regions and is attributed to a large height difference because of the presence of long MAs, which leads to a thermodynamically unstable system due to exposed hydrophobic chains at the ordered/disordered interface.
The gradient of membrane order and fluidity inherent in going from OM to IM in these models suggests an intrabilayer arrangement to regulate the uptake of nutrients, transport of protein, and differential drug interaction profiles. Furthermore, the presence of specific lipids in the mycomembrane fine-tunes the membrane properties, revealing that mycobacteria could modulate the membrane properties in a spatially constrained fashion by altering the synthesis of IMLs or OMLs, respectively. It is emphasized that these findings are derived from simplified protein-free membrane mimics of the Msm cell envelope, thus the role of membrane proteins in regulating the studied membrane properties and hence the complete biological functions of mycobacterial membranes remain to be investigated. Nonetheless, our findings will impact future studies targeted toward understanding transport pathways and the mechanism for lipids, proteins, and antimycobacterial drugs. In addition, crucial insights into the role of structurally dissimilar IMLs and OMLs in maintaining mycobacterial cell membrane integrity and function would facilitate uncovering new potential targets against tuberculosis in specific and lipid-dependent maladies in general.
Author Contributions
S.K. conceived and designed the project. P.A. and A.T.S. conducted experiments for Figs. 2, 3, 4, and 5. M.M. performed experiments for Figs. 4 D and 5. A.S. performed experiments for Fig. 3 E. R.K. contributed to the purification of OM fractions, which was essential for Figs. 2, 3, 4, and 5. S.K., P.A., A.T.S., and M.M. prepared the figures. S.K. wrote the manuscript and interpreted the results. All authors reviewed the manuscript.
Acknowledgments
We acknowledge the central AFM and confocal facility at Indian Institute of Technology Bombay. We thank Prof. M. Daffe, Universite de Toulouse of Centre national de la recherche scientifique for his kind suggestions and initial experiments with high-pressure TLC of extracted lipids. We thank Dr. Ruchika Dadhich for her help with the cryo-TEM data analysis, manuscript assembling, and insights. We acknowledge Prof. T. Fukuma, Prof. Keizuke Miyazawa, and Bio-AFM Summer School of Kanazawa University for enabling Dr. Dadhich to acquire super-resolution AFM images. Abhishek Kalarikkal is gratefully acknowledged for initial optimization of the TLC experiments. Prof. Roland Winter is kindly acknowledged for lending the GUV electroformation cell on a long-term loan. We thank Prof. Supdita Maiti and Mamata Kallianpur for providing access to a two-photon microscope.
Department of Science and Technology-Science and Engineering Research Board grant EMR/2016/005414, Wadhwani Research Centre for Bioengineering, Board of Research in Nuclear Sciences grant 201805BRE01RP04922, Department of Biotechnology Ramalingaswami Fellowship, and Department of Science and Technology-Inspire Faculty Award (to S.K.) financially supported this work.
Editor: Ilya Levental.
Footnotes
P. Adhyapak and A. T. Srivatsav contributed equally to this work.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.01.027.
Supporting Material
References
- 1.Pai M., Behr M.A., Raviglione M. Tuberculosis. Nat. Rev. Dis. Primers. 2016;2:16076. doi: 10.1038/nrdp.2016.76. [DOI] [PubMed] [Google Scholar]
- 2.Jackson M. The mycobacterial cell envelope-lipids. Cold Spring Harb. Perspect. Med. 2014;4:a021105. doi: 10.1101/cshperspect.a021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gago G., Diacovich L., Gramajo H. Lipid metabolism and its implication in mycobacteria-host interaction. Curr. Opin. Microbiol. 2018;41:36–42. doi: 10.1016/j.mib.2017.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegrist M.S., Bertozzi C.R. Mycobacterial lipid logic. Cell Host Microbe. 2014;15:1–2. doi: 10.1016/j.chom.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bansal-Mutalik R., Nikaido H. Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc. Natl. Acad. Sci. USA. 2014;111:4958–4963. doi: 10.1073/pnas.1403078111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jankute M., Cox J.A., Besra G.S. Assembly of the mycobacterial cell wall. Annu. Rev. Microbiol. 2015;69:405–423. doi: 10.1146/annurev-micro-091014-104121. [DOI] [PubMed] [Google Scholar]
- 7.Brennan P.J., Nikaido H. The envelope of mycobacteria. Annu. Rev. Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 8.Chiaradia L., Lefebvre C., Daffé M. Dissecting the mycobacterial cell envelope and defining the composition of the native mycomembrane. Sci. Rep. 2017;7:12807. doi: 10.1038/s41598-017-12718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sartain M.J., Dick D.L., Belisle J.T. Lipidomic analyses of Mycobacterium tuberculosis based on accurate mass measurements and the novel “Mtb LipidDB”. J. Lipid Res. 2011;52:861–872. doi: 10.1194/jlr.M010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Layre E., Sweet L., Moody D.B. A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem. Biol. 2011;18:1537–1549. doi: 10.1016/j.chembiol.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann C., Leis A., Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. USA. 2008;105:3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sezgin E., Levental I., Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varshney P., Yadav V., Saini N. Lipid rafts in immune signalling: current progress and future perspective. Immunology. 2016;149:13–24. doi: 10.1111/imm.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen H., Garton N.J., Barer M.R. Lipid domains of mycobacteria studied with fluorescent molecular probes. Mol. Microbiol. 1999;31:1561–1572. doi: 10.1046/j.1365-2958.1999.01304.x. [DOI] [PubMed] [Google Scholar]
- 15.Lopez D., Koch G. Exploring functional membrane microdomains in bacteria: an overview. Curr. Opin. Microbiol. 2017;36:76–84. doi: 10.1016/j.mib.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez-Rivera F.P., Zhou X., Bertozzi C.R. Visualization of mycobacterial membrane dynamics in live cells. J. Am. Chem. Soc. 2017;139:3488–3495. doi: 10.1021/jacs.6b12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pal R., Hameed S., Fatima Z. Comparative lipidomics of drug sensitive and resistant Mycobacterium tuberculosis reveals altered lipid imprints. 3 Biotech. 2017;7:325. doi: 10.1007/s13205-017-0972-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lahiri N., Shah R.R., Moody D.B. Rifampin resistance mutations are associated with broad chemical remodeling of Mycobacterium tuberculosis. J. Biol. Chem. 2016;291:14248–14256. doi: 10.1074/jbc.M116.716704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ortalo-Magné A., Lemassu A., Daffé M. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J. Bacteriol. 1996;178:456–461. doi: 10.1128/jb.178.2.456-461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olubummo A., Schulz M., Binder W.H. Phase changes in mixed lipid/polymer membranes by multivalent nanoparticle recognition. Langmuir. 2014;30:259–267. doi: 10.1021/la403763v. [DOI] [PubMed] [Google Scholar]
- 21.Dadhich R., Singh A., Kapoor S. Biophysical characterization of mycobacterial model membranes and their interaction with rifabutin: towards lipid-guided drug screening in tuberculosis. Biochim Biophys Acta Biomembr. 2019;1861:1213–1227. doi: 10.1016/j.bbamem.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez S.A., Tricerri M.A., Gratton E. Laurdan generalized polarization fluctuations measures membrane packing micro-heterogeneity in vivo. Proc. Natl. Acad. Sci. USA. 2012;109:7314–7319. doi: 10.1073/pnas.1118288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villeneuve M., Kawai M., Nakahara H. Temperature dependence of the Langmuir monolayer packing of mycolic acids from Mycobacterium tuberculosis. Biochim. Biophys. Acta. 2005;1715:71–80. doi: 10.1016/j.bbamem.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg E.M., Lester D.S., Zidovetzki R. Effects of diacylglycerols and Ca2+ on structure of phosphatidylcholine/phosphatidylserine bilayers. Biophys. J. 1994;66:382–393. doi: 10.1016/s0006-3495(94)80788-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villeneuve M., Kawai M., Nakahara H. Conformational behavior of oxygenated mycobacterial mycolic acids from Mycobacterium bovis BCG. Biochim. Biophys. Acta. 2007;1768:1717–1726. doi: 10.1016/j.bbamem.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Liu J., Barry C.E., III, Nikaido H. Mycolic acid structure determines the fluidity of the mycobacterial cell wall. J. Biol. Chem. 1996;271:29545–29551. doi: 10.1074/jbc.271.47.29545. [DOI] [PubMed] [Google Scholar]
- 27.Kapoor S., Werkmüller A., Winter R. Temperature-pressure phase diagram of a heterogeneous anionic model biomembrane system: results from a combined calorimetry, spectroscopy and microscopy study. Biochim. Biophys. Acta. 2011;1808:1187–1195. doi: 10.1016/j.bbamem.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 28.Huarte N., Carravilla P., Nieva J.L. Functional organization of the HIV lipid envelope. Sci. Rep. 2016;6:34190. doi: 10.1038/srep34190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.García-Sáez A.J., Chiantia S., Schwille P. Effect of line tension on the lateral organization of lipid membranes. J. Biol. Chem. 2007;282:33537–33544. doi: 10.1074/jbc.M706162200. [DOI] [PubMed] [Google Scholar]
- 30.Kuzmin P.I., Akimov S.A., Cohen F.S. Line tension and interaction energies of membrane rafts calculated from lipid splay and tilt. Biophys. J. 2005;88:1120–1133. doi: 10.1529/biophysj.104.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Attwood S.J., Choi Y., Leonenko Z. Preparation of DOPC and DPPC supported planar lipid bilayers for atomic force microscopy and atomic force spectroscopy. Int. J. Mol. Sci. 2013;14:3514–3539. doi: 10.3390/ijms14023514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Manyes S., Oncins G., Sanz F. Effect of temperature on the nanomechanics of lipid bilayers studied by force spectroscopy. Biophys. J. 2005;89:4261–4274. doi: 10.1529/biophysj.105.065581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murthy A.V.R., Guyomarc’h F., Lopez C. The temperature-dependent physical state of polar lipids and their miscibility impact the topography and mechanical properties of bilayer models of the milk fat globule membrane. Biochim. Biophys. Acta. 2016;1858:2181–2190. doi: 10.1016/j.bbamem.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 34.Veatch S.L., Keller S.L. Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys. J. 2003;85:3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korlach J., Schwille P., Feigenson G.W. Characterization of lipid bilayer phases by confocal microscopy and fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. USA. 1999;96:8461–8466. doi: 10.1073/pnas.96.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.