Abstract
Inhibition of virus replication by means of RNA interference has been reported for several important human pathogens, including human immunodeficiency virus type 1 (HIV-1). RNA interference against these pathogens has been accomplished by introduction of virus-specific synthetic small interfering RNAs (siRNAs) or DNA constructs encoding short-hairpin RNAs (shRNAs). Their use as therapeutic antiviral against HIV-1 is limited, because of the emergence of viral escape mutants. In order to solve this durability problem, we tested DNA constructs encoding virus-specific long-hairpin RNAs (lhRNAs) for their ability to inhibit HIV-1 production. Expression of lhRNAs in mammalian cells may result in the synthesis of many siRNAs targeting different viral sequences, thus providing more potent inhibition and reducing the chance of viral escape. The lhRNA constructs were compared with in vitro diced double-stranded RNA and a DNA construct encoding an effective nef-specific shRNA for their ability to inhibit HIV-1 production in cells. Our results show that DNA constructs encoding virus-specific lhRNAs are capable of inhibiting HIV-1 production in a sequence-specific manner, without inducing the class I interferon genes.
Supplementary information
The online version of this article (doi:10.1038/sj.gt.3302786) contains supplementary material, which is available to authorized users.
Keywords: HIV-1, RNA interference, lhRNA
Introduction
RNA silencing or RNA interference (RNAi) is an evolutionary conserved sequence-specific post-transcriptional gene regulation mechanism that plays an important role in cell differentiation and development.1, 2, 3 In addition, RNAi serves as a defence mechanism against invading viruses and transposons.4, 5, 6 RNA interference is triggered by double-stranded RNA (dsRNA) molecules, which are processed in the cytoplasm by the dsRNA-specific endonuclease Dicer into 19–24 nucleotides (nt) small interfering RNAs (siRNAs) or micro-RNAs (miRNAs).7 These si/miRNAs are incorporated into the multiprotein RNA-induced silencing complex (RISC) that guides the recognition and ultimately the cleavage or translational repression of complementary single-stranded RNA, such as messenger RNA or viral genomic RNA.8, 9, 10
RNA interference has been employed to inhibit the replication of a wide range of viruses, including the human immunodeficiency virus type 1 (HIV-1), hepatitis C virus (HCV), hepatitis B virus (HBV), dengue virus, poliovirus, influenza virus A, coronaviruses, herpesviruses and picornaviruses.11 Human immunodeficiency virus type 1 virions contain a single-stranded RNA genome that is a putative RNAi target. After entry into a host cell the genomic RNA is reverse transcribed into dsDNA, which is integrated into the host chromosomal DNA. Newly synthesized unspliced genome-length and spliced subgenomic viral RNAs are possible targets for RNAi in the cytoplasm. It has recently been reported that HIV-1 encodes a suppressor of RNAi, the Tat protein, indicating that HIV-1 replication is controlled by RNAi in human cells.12 Due to its sequence specificity, RNAi is a potentially powerful and selective method for intracellular immunization against HIV-1 infection. RNA interference-mediated suppression of HIV-1 replication has been accomplished by synthetic siRNAs in a transient manner13, 14, 15, 16 and by short-hairpin RNA (shRNA)-expression vectors in stably transfected cells.17, 18, 19 Despite potent inhibition, the use of siRNA/shRNA as a therapeutic antiviral is limited, because of the rapid emergence of HIV-1 escape mutants.19, 20, 21 Minor sequence changes in the target sequence, sometimes even a single point mutation, are sufficient to abrogate the inhibition of virus replication. Strategies to reduce the chance of viral escape include the simultaneous use of multiple siRNAs22, 23 or the use of long-hairpin RNA (lhRNA, a single-hairpin molecule) or long dsRNA (two complementary molecules that form a duplex).16, 24 Another possibility is the use of miRNA-based approaches, which do not require perfect sequence complementarity.25, 26
Several reports describe efficient RNAi induction by lhRNA and long dsRNA as in vitro generated transcripts that are transfected into cells or as gene constructs that produce the transcripts intracellularly. Transfection of pre-implantation mouse embryo cells, undifferentiated embryonic stem cells and embryonic carcinoma cells with in vitro synthesized long dsRNA confers specific gene silencing.27, 28 However, exposure of non-embryonic mammalian cells to dsRNAs longer than 30 basepairs (bp) leads to rapid induction of a specific set of cytokines, including the class I interferons (IFNs).29 During natural virus infections, the IFN response is activated by virus-produced dsRNAs, and acts as an innate defense mechanism. Viruses counter this response by encoding IFN antagonists, which are also responsible for the fact that antiviral IFN therapy is often not successful.30, 31 So far, virus-encoded RNAi suppressor factors, like the HIV-1 Tat protein, do not appear to be able to suppress induced antiviral RNAi. Strong induction of RNAi by intracellular expression of virus-specific dsRNAs is likely to outcompete the inhibiting effects of RNAi suppressors.
Efficient RNAi-mediated gene silencing has been shown in mammalian cells by endogenously expressed long dsRNAs.28, 32, 33 In Chinese hamster ovary (CHO) cells, a DNA construct encoding a 700 bp long dsRNA specifically inhibits luciferase expression in a sequence-specific manner.34 Complete and specific gene silencing was achieved in different mammalian cell types by expression of 500, 800, or even 1000 bp long dsRNAs.32, 35, 36, 37 Interestingly, intact dsRNA could not be detected in these cells, suggesting that it is rapidly processed by Dicer in the cytoplasm. Recently, Ski knockdown mice have been produced using a DNA construct encoding long dsRNA-specific for the murine Ski gene.38 These results suggest that dsRNA is tolerated in mammalian cells, most likely because it is rapidly processed by the RNAi machinery.
Several antiviral approaches using extended dsRNA have been reported in plant and insect cells lacking the innate antiviral IFN response. Although plants and insects lack the IFN response, they also have potent innate antiviral responses, comparable to those in mammals.39 Transient expression of DNA constructs encoding virus-specific dsRNA in plant protoplasts or insect cells partially protects the cells from infection by the homologous virus.40, 41 Stable expression of such constructs in plant or insect cells renders the cells completely resistant or immune to infection.42, 43 In vitro made long dsRNAs have been used to inhibit HIV-1 production under certain conditions without induction of the IFN response.16, 24 We have previously demonstrated potent inhibition of HIV-1 replication in T cells that stably express an shRNA targeted to viral nef gene sequences.19 To test whether endogenously expressed lhRNA and long dsRNA can inhibit HIV-1 at least as potently as sh-nef, we constructed and tested a series of DNA constructs.
Results
RNA interference targets in the human immunodeficiency virus type 1 genome
It has previously been demonstrated that HIV-1 replication can be inhibited by siRNAs and shRNAs directed against viral targets.11, 14, 15, 18, 19, 44 Most of the active siRNAs against HIV-1 are targeted to the early regulatory tat, rev and nef genes.19, 20, 45, 46 Interference with an early stage of the HIV-1 replication cycle may be beneficial. For this reason, the DNA constructs encoding lhRNAs (a single-hairpin molecule) and long dsRNAs (two complementary molecules that form a duplex) were designed to target tat, rev and nef sequences as indicated in Figure 1.
Figure 1.
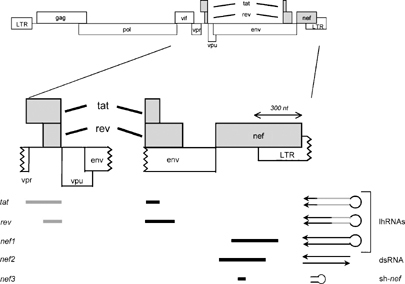
Scheme of the human immunodeficiency virus type 1 (HIV-1) pLAI proviral genome and target sequences used for the design of long-hairpin RNAs (lhRNAs). The target sequences are indicated as bars below the HIV-1 coding regions. lhRNA (300 basepairs (bp)) tat fuses tat exon 1 (gray bar, 5422–5626) and tat exon 2 (black bar, 7972–8017) sequences, rev fuses rev exon 1 (gray bar, 5562–5626) and rev exon 2 (black bar, 7972–8206) and nef1 contains nef-LTR sequences (8519–8818). Double-stranded RNA nef2 is a duplex of two separate, complementary sense and antisense nef sequences (8416–8695). The positive control sh-nef is a 21-bp hairpin consisting of nef sequences (8552–8571).19
Inhibition of human immunodeficiency virus type 1 by in vitro transcribed ds-nef2 RNA and its diced product
We initially tested whether in vitro transcribed and annealed nef2 dsRNA and its in vitro diced product si-nef2, a mixture of nef-specific siRNAs, can inhibit HIV-1. Nef2 dsRNA of 300 bp was diced in vitro to create si-nef2 RNAs of approximately 21 bp (Figure 2a). We cotransfected 500 ng of the HIV-1 molecular clone pLAI with and without 10 ng inhibitory RNA in human embryonic kidney (HEK) 293T cells. DNA of pRL expressing Renilla luciferase was included in the transfection mixtures to monitor cell viability and possible non-specific effects, for example, due to IFN induction by dsRNA. Virus production was measured by CA-p24 enzyme-linked immunosorbent assay (ELISA) in the culture supernatant 3 days after transfection. The amount of virus production without an inhibitory RNA, generally in the 50–250 ng/ml CA-p24 range, was set at 100%. nef2 dsRNA induced a significant decrease in CA-p24 production, but even more pronounced level of inhibition was obtained with diced si-nef2 (Figure 2b). This can be explained by the fact that si-nef2 bypasses the intracellular dicing step, which may be a limiting factor in the RNAi pathway.
Figure 2.
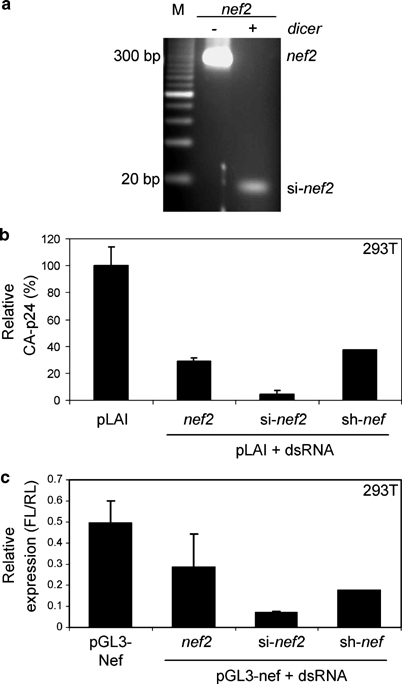
Inhibition of human immunodeficiency virus type 1 (HIV-1) by in vitro transcribed nef2 double-stranded RNA (dsRNA), in vitro diced si-nef2 and sh-nef RNA. (a) 2% MetaPhor agarose gel stained with ethidium bromide, showing the 300 basepairs (bp) nef2 dsRNA and the diced 19 bp si-nef2 products; M, 20 bp marker. (b) Cotransfection of human embryonic kidney (HEK) 293T cells with 10 ng of the indicated RNA, 500 ng pLAI and 2.5 ng pRL as an internal control. Transfections were performed with Lipofectamine 2000 and 1.5 × 105 cells. Virus production was measured in the culture supernatant 2 days after transfection. CA-p24 values are given as percentage of the pLAI production without inhibitory RNA. Standard error bars represent the means of four independent experiments. (c) Cotransfection of HEK 293T cells with 10 ng of the indicated dsRNA, 100 ng pGL3-Nef and 2.5 ng pRL as an internal control. Transfections were performed as described above. After 72 h cells were lysed and firefly and renilla luciferase expression was measured.
One of the hallmarks of the RNAi is its sequence specificity. Therefore, we tested if nef2 dsRNA and its in vitro diced product si-nef2 would inhibit pGL3-Nef reporter, in which 250 nt from the nef2 target sequence was placed downstream of the luciferase reporter gene.21 nef2 dsRNA induced a decrease in luciferase expression, but an even more pronounced level of inhibition was obtained with diced si-nef2 (Figure 2c). The pRL expression was not influenced (results not shown). In fact, in vitro diced si-nef2 is a much more effective inhibitor than the in vitro synthesized short-hairpin inhibitor sh-nef RNA, which was included as a positive control. We next tested the inhibitory potential of nef2 dsRNA in the 1–1000 ng range. At amounts above 10 ng, non-specific decrease of Renilla reniformis luciferase (RL) expression was observed, which is most likely due to IFN induction (results not shown, see also Figure 8). The high level of inhibition obtained with low amounts of dsRNA convinced us to design and test a series of DNA expression plasmids encoding long HIV-1-specific dsRNAs.
Figure 8.
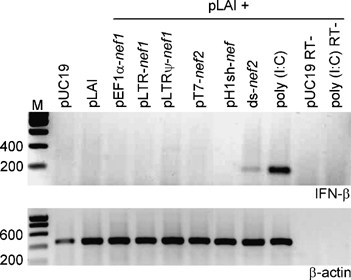
Long-hairpin RNA (lhRNA) constructs do not induce the interferon (IFN) response. Human embryonic kidney (HEK) 293T cells (1.5 × 105) were cotransfected with pLAI and the indicated lhRNA/double-stranded RNA (dsRNA)-expression constructs using Lipofectamine 2000. Construct pT7-nef2 was cotransfected with pT7-pol. Renilla luciferase (pRL) was used as an internal control. The pUC19 plasmid was used as a negative control. In vitro transcribed ds-nef2 RNA and poly (I:C) act as positive controls for IFN-β induction. Two separate transfections were performed, which were processed for either IFN-β mRNA expression or renilla measurement. No significant differences in Renilla expression were measured, except for the toxic treatment with ds-nef2 RNA and poly(I:C) (results not shown). In addition, we measured CA-p24 in the supernatant, which showed the inhibition characteristics described earlier. Total RNA was isolated from the cells 24 h after transfection. The IFN-β expression level was determined by reverse transcriptase-polymerase chain reaction (RT-PCR). β-Actin mRNA expression was analyzed as an internal control. PUC19 RT- and poly (I:C) RT- are control reactions without RT step.
Low-level inhibition of human immunodeficiency virus type 1 by long-hairpin RNA expression plasmids
To make lhRNA constructs, we cloned the HIV-1 tat, rev and nef gene sequences as inverted repeats under the transcriptional control of the constitutive EF1α promoter (Figure 3). These vectors should produce lhRNA, a long-hairpin structure consisting of an approximately 300 bp stem and a 46 nt loop. During transcription of the inverted sequences, an RNA molecule is made, which folds back on itself to form a hairpin structure with a stem of approximately 300 bp. In silico RNA analysis with the Mfold program47 confirms the folding of these extended hairpins (data not shown). We tested inhibition of HIV-1 in several mammalian cell lines (C33A, HEK 293T and Vero). Cotransfection of pLAI with the pEF1α-tat, pEF1α-rev or pEF1α-nef1 vectors resulted in marginal inhibition of HIV-1 production in C33A and HEK 293T cells, and only the pEF1α-tat vector was inhibitory in Vero cells (Figure 4a).
Figure 3.
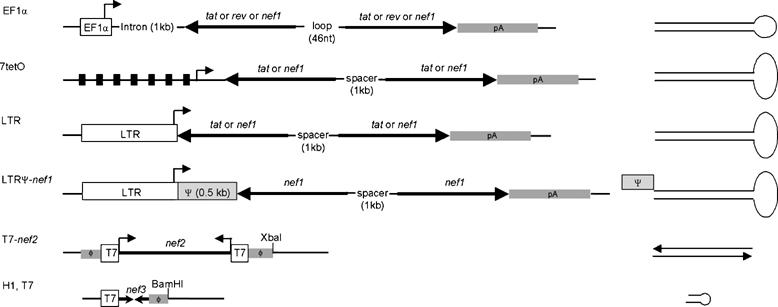
Expression vectors for long-hairpin RNA (lhRNA), double-stranded RNA (dsRNA) and short-hairpin RNA (shRNA). Long-hairpin RNAs were created by cloning the 300-nucleotide (nt) inverted repeats from tat, rev and nef1 (see Figure 1) downstream of the EF1α, 7tetO or long-terminal repeat (LTR) promoters. The 1 kb EF1α intron is positioned downstream of the EF1α promoter. A schematic representation of the final hairpin structures is shown on the right. In pEF1α constructs, the two complementary RNA strands are separated by a 46 nt loop. In the p7tetO and pLTR constructs, the complementary sequences are separated by a 1 kb spacer that contains splice donor and acceptor sites. Vector pLTRΨ-nef1 is a derivative of pLTR-nef1 in which the human immunodeficiency virus type 1 (HIV-1) leader sequence (Ψ, 76–630, marked as a gray box) was inserted. The predicted transcript will have the Ψ domain upstream of the RNA hairpin. All transcripts contain a polyadenylation signal (pA) downstream of the hairpin sequences. Vector pT7-nef2 has 300 basepairs (bp) long double-stranded nef sequences flanked by T7 promoters (T7) and terminators (φ) at both 5′ and 3′ ends. Two separate complementary RNA chains, potentially capable of forming dsRNA, are transcribed from the convergent promoters by T7 RNA polymerase (encoded by expression plasmid pT7-pol). Vectors pT7sh-nef and pH1sh-nef express sh-nef from the T7 and H1 promoters, respectively.
Figure 4.
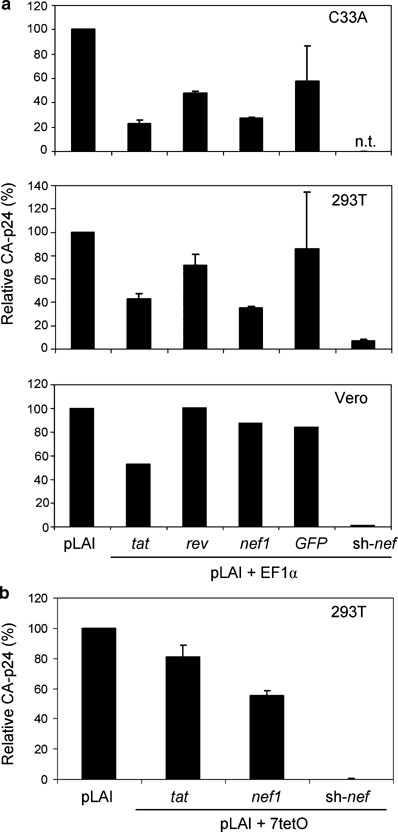
Marginal inhibition of human immunodeficiency virus type 1 (HIV-1) production by pEF1α- and p7tetO-driven long-hairpin RNA (lhRNA) constructs. (a) Cells (C33A, human embryonic kidney (HEK) 239T and Vero) were lipofectamine-transfected with 500 ng pLAI, 500 ng inhibitory construct, 3 ng pCMV-rtTA and 2.5 ng pRL. Vector pEF1α-green fluorescent protein (GFP) was used as a control expressing an irrelevant lhRNA against GFP. Vectors pH1sh-nef and the empty vector were used as negative and positive controls, respectively. Virus production was determined as described in the legend to Figure 2. Standard error bars represent the means of three independent experiments. The sh-nef control construct was not tested (n.t.) in C33A cells. (b) HEK 239T cells were cotransfected with 100 ng pLAI, 100 ng p7tetO-tat, p7tetO-nef1 or pH1sh-nef, 3 ng pCMV-rtTA and 2.5 ng pRL as an internal control. Culture medium was refreshed after 16 h and 1 μg/ml doxycycline was added. Virus production was determined as described in the legend to Figure 2. Standard error bars represent the means of four independent experiments.
We also designed a control pEF1α-green fluorescent protein (GFP) plasmid that expresses a similar extended lhRNA against GFP mRNA. No inhibition of virus production was observed with the control pEF1α-GFP vector, thus providing additional evidence for the specificity of lhRNA-mediated inhibition of HIV-1 production. We previously demonstrated potent inhibition of HIV-1 replication in T cells that stably express an shRNA targeted to viral nef gene sequences.19 Therefore, if lhRNA inhibits HIV-1 potently, it should be at least as active as the sh-nef control. Unlike the results with in vitro synthesized nef2 dsRNA, either diced or not, all lhRNA constructs (tat, rev and nef1) were much less potent inhibitors than the control sh-nef construct, which produces the short hairpin from a polymerase III promoter. Because pEF1α-tat and pEF1α-nef1 were slightly more effective than pEF1α-rev in C33A and 293T cells, we focused on these lhRNAs in the subsequent experiments.
Inducible long-hairpin RNA expression
In order to avoid innate viral responses or possible other side effects and to obtain controllable expression of lhRNA, we placed the expression of the lhRNAs tat and nef1 under control of inducible promoters: (i) the doxycycline (dox) inducible Tet system48 and (ii) the Tat-inducible long-terminal repeat (LTR) promoter/enhancer of HIV-149 (Figure 3). The latter system seems ideally suited to restrict lhRNA expression to cells that are infected by HIV-1, thus providing a unique safety feature. Furthermore, we replaced the 46 bp spacer between the repeats by the 1000 bp EF-1α intron, since it has been shown that this improves the inhibitory potential of lhRNA-encoding DNA constructs.50
The Tet-On system is based on the specific, high-affinity binding of the rtTA trans-activator to the tet operator (tetO) in the presence of dox, triggering transcription of downstream genes. This system has recently been used to express synthetic miRNA precursors in mouse or human genomes, following transduction with retroviral or lentiviral vectors.51 Vector pLAI was cotransfected with p7tetO-tat or p7tetO-nef1 (Figure 3). We also included pCMV-rtTA and the control pRL, and dox was added 4 h later. p7tetO-tat or p7tetO-nef1 conferred no or very poor inhibition of HIV-1 as compared to the sh-nef control (Figure 4b). The potency of the lhRNAs was not increased by varying the ratio of lhRNA-expression vector to pCMV-rtTA or the dox concentration. Moreover, a non-specific decrease of RL expression was observed at higher pCMV-rtTA concentrations, which is probably due to promoter squelching (results not shown).
In order to restrict lhRNA expression to cells that are infected with HIV-1, the inverted tat and nef1 repeats were cloned downstream of the Tat-inducible LTR promoter/enhancer of HIV-1, resulting in plasmids pLTR-tat and pLTR-nef1 (Figure 3). A Tat-inducible Pol II promoter expressing anti-HIV shRNA has been described for inhibition of HIV-1 gene expression in mammalian cells.52 In this setting, lhRNA or shRNA expression is activated in trans by the Tat protein encoded by HIV-1. To avoid self-targeting, we deleted a large part of the U3 region (up to position −179) in the LTR promoter of both LTR expression plasmids that overlaps with the nef coding domain (323 nt). Human embryonic kidney 293T cells were cotransfected with pLAI and pLTR-tat or pLTR-nef1. Both expression plasmids inhibit virus production by approximately 60% (Figure 5a).
Figure 5.
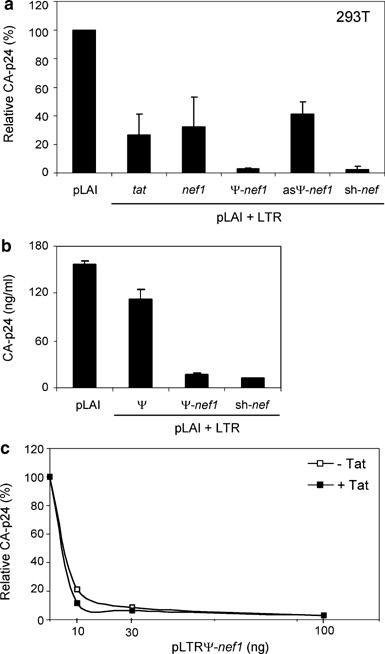
Antiviral long-hairpin RNA (lhRNA) production from the human immunodeficiency virus (HIV)-inducible long-terminal repeat (LTR) promoter. (a) Inhibition of HIV-1 production by lhRNA expressed from the Tat-inducible HIV-1 LTR. Human embryonic kidney (HEK) 239T cells were cotransfected with 100 ng of pLAI and 100 ng pLTR-tat, pLTR-nef1, pLTRΨ-nef1 or pLTRasΨ-nef1. Equal amounts of a pH1sh-nef expression vector and the empty vector were added as positive and negative controls, respectively. (b) Sequence-specific inhibition of HIV-1 production by pLTRΨ-nef1. HEK 239T cells were cotransfected with 100 ng of pLAI and 10 ng pLTRΨ, pLTRΨ-nef1 or pH1sh-nef. On the y-axis, the CA-p24 values (ng/ml) are presented. (c) Effect of the trans-activator protein Tat on the inhibitory effect of pLTRΨ-nef1. pLAI (100 ng) was cotransfected with 0–10–30–100 ng pLTRΨ-nef1 with or without 20 ng pcDNA3-Tat. Virus production was determined as described in the legend to Figure 2. Standard error bars represent the means of five independent experiments.
Directing long-hairpin RNA along viral pathways
The poor inhibitory potency of the different lhRNA constructs could be due to expression problems, but the lhRNA may also encounter difficulties in entering or proceeding along the RNAi pathway. In order to increase the efficacy of the lhRNA molecules, we cloned the HIV-1 leader sequence (Ψ) between the LTR and nef1 in pLTRΨ-nef1 (Figure 3). Control plasmid pLTRasΨ-nef1 was made with the Ψ element inserted in antisense orientation. Previously, we reported that sequences from the 5′ untranslated leader of the HIV-1 genome, such as the RNA dimerization signal, can be used to inhibit HIV expression in trans.53 We presumed that Ψ should bring the antiviral RNA along viral pathways, conferring a stronger inhibitory effect. Indeed, in HEK 293T cells transfected with pLAI and pLTRΨ-nef1, HIV-1 production was almost completely inhibited (Figure 5a). The level of inhibition conferred by pLTRΨ-nef1 is comparable to that of the positive control sh-nef. Inhibition is specific and not due to a more general cell toxicity problem because no significant decrease in RL expression was observed (results not shown). Control plasmid pLTRasΨ-nef1 was much less effective in inhibiting HIV-1 production, with a potency comparable to that of the original pLTR-nef1 construct. Plasmid pLTRΨ, in which the nef1 sequence has been deleted, failed to inhibit HIV-1 production (Figure 5b). This result indicates that the presence of the Ψ element enhances lh-nef1-mediated inhibition of HIV-1, but the presence of the nef1 sequence is essential for the potent effect of pLTRΨ-nef1.
Transcript Ψ-nef1 expression from the LTR promoter is induced by pLAI-encoded Tat protein. Because pLAI gene expression is strongly inhibited by pLTRΨ-nef1, a negative feedback loop may have been established, which leads to an underestimation of the inhibitory potential of pLTRΨ-nef1. We therefore added a Tat-expression plasmid (pcDNA3-Tat) in trans to secure pLTRΨ-nef1 expression. Human embryonic kidney 293T cells were transfected with 100 ng pLAI and 10–100 ng pLTRΨ-nef1 with or without pcDNA3-Tat. As shown in Figure 5c, pronounced HIV-1 inhibition was obtained with as little as 10 ng pLTRΨ-nef1. The presence of additional Tat did not significantly improve the inhibition conferred by pLTRΨ-nef1, indicating that the background LTR promoter activity produces sufficient amounts of the inhibitory transcript.
We next wanted to test if the potent pLTRΨ-nef1 construct was able to inhibit HIV-1 variants that escaped from the sh-nef inhibitor. We described previously a series of viral escape variants with mutations or deletions in the targeted nef sequence.19 We selected mutant R1 with a 106 nt deletion that includes the complete sh-nef target sequence and mutant R3′ with two point mutations in the target (Table 1). Human embryonic kidney 293T cells were cotransfected with 200 ng HIV-1 molecular clone pLAI (wild-type, R1 or R3′) and 40 ng pLTRΨ-nef1 or pH1sh-nef plasmid. Only the wild-type pLAI construct was equally inhibited by the pLTRΨ-nef1 and sh-nef constructs. The deletion mutant R1 is completely resistant to sh-nef, but was potently inhibited by pLTRΨ-nef1. Mutant R3′ partially escapes from sh-nef, but is potently inhibited by pLTRΨ-nef1.
Table 1.
lhRNA inhibits shRNA-escape viruses
| Virus | nef3 target | sh-nef inhibitiona (%) | lh-nef1 inhibitionb (%) |
|---|---|---|---|
| pLAI (WT) | GTGCCTGGCTAGAAGCACA | 94 | 92 |
| R1c | – – – – – – – – – – – – – – – – – –– | 0 | 83 |
| R3′d | GTACCTGGCGAGAAGCACA | 28 | 88 |
Abbreviations: HEK, human embryonic kidney; HIV-1, human immunodeficiency virus type 1.
lhRNA, long-hairpin RNA; shRNA, short-hairpin RNA; WT, wild type.
aHEK 293T cells were cotransfected with 200 ng HIV-1 molecular clone pLAI (WT, R1 or R3′) and 40 ng pH1sh-nef plasmid.
bHEK 293T cells were cotransfected with 200 ng HIV-1 molecular clone pLAI (WT, R1 or R3′) and 40 ng pLTRΨ-nef1 plasmid.
cDeletion of complete target sequence.
dBold and underlined residues cause viral escape.
Inhibition of human immunodeficiency virus type 1 by cytoplasmically expressed ds-nef2
RNA interference is mainly a cytoplasmic process,54 and putative problems in nuclear export of lhRNAs may thus hamper their inhibitory activity. In order to test whether there is a difference between the RNAi-inducing capacity of dsRNAs produced in the nucleus or cytoplasm of cells, we expressed dsRNA targeted to the nef-coding domain (nef2 in Figure 1) using the cytoplasmic T7 RNA polymerase system. To generate pT7-nef2, we cloned a 300 bp nef-specific DNA fragment between convergent T7 promoters and terminators (Figure 3). In addition, the control expression plasmid pT7sh-nef was constructed. Cytoplasmic transcription of pT7-nef2 and pT7sh-nef is performed by T7-polymerase encoded by pT7-pol. Expression of the firefly luciferase (FL) reporter from the T7 promoter (pT7-luc) was measured as a positive control in all transfection experiments with pT7-pol (Figure 6a). Human embryonic kidney 293T cells were transfected with pLAI and pT7-nef2 or pT7sh-nef, with and without pT7-pol. Virus production was measured in the culture supernatant 3 days after transfection by CA-p24 ELISA. In the absence of T7-polymerase, pT7-nef2 and pT7sh-nef confer a low level of inhibition on virus production probably due to leaky RNA expression from the plasmids (Figure 6b). With T7-polymerase, pT7-nef2 and pT7sh-nef confer up to 90% inhibition of HIV-1 production (Figure 6b). This level of inhibition is comparable to that obtained with the sh-nef control expression plasmid. Renilla luciferase expression was not affected (Figure 6b, gray bars). To demonstrate that the effect of pT7-nef2 is sequence-specific, we cotransfected it with the pGL3-Nef reporter (Figure 6c). In the presence of T7-polymerase, pT7-nef2 potently inhibited luciferase expression.
Figure 6.
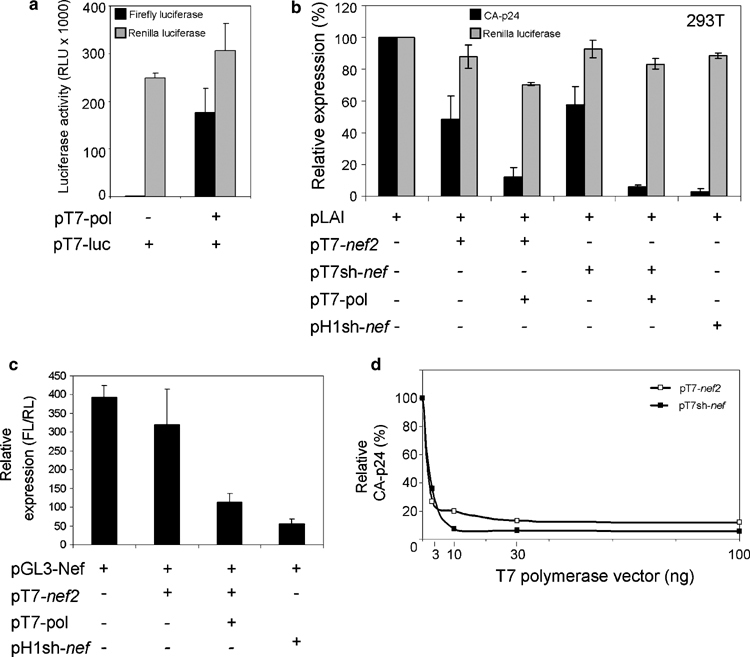
Inhibition of human immunodeficiency virus type 1 (HIV-1) by cytoplasmically expressed long double-stranded RNA (dsRNA). (a) pT7-luc (100 ng) construct was cotransfected in human embryonic kidney (HEK) 293T cells with or without 30 ng pT7-pol. At 2 days after transfection, cells were lysed and the expression of firefly luciferase was measured. (b) pT7-nef2 (100 ng) and pT7sh-nef vectors were linearized 3′ of the T7 termination signal with XbaI and BamHI, respectively, and cotransfected with 100 ng pLAI, 30 ng pT7-pol and 2.5 ng pRL in HEK 293T cells. Two separate complementary RNA chains, potentially forming dsRNA, are transcribed in the cell from convergent T7 promoters. Equal amounts of a pH1sh-nef expression vector and the empty vector were added as positive and negative controls, respectively. Cotransfections were also performed without pT7-pol to check for non-specific effects of the T7 plasmids. Virus production was determined as described in the legend to Figure 2. Standard error bars represent the means of four independent experiments. Cells were lysed 2 days after transfection to measure Renilla luciferase. (c) Sequence-specific inhibition of the pGL3-Nef reporter, containing the 250-nucleotide (nt) nef2 target sequence downstream of the luciferase coding domain, by pT7-nef2. HEK 239T cells were cotransfected with 100 ng of pGL3-Nef, 100 ng pT7-nef2, 30 ng pT7-pol and 2.5 ng pRL. pH1sh-nef (10 ng) expression vector and the empty vector were added as positive and negative controls, respectively. (d) Titration of T7 polymerase. pLAI (100 ng) was cotransfected with increasing amounts (0–3–10–30–100 ng) of pT7-pol and 100 ng pT7-nef2 or pT7sh-nef.
We next tested the inhibitory potential of both pT7-nef2 and pT7sh-nef plasmids in the presence of an increasing amount (5–100 ng) of pT7-polymerase. The inhibition was consistently strong even at low amounts (5 ng) of the pT7-pol plasmid (Figure 6d). When more than 100 ng pT7-pol was used in the transfection experiments, a non-specific decrease in RL reporter expression was observed. A high amount of T7-polymerase accumulating in the cytoplasm of cells may be toxic or high amounts of dsRNA accumulating in the cytoplasm of cells may induce the innate antiviral IFN response.
To measure the RNA expression levels of selected lhRNA-expression constructs, we performed a reverse transcriptase-polymerase chain reaction (RT-PCR) assay with nef-specific primers. All constructs expressed nef RNA (Figure 7). Notably, pT7-nef2 produced low amounts of RNA even in the absence of the pT7pol inducer. This can explain the slight inhibition of gene expression obtained with pT7-nef2 in the absence of pT7pol (Figure 6b and c). No nef-specific fragment was detected in mock-transfected cells or in cells transfected with poly(I:C).
Figure 7.
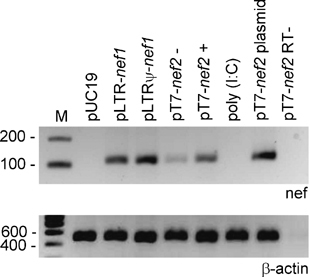
In vivo RNA expression from long-hairpin RNA (lhRNA) constructs. HEK 293T cells were transfected with 500 ng of the indicated lhRNA/double-stranded RNA (dsRNA)-expression constructs using Lipofectamine 2000. Constructs pLTR-nef1 and pLTRΨ-nef1 were cotransfected with 30 ng pTat. Construct pT7-nef2 was cotransfected with 100 ng pT7-pol. The pUC19 plasmid and poly (I:C) were used as negative controls and pT7-nef2 plasmid DNA was used as a positive control. Total RNA was isolated from the cells 48 h after transfection. The nef expression level was determined by reverse transcriptase-polymerase chain reaction (RT-PCR) with primers Nef-B/X and antiU3-att, which create a 110 basepairs (bp) amplification product. β-Actin mRNA expression was analyzed as an internal control. pT7-nef2 RT− is a control reaction without RT step.
Endogenously expressed long-hairpin RNA and long double-stranded RNA do not induce the interferon response
It has been reported that introduction of dsRNAs longer than 30 bp in mammalian cells induces the innate antiviral IFN response. We determined IFN-β induction by RT-PCR in HEK 293T cells upon transfection with in vitro made nef2 dsRNA and DNA constructs encoding pEF1α-nef1, pLTR-nef1, pLTRΨ-nef1, pT7-nef2 or pH1sh-nef. Transfection of 1 μg poly(I:C), a known inducer of IFN and other cytokines, was used as a positive control. The lhRNA- and sh-nef-expression plasmids did not induce detectable levels of IFN-β mRNA (Figure 8). Both nef2 dsRNA and poly(I:C) controls induced high amounts of the IFN-β mRNA.
Discussion
The use of synthetic siRNAs or shRNA-expression plasmids as inducers of RNAi-based therapy against HIV-1 faces the major obstacle of the emergence of virus escape variants. Similar to the combined use of antivirals in highly active antiretroviral therapy (HAART), one could design an RNAi therapy with extended lhRNA/dsRNA. Endogenous expression of two long complementary RNAs, with the potential to form an extended dsRNA duplex, leads to specific suppression of gene expression in mammalian cells.32, 34, 35, 37 Interestingly, full-length dsRNAs could not be detected, suggesting that processing by Dicer precludes their accumulation in the cytoplasm. Long dsRNAs are indeed processed in vitro by the RNAi machinery into multiple active siRNAs.55, 56 Several lines of evidence show that lhRNAs induce gene silencing by the RNAi mechanism. Inhibition of Dicer abrogated the gene silencing induced by lhRNA against GFP, indicating that silencing was mediated by RNAi.32 These results suggest that long dsRNA is well tolerated in mammalian cells, most likely because it is processed rapidly by the RNAi machinery. It is also relevant to mention that mammalian cells may naturally produce dsRNA derived from repetitive and transposable elements.57, 58 A recently performed bioinformatics study revealed the presence of at least 4520 full-length transcripts, which form sense–antisense gene pairs in the human genome.59
We designed a set of anti-HIV-1 lhRNA/dsRNA-expression constructs and compared their ability to inhibit virus production with a very potent shRNA-based inhibitor that we described previously.19 Ideally, a single lhRNA should provide more potent inhibition of HIV than an shRNA. An additional advantage of lhRNA is that it does not require predetermination of optimal shRNAs and HIV-1 target sequences because multiple effective siRNAs will be produced. A potential disadvantage of the use of lhRNA as therapeutic is that the multiple siRNAs are more likely to cause off-target effects.
Long-hairpin RNAs expressed from constitutive (EF1α) and inducible (7tetO, HIV-1 LTR) promoters inhibited HIV-1 production marginally, but we demonstrated potent and specific HIV-1 silencing with modified DNA constructs. The most active constructs either link the lhRNA to viral RNA sequences (Ψ) or express the long dsRNA directly in the cytoplasm, suggesting that translocation of dsRNA from the nucleus to the cytoplasm is a crucial step for these molecules to enter the cytoplasmic RNAi pathway. It is widely accepted that RNAi is a cytoplasmic process, as most protein components of the RNAi pathway, including Argonaute 2, TRBP and Dicer, assemble and function in the cytoplasm.54, 60, 61, 62, 63, 64 Putative problems in nuclear export of lhRNA may thus hamper its inhibitory activity. Modification of the lhRNA by addition of the HIV-1 leader sequence (Ψ) or direct cytoplasmic expression of long dsRNA by the T7 polymerase created potent inhibitors. The viral Ψ sequence may provide the transcript with a non-self signature and thereby boost RNAi. It has previously been suggested that the inhibitory effect of Ψ-containing transcripts is sequence-specific, possibly due to premature formation of RNA dimers,53 but the exact mechanism of inhibition remains to be elucidated.
A major advantage of an lhRNA inhibitor over shRNA is the reduced chance of viral escape because a larger segment of the viral genome is targeted, as has been shown recently.65 As an initial test of this idea, we used two HIV-1 mutants that escaped from sh-nef by two point mutations or complete deletion of the target sequence.19 These escape mutants could be inhibited by the lhRNA construct pLTRΨ-nef1, confirming the hypothesis that lhRNA may delay or prevent the evolution of HIV-1 escape variants.
We showed that endogenously expressed lhRNA and dsRNA do not activate the innate antiviral response. A similar result has recently been described in literature.33, 66 Exposure of cells to a 50 bp in vitro synthesized dsRNA induces the production of class I IFNs, but not when such molecules are expressed in the cell from a DNA construct with the U6 promoter. Modification of lhRNAs expressed from a Pol III promoter by inclusion of multiple G:U wobbles induced RNAi without any non-specific effects.66 In fact, most reports on IFN induction by long dsRNAs in mammalian cells are based on transfection of cells with in vitro synthesized dsRNAs.40, 67 Apparently, endogenously produced dsRNA is less active than exogenous dsRNA in inducing the IFN response. This finding may have a major impact on the further development of RNAi-based antiviral strategies.
RNA interference-based gene therapy against HIV-1 seems to be a viable option, either as mono-therapy or combined with traditional drug therapy.22, 68 The outgrowth of RNAi-resistant virus mutants presents a major obstacle for all sequence-specific inhibitory strategies. The simultaneous targeting of multiple conserved targets by lhRNA may confer increased robustness to future RNAi therapies. We will focus on optimization of the DNA constructs encoding HIV-1-specific lhRNAs, for example, the type of promoter used and the structure of the lhRNA. We will also test stable expression of these constructs in HIV-susceptible cells. These cell lines will be extensively tested for the emergence of RNAi-resistant virus variants.
Materials and methods
DNA constructs and RNA transcripts
The full-length HIV-1 molecular clone pLAI69 was used to produce wild-type virus and to study inhibition by lhRNAs directed against the tat, rev or nef sequences. A detailed description of the construction of all lhRNA-expression constructs is available as Supplementary information. Plasmid pcDNA3-T7pol, expressing bacteriophage T7 polymerase (pT7-pol, a kind gift of Dr Jean-Marc Jacque, University of Massachusetts Medical School, Worcester, MA, USA), pGL3-Nef and pH1sh-nef have been described previously.14, 19
The Mfold program47 (http://www.bioinfo.rpi.edu/applications/mfold) was used to check the correct folding of extended hairpins.
In vitro transcription and dicing of ds-nef2 RNA
The ds-nef2 RNA was in vitro transcribed with the Megashortscript T7 transcription kit (Ambion, Austin, TX, USA) from the nef2 PCR template that contains convergent T7 promoters and terminators. The sh-nef RNAi inducer19 was transcribed in vitro from the BamHI-linearized pT7sh-nef vector. ds-nef2 RNA (1 μg) was diced in vitro at 37°C for 18–20 h using recombinant Dicer enzyme (Stratagene, Cedar Creek, TX, USA). The full-length ds-nef2 RNA, the cleaved si-nef2 (a mix of siRNAs derived from ds-nef2) and sh-nef were purified through MicroSpin G-25 column (Amersham Biosciences, Piscataway, NJ, USA). To remove undigested dsRNA from the dicing reaction, the si-nef2 was purified further on a Microcon YM-100 column (Millipore, Billerica, MA, USA). si-nef2 RNA was analyzed on 2% MetaPhor (BMA, Sanver Tech, The Netherlands) alongside a 20 bp DNA marker (Gensura, San Diego, CA, USA).
Cell culture and transfections
Human embryonic kidney 293T, C33A cervix carcinoma and African green monkey kidney Vero cells were grown as a monolayer in Dulbecco's modified Eagle's medium (DMEM) (Gibco BRL, Carlsbad, CA, USA) supplemented with 10% fetal calf serum (FCS) (Hybond, Escondido, CA, USA), minimal essential medium with non-essential amino acids, penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37°C and 5% CO2. At 1 day before transfection, cells were trypsinized, resuspended in DMEM and seeded in 24-well plates at a density of 1.5 × 105 cells per well. Cells were cotransfected with 100–500 ng pLAI and 1–1000 ng in vitro transcribed ds-nef2 RNA and in vitro diced si-nef2 or 10–500 ng lhRNA (tat, rev, nef1) expression constructs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Virus production was determined by measuring the CA-p24 levels in the culture supernatant by ELISA (Abbott, Abbott Park, IL, USA) as described previously.70 For firefly luciferase measurements, cells were cotransfected with 100 ng pGL3-Nef and 10 ng in vitro transcribed ds-nef2 RNA and in vitro diced si-nef2 or 100 ng pT7-nef2 and 100 ng pT7-pol. In all experiments, vector pRL (2.5 ng) (Promega, Madison, WI, USA), expressing RL under the control of the CMV promoter, was added to the transfection mix as a control for variation in transfection efficiency and cell viability. Equal amounts of the pH1sh-nef expression vector or the empty vector were added as positive and negative controls, respectively. Vectors pT7-luc and pT7-pol were cotransfected in equimolar amounts (100 ng) and FL reporter expression was measured. Briefly, cells were lysed 48–72 h after transfection in 150 μl 1 × passive lysis buffer (Promega) by shaking for 30 min at room temperature. The cell lysate was centrifuged for 5 min at 4000 r.p.m. and firefly and renilla luciferase expression was measured in 10 μl supernatant with the dual-luciferase reporter assay system or Renilla luciferase assay system (Promega).
Interferon assay
Induction of the IFN system was measured by a sensitive RT-PCR on the IFN-β mRNA.71 RNA was isolated from HEK 293T cells with the mirVana miRNA isolation kit (Ambion) 24 h after transfection with the long dsRNA constructs. First-strand cDNA was reverse transcribed from approximately 1 μg RNA with random hexamer primers (Invitrogen) using the MMLV-RT enzyme (Invitrogen) according to the manufacturer's instructions. Approximately 200 ng cDNA was PCR amplified with primers IFN-βF and IFN-βR using standard conditions. Amplification of the β-actin gene was used as an internal control. Transfection of 1 μg poly(I:C) (Amersham Pharmacia Biotech, Piscataway, NJ, USA), a synthetic inosine/cytosine polymer that mimics viral dsRNA, was used as a positive control for IFN-β induction.
RNA of transfected cells was subjected to PCR with nef-specific primers Nef-B/X and AntiU3-att, yielding a 110 bp fragment (Supplementary Table 1).
Supplementary information
Acknowledgements
RNAi research in the Berkhout lab is sponsored by NWO-CW (TOP grant) and Senter-NOVEM (TS grant with Viruvation). We thank Stephan Heynen for performing CA-p24 ELISA; Dr P Midoux (Centre de Biophysique Moleculaire, Orleans, France) for providing us with the pT7-luc plasmid; Dr Jean-Marc Jacque (University of Massachusetts Medical School, USA) for providing the pcDNA3-T7pol expressing vector and Dr E de Jong (University of Amsterdam, The Netherlands) for the kind gift of IFN-β primers.
Footnotes
Supplementary Information accompanies the paper on Gene Therapy website (http://www.nature.com/gt)
References
- 1.Cullen BR. Induction of stable RNA interference in mammalian cells. Gene Therapy. 2006;13:503–508. doi: 10.1038/sj.gt.3302656. [DOI] [PubMed] [Google Scholar]
- 2.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 4.Plasterk RHA. RNA silencing: the genome's immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- 5.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soifer HS, Zaragoza A, Peyvan M, Behlke MA, Rossi JJ. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res. 2005;33:846–856. doi: 10.1093/nar/gki223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 9.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 10.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/S0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 11.Haasnoot PCJ, Cupac D, Berkhout B. Inhibition of virus replication by RNA interference. J Biomed Sc. 2003;10:607–616. doi: 10.1007/BF02256311. [DOI] [PubMed] [Google Scholar]
- 12.Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Capodici J, Kariko K, Weissman D. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J Immunol. 2002;169:5196–5201. doi: 10.4049/jimmunol.169.9.5196. [DOI] [PubMed] [Google Scholar]
- 14.Jacque JM, Triques K, Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 16.Park WS, Miyano-Kurosaki N, Hayafune M, Nakajima E, Matsuzaki T, Shimada F. Prevention of HIV-1 infection in human peripheral blood mononuclear cells by specific RNA interference. Nucleic Acids Res. 2002;30:4830–4835. doi: 10.1093/nar/gkf627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson J, Banerjea A, Akkina R. Bispecific short hairpin siRNA constructs targeted to CD4, CXCR4, and CCR5 confer HIV-1 resistance. Oligonucleotides. 2003;13:303–312. doi: 10.1089/154545703322616989. [DOI] [PubMed] [Google Scholar]
- 18.Boden D, Pusch O, Silbermann R, Lee F, Tucker L, Ramratnam B. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res. 2004;32:1154–1158. doi: 10.1093/nar/gkh278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berkhout B. RNA interference as an antiviral approach: targeting HIV-1. Curr Opin Mol Ther. 2004;6:141–145. [PubMed] [Google Scholar]
- 23.Brake O, Berkhout B. A novel approach for inhibition of HIV-1 by RNA interference: counteracting viral escape with a second generation of siRNAs. J RNAi Gene Silencing. 2005;1:56–65. [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto T, Omoto S, Mizuguchi M, Mizukami H, Okuyama H, Okada N. Double-stranded nef RNA interferes with human immunodeficiency virus type 1 replication. Microbiol Immunol. 2002;46:809–817. doi: 10.1111/j.1348-0421.2002.tb02768.x. [DOI] [PubMed] [Google Scholar]
- 25.Yeung ML, Bennasser Y, Myers TG, Jiang G, Benkirane M, Jeang KT. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology. 2005;2:81. doi: 10.1186/1742-4690-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337:1214–1218. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- 27.Wianny F, Zernicka-Goetz M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- 28.Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMillan NA, Chun RF, Siderovski DP, Galabru J, Toone WM, Samuel CE. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virol. 1995;213:413–424. doi: 10.1006/viro.1995.0014. [DOI] [PubMed] [Google Scholar]
- 31.Cai R, Carpick B, Chun RF, Jeang KT, Williams BR. HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch Biochem Biophys. 2000;373:361–367. doi: 10.1006/abbi.1999.1583. [DOI] [PubMed] [Google Scholar]
- 32.Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci USA. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto S, Miyagishi M, Akashi H, Nagai R, Taira K. Analysis of double-stranded RNA-induced apoptosis pathways using interferon-response noninducible small interfering RNA expression vector library. J Biol Chem. 2005;280:25687–25696. doi: 10.1074/jbc.M412784200. [DOI] [PubMed] [Google Scholar]
- 34.Ui-Tei K, Zenno S, Miyata Y, Saigo K. Sensitive assay of RNA interference in Drosophila and Chinese hamster cultured cells using firefly luciferase gene as target. FEBS Lett. 2000;479:79–82. doi: 10.1016/S0014-5793(00)01883-4. [DOI] [PubMed] [Google Scholar]
- 35.Diallo M, Arenz C, Schmitz K, Sandhoff K, Schepers U. Long endogenous dsRNAs can induce complete gene silencing in mammalian cells and primary cultures. Oligonucleotides. 2003;13:381–392. doi: 10.1089/154545703322617069. [DOI] [PubMed] [Google Scholar]
- 36.Yi CE, Bekker JM, Miller G, Hill KL, Crosbie RH. Specific and potent RNA interference in terminally differentiated myotubes. J Biol Chem. 2003;278:934–939. doi: 10.1074/jbc.M205946200. [DOI] [PubMed] [Google Scholar]
- 37.Tran N, Raponi M, Dawes IW, Arndt GM. Control of specific gene expression in mammalian cells by co-expression of long complementary RNAs. FEBS Lett. 2004;573:127–134. doi: 10.1016/j.febslet.2004.07.075. [DOI] [PubMed] [Google Scholar]
- 38.Shinagawa T, Ishii S. Generation of Ski-knockdown mice by expressing a long double-strand RNA from an RNA polymerase II promoter. Genes Dev. 2003;17:1340–1345. doi: 10.1101/gad.1073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 40.Caplen NJ, Zheng Z, Falgout B, Morgan RA. Inhibition of viral gene expression and replication in mosquito cells by dsRNA-triggered RNA interference. Mol Ther. 2002;6:243–251. doi: 10.1006/mthe.2002.0652. [DOI] [PubMed] [Google Scholar]
- 41.Qi Y, Zhong X, Itaya A, Ding B. Dissecting RNA silencing in protoplasts uncovers novel effects of viral suppressors on the silencing pathway at the cellular level. Nucleic Acids Res. 2004;32:e179. doi: 10.1093/nar/gnh180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waterhouse PM, Graham MW, Wang MB. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adelman ZN, Sanchez-Vargas I, Travanty EA, Carlson JO, Beaty BJ, Blair CD. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J Virol. 2002;76:12925–12933. doi: 10.1128/JVI.76.24.12925-12933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez MA, Clotet B, Este JA. RNA interference of HIV replication. Trends Immunol. 2002;23:559–561. doi: 10.1016/S1471-4906(02)02328-1. [DOI] [PubMed] [Google Scholar]
- 45.Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 46.Arteaga HJ, Hinkula J, Dijk-Hard I, Dilber MS, Wahren B, Christensson B. Choosing CCR5 or Rev siRNA in HIV-1. Nat Biotechnol. 2003;21:230–231. doi: 10.1038/nbt0303-230. [DOI] [PubMed] [Google Scholar]
- 47.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berkhout B, Silverman RH, Jeang KT. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 50.Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM. Total silencing by intron-spliced hairpin RNAs. Nature. 2000;407:319–320. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- 51.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci USA. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Unwalla HJ, Li MJ, Kim JD, Li HT, Ehsani A, Alluin J. Negative feedback inhibition of HIV-1 by TAT-inducible expression of siRNA. Nat Biotechnol. 2004;22:1573–1578. doi: 10.1038/nbt1040. [DOI] [PubMed] [Google Scholar]
- 53.Berkhout B, van Wamel JL. Inhibition of human immunodeficiency virus expression by sense transcripts encoding the retroviral leader RNA. Antiviral Res. 1995;26:101–115. doi: 10.1016/0166-3542(94)00068-J. [DOI] [PubMed] [Google Scholar]
- 54.Zeng Y, Cullen BR. RNA interference in human cells is restricted to the cytoplasm. RNA. 2002;8:855–860. doi: 10.1017/S1355838202020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 56.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 57.Lehner B, Williams G, Campbell RD, Sanderson CM. Antisense transcripts in the human genome. Trends Genet. 2002;18:63–65. doi: 10.1016/S0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- 58.Wang Q, Carmichael GG. Effects of length and location on the cellular response to double-stranded RNA. Microbiol Mol Biol Rev. 2004;68:432–452. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 60.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 62.Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 63.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe T, Sudoh M, Miyagishi M, Akashi H, Arai M, Inoue K et al. Intracellular-diced dsRNA has enhanced efficacy for silencing HCV RNA and overcomes variation in the viral genotype. Gene Therapy 2006, (Epub ahead of print). [DOI] [PubMed]
- 66.Akashi H, Miyagishi M, Yokota T, Watanabe H, Hino T, Nishina K. Escape from the interferon response associated with RNA interference using vectors that encode long modified hairpin-RNA. Mol BioSys. 2005;1:382–390. doi: 10.1039/b510159j. [DOI] [PubMed] [Google Scholar]
- 67.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolkowicz R, Nolan GP. Gene therapy progress and prospects: novel gene therapy approaches for AIDS. Gene Therapy. 2005;12:467–476. doi: 10.1038/sj.gt.3302488. [DOI] [PubMed] [Google Scholar]
- 69.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-L. [DOI] [PubMed] [Google Scholar]
- 70.Jeeninga RE, Hoogenkamp M, Armand-Ugon M, de Baar M, Verhoef K, Berkhout B. Functional differences between the long terminal repeat transcriptional promoters of HIV-1 subtypes A through G. J Virol. 2000;74:3740–3751. doi: 10.1128/JVI.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lebre MC, Antons JC, Kalinski P, Schuitemaker JH, van Capel TM, Kapsenberg ML. Double-stranded RNA-exposed human keratinocytes promote Th1 responses by inducing a Type-1 polarized phenotype in dendritic cells: role of keratinocyte-derived tumor necrosis factor alpha, type I interferons, and interleukin-18. J Invest Dermatol. 2003;120:990–997. doi: 10.1046/j.1523-1747.2003.12245.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


