Abstract
The nucleocapsid (N) protein of severe acute respiratory syndrome-coronavirus (SARS-CoV) is a major virion structural protein. In this study, two epitopes (N1 and N2) of the N protein of SARS-CoV were predicted by bioinformatics analysis. After immunization with two peptides, the peptides-specific antibodies were isolated from the immunized rabbits. The further experiments demonstrated that N1 peptide-induced polyclonal antibodies had a high affinity to bind to E. coli expressed N protein of SARS-CoV. Furthermore, it was confirmed that N1 peptide-specific IgG antibodies were detectable in the sera of severe acute respiratory syndrome (SARS) patients. The results indicated that an epitope of the N protein has been identified and N protein specific Abs were produced by peptide immunization, which will be useful for the study of SARS-CoV.
Keywords: severe acute respiratory syndrome-coronavirus, necleocapsid protein, epitope, polyclonal antibody, antiserum
INTRODUCTION
An outbreak of atypical pneumonia, referred to as SARS and first identified in Guangdong Province, China, has spread to several countries1. The mortality of this disease is higher and therefore few hundreds of people have died from the disease. In the absence of effective vaccines and drugs, infectious diseases such as SARS that is spread by the respiratory route must be taken very seriously. It is very important that public health officials are able to identify people infected with SARS at early stages of the disease and isolate them for an appropriate time, thus the cycle of transmission can be broken. Voluntary isolation and quarantine are of great inconvenience for a lot of people, but they are currently our best tools to save lives2. An accurate diagnosis method for SARS is urgent needed and it will be great helpful for people against SARS3, 4, 5.
A novel coronavirus has been identified in the patients of SARS. SARS-CoV is enveloped, positive-sense, ssRNA virus. The genome of SARS-CoV is 29,727 nucleotides in length, has 11 open reading frames, and the genome organization is similar to that of other coronaviruses. The structural proteins of SARS-CoV contain four proteins including the surface spike (S) glycoprotein, the small membrane (M) protein, the envelope (E) glycoprotein and the nucleocapsid (N) protein6, 7.
Based on available information of other coronaviruses, nucleocapsid (N) protein has multiple functions that are involved in providing nuclear-import signal, interfering cell process, virus replication and RNA package. In addition, N protein in many coronaviruses is highly conserved, immunogenic, and abundantly expressed during infection8, 9, 10, 11, 12. These features make it to be a suitable candidate for raising neutralizing antibodies and diagnostic applications.
In order to identify the epitopes of the N protein and to be able to quickly make specific antibodies against the N protein of SARS, in this study, two peptides were synthesized according to the sequences of the N protein, and the polyclonal antibodies were raised in the peptide-immunized rabbits. Our data showed that the antibodies were capable of binding to the N protein of SARS-CoV. It was interesting to observe that N1 peptide was also able to bind to IgG antibody from the sera of SARS patients. Our work was the first time to identify an epitope of the N protein of SARS-CoV, the antibodies induced by the peptide will be useful for the SARS diagnosis or analysis of N protein functions.
MATERIALS AND METHODS
Animals
New Zealand rabbits were purchased from Shanghai Laboratory Animal Center, Chinese Academy of Sciences. Animals were kept in conventional conditions and were handled in compliance with Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences Chinese Academy of Sciences (Shanghai, China) guidelines for animal care and use.
Peptide design and synthesis
Peptides (N1 and N2) derived from the sequences of SARS putative nucleocapsid protein (NC-004718, Genbank) were synthesized using conventional solid-phase chemistry as described previously13 and purified by GL Biochem (Shanghai) Ltd. These sequences (amino acid residues 21-44, PTDSTDNNQNGGRNGARPKQRRPQ, N1 peptide, and amino acid residues 138-160, GALNTPKDHIGTRNPNNNAATVL, N2 peptide) were chosen as highly antigenic sites by computer prediction with DNAstar (http://www.dnastar.com/). The synthetic peptides were covalently conjugated to bovine serum albumin (BSA, Sigma) by N-(3-Dimethylaminopropyl)- N′-ethylcarbodiimide hydrochloride (EDAC) (Sigma).
Immunization and raising polyclonal antibodies
Rabbits were immunized s.c. with 200 μg of peptide-BSA in 0.4 ml emulsion 1/1 v/v with complete Freund's adjuvant (CFA) that had been supplemented with Mycobacterium tuberculosis to the final concentration of 1 mg/ml and were injected subcutaneously at multiple sites on the back of rabbits. Three weeks later, booster injection was done with freshly prepared emulsion of the conjugate and Freund's incomplete adjuvant. Blood was drawn from the rabbits at five weeks following immunization, the blood was allowed to clot at 4°C, and the antiserum was recovered by centrifugation14.
Prokaryotic expression and purification of SARS-CoV nucleocapsid protein
The full length of SARS nucleocapsid gene (1266bp) was subcloned into pQE30 His-tag expression vector (the detailed description will be published elsewhere). The 6XHis residues at the N terminus of the vector facilitated the purification of the expressed SARS N protein using Immobilized Metal Affinity Chromatography (IMAC). The recombinant plasmid was transformed into M15 E. coli cells for expression of the N protein. A freshly isolated colony was chosen and incubated overnight in liquid LB medium containing ampicillin (50 μg/ml) and kanamycin (25 μg/ml) at 37°C. The overnight culture was diluted 1:10 in the same LB medium and grown at 37°C until A600 reached 0.6–0.8 absorbance units. The expression was then induced at 30°C for about 4 h by the addition of 0.5 mM isopropyl-b-Dthiogalactopyranoside (IPTG). The recombinant N protein was purified using Ni-NTA agarose (Qiagen) according to the procedure provided by the manufacturer. The purified N protein was then subjected to SDS-PAGE.
Western blotting
Purified N protein was loaded in SDS-sample buffer [50 mM Tris-HCl (pH 6.8), 2% w/v SDS, 10% glycerol, 100 mM DTT and 0.1% w/v bromophenol blue], then subjected to electrophoresis on a 12% polyacrylamide gel followed by electrotransfer to PVDF membranes(Bio-rad). After blocking with 3% gelatin solution, the membrane was incubated with anti-peptide rabbit serum (1/1000 dilution) for one hour and the bound antibodies were detected by the use of HRP-conjugated goat anti-rabbit IgG (H+L) antibodies (Southern Biotech) and ECL detection reagents (Amersham Pharmacia Biotech) were used to develop the film.
ELISA analysis
The titers of the antisera from rabbits were determined by ELISA as described previously15. In brief, 96-well microtiter plates (Corning Costar) were coated with the tested BSA-peptide conjungates in 0.1M carbonate buffer (pH 9.6) (5μg/ml, 50μl/well) at 4°C overnight. After blocking with PBS containing 3% gelatin/0.1% Tween 20, and the plates were incubated with diluted rabbit antisera at various concentrations at 37°C for 2 h. Bound antibodies were determined with HRP-coupled goat anti-rabbit IgG (Bio-rad) and the OD values were measure by microplate autoreader (Bio-tek) at 450 nm. Nine SARS patients (clinical diagnosed) and 4 health controls were selected and their sera were collected. All the sera from patients were confirmed IgG positive by an ELISA with whole virus lysate as coating antigen and immunoflurorences assay (IFA). The reactivities of the peptides (N1 and N2) to the sera were measured by ELISA. The protocol was similar as mentioned above. The sera dilution was 1:10 and HRP-coupled goat anti-human IgG (Bio-rad) was used to detect IgG antibodies in the sera. All the experiments that were in volved in using the patients' sera were carried on in Bio-safety Level 3 laboratory.
RESULTS
The N1 and N2 peptides showed a similar antigenicity in immunized rabbits
Based on the computer software analysis, the Antigenic Index (AI) was similar between N1 and N2 peptides. So it was expected that both of them were capable of inducing Abs. When sera were analyzed by ELISA, the anti-serum titers were similar between two peptides-immunized rabbits (Tab 1). The data showed that both of N1 and N2 peptides were antigenic and induced the peptide-specific antibodies in the rabbits.
Table 1.
ELISA determination of antibody titers in rabbit antiserum after immunization with N1- and N2-BSA.
| Coated antigen | ||||
|---|---|---|---|---|
| Peptides | Serum sample | Neutralized with | Peptide-BSA | BSA |
| Pre-immunization | None | 4,000* | <1,000 | |
| N1 | Post-immunization | BSA | >128,000 | 1,000 |
| Post-immunization | None | >128,000 | 128,000 | |
| Pre-immunization | None | 2,000 | <1,000 | |
| N2 | Post-immunization | BSA | >128,000 | 1,000 |
| Post-immunization | None | 128,000 | 16,000 | |
*The antiserum dilution were used in ELISA determination.
N1 peptide-induced polyclonal antibody had a higher affinity to expressed N-protein of SARS-CoV
In order to determine whether N1 and N2 peptide-induced polyclonal Abs bind to N protein of SARS-CoV, the expressed full length N protein (422 amino acids) was served as target protein and the binding activity of N protein to antibodies was measured with western blot analysis. Our data demonstrated that the purified E.coli expressed N protein (47 kD) was clearly observed in SDS-PAGE electrophoresis analysis (Fig 1). The E.coli expressed glutathion-S-transferase (GST) protein was selected as unrelated protein control (Fig 1) and the binding activity of both N protein and GST protein (26 kD) to the anti-sera was analyzed by western blotting (Fig 2). The results showed that N1 peptide-induced antiserum had a strong binding signal to the N protein and the anti-N2 serum revealed relatively low signal. Whereas there are no signal in GST controls and normal rabbit serum control. The results suggested that N1-peptide was a good epitope of the N protein of SARS-CoV. The N1-peptide induced Ab was capable of binding to the expressed N protein of SARS-CoV.
Figure 1.
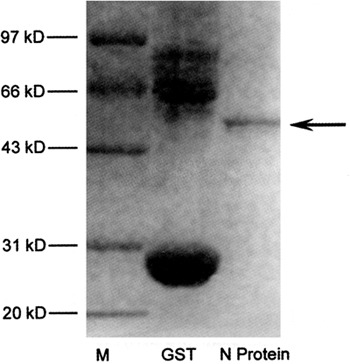
SDS-PAGE of the purified N protein of SARS-CoV and GST control protein. Lane 1: low molecular protein standards. Lane 2: control GST protein (with 26 kD) and Lane 3: the purified N protein of SARS-CoV (with 47 kD). SDS-PAGE was performed on a 12% polyacrylamide gel and stained using Coomassie brilliant blue.
Figure 2.
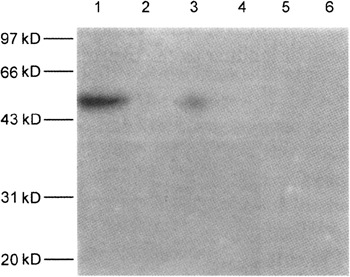
The peptide-induced antibodies were capable of binding to N protein of SARS-CoV measured by western blot analysis. Lanes 1, 3 and 5 with loading of the purified N protein of SARS-CoV. Lanes 2, 4, 6 with loading of GST control protein. Lanes 1-2 were blotted with N1-peptide immunized rabbit serum. Lane 3-4 with N2-peptide immunized rabbit serum and Lane 5-6 with normal rabbit serum. The N protein and control GST protein were subjected to SDS-PAGE, then were transferred to PVDF membrane. The blot was probed with the antiserum (1:1000), and the immunological complex was detected with HRP-conjugated goat-anti rabbits IgG (Southern Biotech).
N1 peptide was capable of binding to IgG from SARS patients
Since N1 peptide was a good epitope of the N protein of SARS-CoV, it was reasonable to ask whether N1 peptide has a capacity to binding to human IgG from SARS patients. To address our question, N1 and N2 peptides served as an antigen were used to screen the sera of nine SARS patients (clinical diagnosed) and four health people by ELISA. It was interested to observe that three of nine (33%) SARS patients had positive response to N1 peptide, whereas N2 peptide showed negative response to all tested sera (Tab 2). At mean time, four health controls also had a negative response to both peptides.
Table 2.
N1- and N2-peptide specific antibodies (total IgG) were detected in the sera from SARS patients and normal health controls by ELISA.
| Coated Ag | Health controls No. | SARS patients No. (clinical diagnosis) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| N1-BSA | − | − | − | − | + | − | − | + | − | + | − | − | − |
| N2-BSA | − | − | − | − | − | − | − | − | − | − | − | − | − |
Notice: serum dilution is 1:10. + means positive response. − means negative response.
DISCUSSION
In this study, we were the first time to identify an epitope (N1) of the N protein of SARS-CoV, which will be helpful to study the functions of the N protein.
First, two peptides (N1 and N2) were selected from the sequences of N protein of SARS-CoV by computer software analysis. Theoretically, both of peptides were good antigens based on AI. When antisera were analyzed, the peptide-specific antibodies were detectable in N1 and N2 peptide-immunized rabbit and the titers of Abs were at similar levels.
Secondly, whether that synthetic peptide closely mimics the native structure of target protein is critical for peptide application. Our data showed that only N1 peptide-induced antibodies had a high affinity with the N protein, indicating N1 peptide was a good epitope of the N protein. In addition, N1 peptide-induced antibodies will be useful for the study of the N protein. The data also suggested that only limited synthetic peptides might induce good antibodies that will bind to native protein.
To further confirm whether the differences of the two peptides antibodies to bind to the expressed N protein reflected really the difference of their antigenicities in vivo, two peptides in responding to the IgG from the sera of SARS patients were measured by ELISA. It is obviously observed that the N1 peptide (20 amino acids in length) from SARS-CoV was the good epitope to induce antibodies in SARS patients. Although only 33% (3/9) of the patients were positive, it was reasonable rate. We have also observed that when the N protein of SARS-CoV (422 amino acids in length) was used as an antigen to determine the specific-antibodies of sera from SARS patients, only 40% positive response could be detected in SARS patients who had been confirmed by the clinical diagnosis (Rui-Fu Yang, unpublished data). Our work was the first time to identify an epitope of the N protein of SARS-CoV, the antibodies induced by the peptide will be useful for the SARS diagnosis or analysis of N protein functions.
Acknowledgements
This work was supported by the grant of Shanghai Science and Technology Committee (No. 03DZ19113), National Key Basic Research Program of China (No. 2001CB510006), 863 project (No. 2001AA231011) and a specific project against SARS from Chinese Academy of Sciences.
Accession codes
Accessions
GenBank/EMBL/DDBJ
Footnotes
Ying LIN, Xu SHEN, Rui Fu YANG and Yi Xue LI: These authors contributed equally to the work.
Contributor Information
Jia Rui WU, Email: wujr@sunm.shcnc.ac.cn.
Bing SUN, Phone: 0086-21-54921376 FAX: 0086-21-54921011, FAX: (, Email: bsun@sibs.ac.cn.
References
- 1.Ksiazek TG, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1947–59. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Bloom BR. Lessons from SARS. Science. 2003;300:701. doi: 10.1126/science.300.5620.701. [DOI] [PubMed] [Google Scholar]
- 3.Lipsitch M . Transmission dynamics and control of severe acute respiratory syndrome. Published online 23 May 2003; 10.1126/Science.1086616 [DOI] [PMC free article] [PubMed]
- 4.Steven Riley .Transmission dynamics of the aetiological agent of severe acute respiratory syndrome (SARS) in Hong Kong: The impact of public health interventions. Published online 23 May 2003; 10.1126/Science.1086478. [DOI] [PubMed]
- 5.Rota RA. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–9. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Marra MA. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 7.Holmes KV, Enjuanes L. Perspective: The SARS coronavirus: A postgenomic era. Science. 2003;300:1377–8. doi: 10.1126/science.1086418. [DOI] [PubMed] [Google Scholar]
- 8.Kuo L, Masters PS. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J Virol. 2002;76(10):4987–99. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narayanan K, Chen CJ, Maeda J. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J Virol. 2003;77(5):2922–7. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu L, Liu W, Schnitzlein WM. Study of protection by recombinant fowl poxvirus expressing C-terminal nucleocapsid protein of infectious bronchitis virus against challenge. Avian Dis. 2001;45(2):340–8. doi: 10.2307/1592973. [DOI] [PubMed] [Google Scholar]
- 11.Hiscox JA, Wurm T, Wilson L. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J Virol. 2001;75(1):506–12. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu C, Kokuho T, Kubota T. DNA mediated immunization with encoding the nucleoprotein gene of porcine transmissible gastroenteritis virus. Virus Res. 2001;80(1–2):75–82. doi: 10.1016/S0168-1702(01)00333-1. [DOI] [PubMed] [Google Scholar]
- 13.Wu YD, Wang YP, Zhong CP, Li YC, Li XY, Sun B . The suppressive effect of triptolide on experimental autoimmune uveoretinitis by down-regulating Th1-type response. International Immunopharmocology 2003 (in press). [DOI] [PubMed]
- 14.Wu YD, Lin Y, Hou WQ, Wang YP, Sun B. IFN-g up-regulation of IL-12 beta 2 receptor is associated with Experimental Autoimmune Uveitis (EAU) in susceptible B10.A mice, but not in EAU resistant BALB/c mice. Journal of Neuroimmunology. 2003;137:154–63. doi: 10.1016/S0165-5728(03)00071-7. [DOI] [PubMed] [Google Scholar]
- 15.Hou WQ, Wu YD, Sun SH, Shi MD, Sun Y, Yang CH, Pei G, Gu YD, Zhong CP, Sun B. Perterssis toxin enhaces Th1 responses by stimulation of dendritic cells. Journal of Immunology. 2003;170:1728–36. doi: 10.4049/jimmunol.170.4.1728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


