Abstract
To explore the potential of using non-human coronaviruses for cancer therapy, we first established their ability to kill human tumor cells. We found that the feline infectious peritonitis virus (FIPV) and a felinized murine hepatitis virus (fMHV), both normally incapable of infecting human cells, could rapidly and effectively kill human cancer cells artificially expressing the feline coronavirus receptor aminopeptidase N. Also 3-D multilayer tumor spheroids established from such cells were effectively eradicated. Next, we investigated whether FIPV and fMHV could be targeted to human cancer cells by constructing a bispecific single-chain antibody directed on the one hand against the feline coronavirus spike protein – responsible for receptor binding and subsequent cell entry through virus–cell membrane fusion – and on the other hand against the human epidermal growth factor receptor (EGFR). The targeting antibody mediated specific infection of EGFR-expressing human cancer cells by both coronaviruses. Furthermore, in the presence of the targeting antibody, infected cancer cells formed syncytia typical of productive coronavirus infection. By their potent cytotoxicity, the selective targeting of non-human coronaviruses to human cancer cells provides a rationale for further investigations into the use of these viruses as anticancer agents.
Keywords: cancer, coronavirus, virotherapy, targeting, EGFR, bispecific single-chain
Introduction
Replicative oncolytic viruses represent new agents with potential utility in cancer therapy. They are aimed to effectively eradicate tumor cells without affecting normal tissues. Several different DNA and RNA viruses are currently being evaluated for their efficacy and selectivity toward cancer cells.1, 2 So far, there are no reports describing the potential use of coronavirus-based oncolytic agents in cancer virotherapy, despite a number of features that make coronaviruses potentially attractive for this purpose.
Coronaviruses are positive-strand RNA viruses consisting of a nucleocapsid, which contains the approximately 30 kb genome and the nucleocapsid (N) protein, and which is surrounded by an envelope carrying three membrane proteins, spike (S), envelope (E), and matrix (M). Of these, the spike glycoprotein S is responsible for virus entry and syncytia formation, as it binds to the cellular receptor and induces membrane fusion.3, 4, 5 The specific interaction between the amino-terminal spike protein domain S1 and its cognate receptor6, 7, 8 induces conformational changes in the spike protein domain S2 that trigger the membrane fusion process. These conformational changes require physical interaction between two heptad repeat (HR) regions, HR1 and HR2, that occur in the S2 domain. Prevention of this interaction by using peptides that correspond to these HR1 and HR2 regions can block membrane fusion, by binding of the peptides to their respective counterparts.9, 10, 11 Most coronaviruses exhibit strict species specificity, as determined by the spike–receptor interaction.12, 13, 14 The coronavirus feline infectious peritonitis virus (FIPV), for instance, selectively infects and induces syncytium formation of feline cells via its receptor feline aminopeptidase N (fAPN).6 Likewise, the recombinant felinized mouse hepatitis virus (fMHV),15 a derivative of mouse hepatitis virus (MHV) carrying a chimeric spike of which the ectodomain is from the FIPV spike protein, also infects and fuses only feline cells through the fAPN molecule. As a consequence of their species restricted tropism, FIPV and MHV are nonpathogenic to human cells. However, once the tropism barrier is alleviated, coronaviruses can replicate in cells of different species.15, 16 Thus, FIPV and MHV may potentially be converted into specific oncolytic agents for the treatment of human cancer if their spike protein would recognize a receptor on human tumor cells.
Toward developing recombinant coronaviruses with a selective human tumor tropism, we first set out to investigate the effectiveness of FIPV and MHV in eradicating, in vitro, human cancer cells expressing the virus receptor. To allow comparison of the two viruses in the same cells, we assessed FIPV and, instead of MHV, the chimeric coronavirus fMHV for their ability to kill human cancer cells artificially expressing fAPN. Subsequently, we investigated whether FIPV and fMHV could be targeted to human cancer cells expressing the epidermal growth factor receptor (EGFR), a molecule commonly overexpressed on many types of cancer cells17 and associated with poor prognosis and response to cancer therapy.18, 19 For this purpose, we prepared a bispecific single-chain variable fragment (scFv) antibody, binding on one side to the FIPV and fMHV spike protein and on the other side to the human EGFR. The antibody indeed functioned as a specific targeting device. Our findings justify the further development of coronaviruses as oncolytic agents.
Results
Infection and killing of human cancer cells expressing fAPN by FIPV and fMHV
To determine whether non-human coronaviruses can infect and kill human cancer cells when the host species barrier determined by specific receptor recognition is alleviated, FIPV and fMHV were redirected to human cancer cells via the virus receptor fAPN. First, the susceptibility of the tumor cell lines Caco-2, OVCAR-3, HCT-8, HeLa, HepG2, and WiDr to FIPV and fMHV was successfully demonstrated by transient transfection of these cells with an fAPN expression plasmid followed by virus inoculation (data not shown). Control inoculations on cells not transfected with the fAPN expression construct did not show any detectable FIPV or fMHV infection, confirming that infection required the appropriate receptor expression.
Studies of FIPV and fMHV propagation, cytotoxicity, and syncytia formation require host cells with stable receptor expression. Therefore, HeLa and OVCAR-3 cell lines stably expressing fAPN were produced. FIPV and fMHV were tested for their growth characteristics in these HeLa-fAPN and OVCAR-fAPN cells (Figure 1a). Evidently, infection of fAPN-expressing human cancer cells with the viruses FIPV and fMHV resulted in a typical rapid production of progeny virus.
Figure 1.
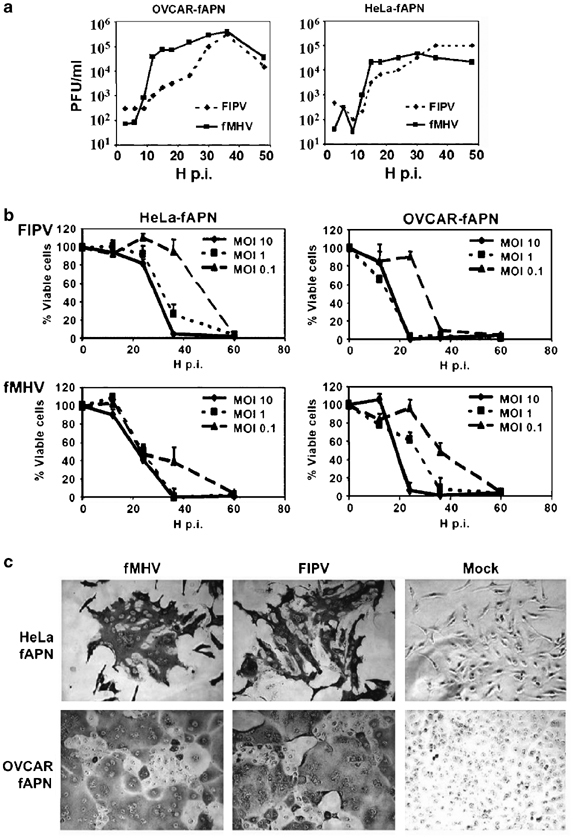
Propagation and cytotoxicity of FIPV and fMHV in human cancer cells expressing fAPN. (a) HeLa-fAPN and OVCAR-fAPN cells were inoculated at an MOI of 5 with FIPV or fMHV for 1 h as indicated and cultured for up to 48 h. At several time points p.i., the virus titer in the culture medium was determined by end point titration on FCWF-4 cells. The data are from a representative experiment. (b) Cell viability of HeLa-fAPN and OVCAR-fAPN monolayers after inoculation with various multiplicities of FIPV and fMHV, measured at several time points after infection by WST-1 conversion assay. Results are depicted as the percentage of viable cells in infected relative to mock-infected cells. The data shown are the means+standard deviations of a representative experiment performed in triplicate. (c) Immunohistochemical staining for coronavirus proteins on HeLa-fAPN and OVCAR-fAPN cells 15 h after infection with FIPV or fMHV at MOI 1.
To determine the potency of FIPV and fMHV to kill human cancer cells, HeLa-fAPN and OVCAR-fAPN cells were inoculated at various multiplicities of FIPV or fMHV and the cell viability was measured at several time points after infection. FIPV and fMHV efficiently killed both types of cells in a dose-dependent manner. The onset of cell death was found to occur as early as 12–24 h after inoculation. Following infection at a multiplicity of infection (MOI) of 10, both viruses completely eliminated the OVCAR-fAPN and HeLa-fAPN monolayers within 24 and 36 h, respectively. Infections at lower MOI apparently required multiple rounds of infection resulting in the death of all or nearly all cells within 60 h (Figure 1b). Furthermore, infected cells clearly showed membrane fusion typical of coronavirus replication (Figure 1c).
fMHV-mediated eradication of human cancer cells in an in vitro multilayer tumor model
We also determined whether non-human coronaviruses were able to eradicate human cancer cells in an in vitro solid tumor model. Multilayer tumor spheroids offer a useful 3-D model for assessing virus-mediated eradication of tumor tissue. This model has already been employed to study the potency of oncolytic adenoviruses and adeno-associated viruses.20, 21 OVCAR-fAPN spheroids were established and inoculated with fMHV at 5 × 104 plaque forming units (PFU)/spheroid. For comparison, the oncolytic effect of adenovirus type 5 (Ad5) at 5 × 108 PFU/spheroid was also measured. At several days post-inoculation (p.i.), the cell viability was determined (Figure 2a). At day 5 p.i., a clear decrease in viability was observed for spheroids inoculated with either virus. After 2 days, the OVCAR-fAPN spheroids infected with 5 × 104 PFU fMHV were essentially destroyed, whereas spheroids infected with 5 × 108 PFU Ad5 were only partially eradicated. It should, however, be noted that the cytolytic and entry mechanisms of coronaviruses and adenoviruses differ distinctly, making a direct comparison not entirely valid. The results were confirmed by light microscopic analysis (Figure 2b).
Figure 2.
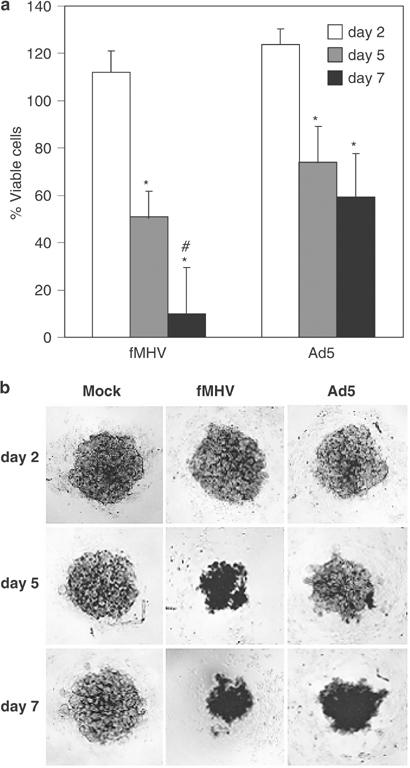
fMHV efficiently kills OVCAR-fAPN multilayer tumor spheroids. (a) Spheroids were inoculated with fMHV (5 × 104 PFU/spheroid) or Ad5 (5 × 108 PFU/spheroid) or mock inoculated and cultured for 2, 5, and 7 days, after which the cell viability was measured by WST-1 conversion assay. Results are depicted as the percentage of viable cells in infected relative to mock-infected control spheroids. The data shown are the means+standard deviations of a representative experiment performed in triplicate. *Significant difference between day 2 and day 5 or 7, P<0.01; #significant difference between the cell death on day 7 mediated by fMHV compared to the cell death on day 7 mediated by Ad5, P<0.05. (b) Representative images of OVCAR-fAPN multilayer spheroids inoculated with fMHV, Ad5, or mock inoculated, and cultured for 2, 5, or 7 days.
Bispecific antibody-mediated coronavirus infection of human cancer cells
Having established that FIPV and fMHV can infect and destroy cancer cells once the entry barrier has been overcome, we wanted to develop a general method to target these viruses to a suitable antigen expressed on such cells. To this end, we constructed the bispecific scFv 23F-425, which combines the antigen binding domains from antibodies 23F8.1 and 425, recognizing the FIPV S protein and EGFR, respectively. The protein was produced by expression in eucaryotic cells. Its synthesis and secretion were verified by radiolabeling followed by immunoprecipitation from the cell lysate and culture medium using an anti-Myc antibody (Figure 3). The results clearly show the synthesis and secretion of the approximately 58 kDa bispecific single-chain molecules.
Figure 3.
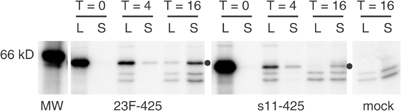
Analysis of synthesis and secretion of expressed scFv 23F-425. The Myc-tagged bispecific protein 23F-425 was radiolabeled in OST7-1 producer cells and analyzed by immunoprecipitation using an anti-Myc antibody. At 0, 4, and 16 h after labeling, equal volume fractions of cell lysate (L) and culture medium (S) were analyzed. The Myc-tagged bispecific scFv s11-42539, 40 was included in the experiment for comparison. The position of the scFv’s is indicated by a dot. At the right, a control analysis of cells transfected without DNA indicates the nonspecific nature of some proteins.
To investigate whether scFv 23F-425 could serve as an adapter molecule for FIPV and fMHV infection via human EGFR, cultures of human cancer cell lines of different tissue origin with confirmed expression of EGFR (Figure 4) were inoculated with similar amounts of FIPV or fMHV in the presence or absence of the bispecific antibody. After 1 h at 37°C, the inoculum was replaced by regular culture medium and incubation of the cells was continued for 24 h. The cells were immunostained for coronavirus protein expression. As can be seen in Figure 4, all cell lines tested had become infected with FIPV and fMHV in the presence of scFv 23F-425. In contrast, none of the cells stained positive after inoculation with FIPV or fMHV that had been preincubated with mock control supernatant (data not shown). Similarly, no positive staining was observed when human cancer cells had been inoculated with the control virus MHV in the presence of scFv 23F-425 (data not shown).
Figure 4.
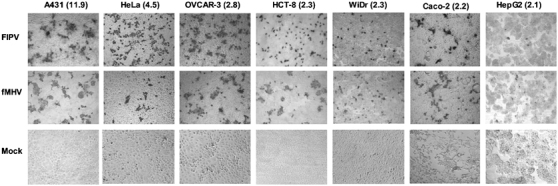
Targeted coronavirus infection of human cancer cells. Cells were inoculated (MOI of 5) with preincubated mixtures of FIPV or fMHV (control: no virus) and the bispecific adapter protein scFv 23F-425. The cells were stained for coronavirus proteins 24 h after inoculation. All cell lines tested showed susceptibility to EGFR-targeted FIPV or fMHV. Values in parentheses indicate the relative EGFR expression (relative geometric mean fluorescence (GMF)) as determined for each cell line by FACS analysis and expressed as the ratio of the GMF values obtained in the presence and absence of the anti-EGFR antibody.
Differences in infection efficiency were observed between different cell lines, with the EGFR-high A431 cells showing the highest susceptibility to EGFR-targeted coronavirus infection and the EGFR-low HepG2 cells being the most poorly infected. On most cell lines, HeLa cells being the exception, EGFR-targeted FIPV exhibited a similar infection efficiency as EGFR-targeted fMHV. Interestingly, infected cells formed syncytia typical for productive coronavirus infection. The formation of infectious progeny virus was confirmed by monitoring – through titration on FCWF-4 cells – the increase in viral titers in the medium of A431 cells after inoculation with FIPV or fMHV. Typical growth curves were obtained, but only after the A431 cells had been inoculated in the presence of bispecific scFv 23F-425 (Figure 5).
Figure 5.
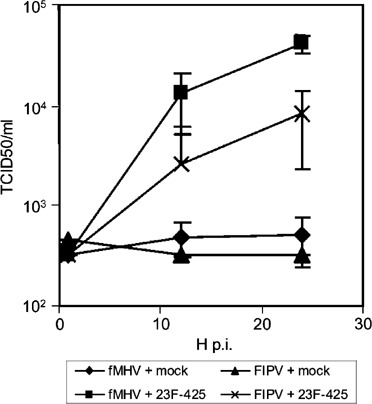
Bispecific scFv 23F-425-mediated infections of A431 cells by FIPV and fMHV result in the production of infectious progeny virus. FIPV and fMHV were preincubated in the presence or absence of scFv 23F-425 and inoculated on EGFR-expressing A431 cells. After 1 h, the cells were washed three times and incubation was continued. At several time points, samples were taken from the cell culture medium, which were subsequently titrated on feline FCWF-4 cells to determine the amount of virus produced. Productive infection of A431 cells was evident with both viruses but only after inoculation in the presence of the bispecific antibody. All data shown represent the average and standard deviation of an experiment performed in triplicate.
Bispecific antibody-mediated coronavirus infection is human EGFR specific
To confirm that the infections of FIPV and fMHV established by scFv 23F-425 were indeed mediated by the human EGFR protein, we tested the mouse fibroblast cell line NIH 3T3 and its human EGFR-expressing derivative NIH 3T3-hEGFR22 for their susceptibility to FIPV and fMHV in the presence and absence of scFv 23F-425. As illustrated in Figure 6a, positive immunostaining for coronavirus proteins was only obtained with NIH 3T3-hEGFR cells inoculated in the presence of scFv 23F-425. This result demonstrated that FIPV and fMHV infection of these otherwise refractory NIH 3T3 cells required both the adapter molecule and expression of human EGFR.
Figure 6.
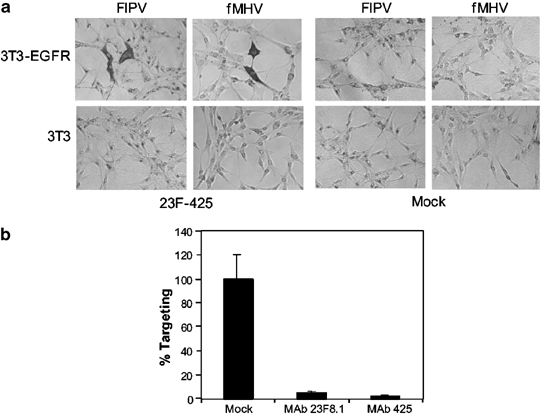
EGFR-specific coronavirus targeting. (a) NIH 3T3-hEGFR and NIH 3T3 cells were inoculated with FIPV or fMHV in the presence or absence of the bispecific adapter protein scFv 23F-425. After 1 h at 37°C, the inoculum was replaced by regular culture medium and incubation of the cells was continued. The cells were stained for coronavirus proteins 24 h after inoculation. Only NIH 3T3-hEGFR cells inoculated with FIPV or fMHV in the presence of the bispecific scFv 23F-425 stained positive. (b) Compared to EGFR-targeted infections of HeLa cells in the absence of blocking antibody, virus preincubation with anti-spike MAb 23F8.1, or preincubation of the cells with anti-EGFR MAb 425, decreased the amount of infected cells. The data represent the average percentage of infected cells relative to cells inoculated in the absence of blocking antibody. The error bars show the standard deviation of an experiment performed in triplicate.
Further evidence for a specific interaction between the bispecific scFv 23F-425 and human EGFR was obtained by studying the effect of the EGFR antibody 425 on the scFv 23F-425-mediated coronavirus infection. HeLa cells were incubated with the 425 monoclonal antibody prior to inoculation of fMHV in the presence of scFv 23F-425. Figure 6b shows that infection was blocked almost completely, confirming that a direct interaction with the EGFR is required. Similarly, the necessity of an interaction between the FIPV spike protein and scFv 23F-425 was confirmed. We incubated fMHV with and without anti-FIPV spike monoclonal antibody 23F8.1 before adding scFv 23F-425 and inoculating HeLa cells. Again, the anti-S antibody inhibited infection, demonstrating that scFv 23F-425 has to bind to the virus spike protein in order to function as a targeting adaptor (Figure 6b).
Bispecific antibody-mediated syncytia formation of coronavirus-infected cancer cells
Several human cancer cell lines that were infected with EGFR-targeted FIPV or fMHV showed cell–cell fusion typical for coronaviruses (see Figure 4). Syncytium formation is an important determinant for cytotoxicity and spread of coronaviruses. Therefore, it was important to investigate if syncytium formation between infected and neighboring cells resulted from undefined interactions or from specific bridging by scFv 23F-425. To this end, A431 cells were inoculated with fMHV in scFv 23F-425-containing medium for 2 h and subsequently cultured for another 22 h in the presence or absence of scFv 23F-425. Thus, scFv 23F-425 was either present continuously to support infection and syncytium formation or only briefly to mediate targeted infection. Removal of scFv 23F-425 after 2 h significantly reduced syncytium formation by approximately four-fold from 40.8±4.1 to 11.2±2.2 nuclei per syncytium (the data represent the average amount of nuclei per syncytium measured in three independent experiments; ± indicates the standard deviation). Hence, scFv 23F-425 did not only promote fusion between the fMHV envelope and target cells, but also fusion between fMHV-infected and neighboring cells.
Targeted coronavirus infection and syncytium formation require the native spike membrane fusion function
Coronaviruses use a class I membrane fusion mechanism,11 common to a number of enveloped virus families,23 in which HR regions occurring in the spike protein play an instrumental role. The mechanism involves conformational changes in the spike protein subsequent to receptor binding, resulting in an interaction of the HR1 and HR2 domains, which is necessary to drive the membrane fusion process. This process can be inhibited specifically using peptides corresponding to the HR domains as we showed recently for MHV.11 In order to investigate if the bispecific antibody-targeted FIPV and fMHV infections and their induction of syncytia also depend on those conformational rearrangements in the viral spike protein, we tested the sensitivity of these processes to HR-derived peptide. To this end, we prepared a peptide corresponding to the HR1 region of the FIPV S protein and initially studied this fHR1 peptide for its ability to block FIPV and fMHV infection of feline FCWF-4 cells, which it did. As illustrated for fMHV (Figure 7a and b), addition of fHR1 during or after inoculation of feline FCWF-4 cells abrogated infection and syncytium formation, respectively. Next, we determined whether scFv 23F-425-mediated infection and syncytium formation are also sensitive to the fHR1 peptide. To this end, fMHV was targeted toward EGFR on A431 cells in the presence or absence of fHR1. As can be seen in Figure 7c and d, addition of the peptide reduced both scFv 23F-425-mediated infection and syncytium formation. Thus, both processes seem to utilize a membrane fusion process similar to that of the native coronavirus.
Figure 7.
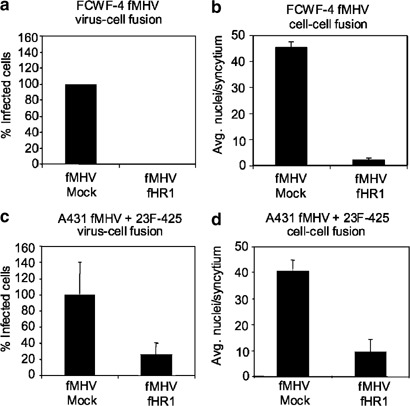
Effect of fHR1 on fMHV-mediated fusion processes. (a) Preincubation of fMHV with fHR1 blocked the infection of feline FCWF-4 cells. (b) In addition, the formation of syncytia was inhibited when fHR1 peptide was added to the culture medium 1 h after infection of FCWF-4 cells. (c) fHR1 also caused a significant reduction in the number of infected human A431 cancer cells when inoculated with a preincubated mixture of fMHV and scFv 23F-425 in the presence of 0.5 μM fHR1. (d) Also the cell–cell fusion was inhibited by fHR1 when peptide and scFv 23F-425 were added to the medium of A431 cells that had been infected with a preincubated mixture of fMHV and scFv 23F-425. All data shown represent the average and standard deviation of an experiment performed in triplicate.
Discussion
Despite tremendous research efforts over the last decades into the nature of the disease and its causes, and despite the significant new insights acquired, cancer remains one of the most common causes of death. Actually, treatment still relies for a major part on classical approaches such as surgery, radiotherapy, and chemotherapy. Clearly, novel and creative methods are needed to complement the conventional treatment options. Recently, the use of viruses as potential tools for anticancer therapy has gained considerable interest.1, 2 In this first exploratory study, we demonstrate two important features that make coronaviruses attractive for this purpose, that is, the ability to target these viruses to human tumor cells and their subsequent infection and eradication of these cells.
The non-human coronaviruses FIPV and fMHV appeared to possess a strong capacity to kill human cancer cells once they are able to enter these cells through an artificially expressed receptor, consistent with earlier observations with MHV by Gallagher and co-workers.6, 24 The observed rapid and efficient eradication of cancer cells can probably be attributed to two coronaviral features. First, coronaviruses are positive-strand RNA viruses that exhibit a fast, cytoplasmic transcription process leading to rapid virus protein synthesis and progeny virus production. Second, the ability of coronaviruses to induce syncytia between infected and noninfected neighboring cells amplifies their cytotoxicity. We found that FIPV and fMHV retained these properties in human cancer cells expressing the virus receptor, thus enabling a rapid cytotoxic spread of the virus to surrounding noninfected cancer cells. Coronaviruses share some of these attractive characteristics with other enveloped viruses such as a fusogenic mutant of herpes simplex virus25 and the live attenuated Edmonston B vaccine strain of measles virus.26 However, in contrast to these viruses, the coronaviruses FIPV and fMHV are normally incapable of infecting human cells, due to their restricted tropism. Thus, their native tropism does not need to be abolished in order to specifically limit their access, hence cytotoxicity, to human cancer cells.
The ability to deliberately target viruses to preselected cells is a tremendous challenge with far-reaching implications for all kinds of therapeutic applications. Although we were able to demonstrate the principle of retargeting of coronaviruses by exchanging spike ectodomains,15, 16 neither the detailed structural information nor the knowledge and technology required to purposely redesign the spike for binding to any given antigen are presently available. Therefore, as an alternative and also potentially versatile tool, we embarked on the development of a bispecific adapter. Thus, to redirect the non-human coronaviruses FIPV and fMHV to EGFR-expressing cells, we constructed the bispecific antibody molecule scFv 23F-425 that binds to both the feline spike and EGFR. The latter was chosen for its frequent overexpression on human cancer cells. Inoculation of FIPV and fMHV onto a number of different EGFR-expressing human cancer cell lines of various tissue origins in the presence of scFv 23F-425 resulted in infection, replication, and subsequent formation of syncytia. The targeted infection was completely dependent on the presence of the antibody and its efficiency generally correlated with the levels of EGFR expression on the cancer cells. These observations are similar to native coronavirus infections where virus–cell and cell–cell fusion efficiencies also correlate with host cell receptor density.24 The results imply that the bispecific antibody-mediated targeting approach can in principle be applied to direct coronaviruses to any cell surface antigen for which an appropriate antibody (ie hybridoma cell line) is available.
The successful application of bispecific adapters for viral tumor therapy will depend, among others, on cell surface antigens that are – preferably – unique to the tumor. Useful tumor-specific markers have not yet been identified and future work should reveal whether they occur or can be specifically induced in tumor cells. Interestingly, for the attenuated measles virus, it was recently described that receptor density could also be a determinant of preferential tumor killing.27 This indicates that even overexpression of certain receptors on tumor cells may result in tumor selective infection and subsequent cell killing. This may also be of importance for the targeting of coronaviruses, since similar receptor density dependence has been described to be important for coronavirus infection and syncytia formation.24
The application also depends on the sufficient, local presence of the adapter. This might be achieved by incorporating the adapter gene sequence into the viral genome to have the virus produce its own targeting device. We have demonstrated this principle recently for the targeting of conditionally replicating adenoviruses. Thus, adenoviruses expressing a bispecific scFv for targeting to EGFR showed enhanced oncolytic replication in EGFR-positive, adenovirus receptor-negative cancer cells.28 Foreign gene expression can also be achieved with coronaviruses as we showed recently after inserting different reporter genes at various positions in the MHV genome.29
Coronaviruses exhibit high mutation rates and are prone to recombination. Their application in adapter-mediated targeting to tumors will thus raise serious safety questions, particularly regarding the possibility of generating viruses that acquired the capacity to infect human cells independent of targeting devices. These questions will have to be addressed. Several options to reduce the risks already exist. One is the use of coronaviruses lacking specific virulence genes; as we showed recently for MHV30 and FIPV,31 such viruses are strongly attenuated in their natural host. Another option is to combine deletion of nonessential virulence genes with genomic rearrangement; reorganization of the order of the structural protein genes, found for MHV to be tolerated without loss of viability,32 will reduce the risk of generating viable viruses by recombination with circulating field viruses.
Entry of coronaviruses into cells normally requires binding of the spike to the receptor followed by a series of structural rearrangements in the S protein that eventually lead to the merging of viral and cellular membranes. It appeared that the bispecific antibody-mediated entry, as well as the antibody-mediated induction of syncytia, uses this same fusion mechanism as was illustrated most clearly by the inhibitory effect of the HR1-derived peptide. It will be interesting to find out how this fusion process actually takes place and whether, for instance, the necessary conformational changes in the S protein are induced by its interaction with the antibody or triggered by the binding to the EGFR. Alternatively, fusion may not be mediated by the spikes that effect the binding but, rather, by the conformational changes induced in the ‘free’ spikes upon interaction with undefined molecules on the target cells or by the particular conditions experienced when the virus is taken up in endosomes. Evidence for the latter mechanism might be obtained by studying whether infection can also be achieved when using a bispecific antibody not binding the virus through the S protein but through one of the other envelope proteins, M or E, as these are not involved in the fusion process.
An elegant application of the targeting principle has recently been described for another enveloped virus. In measles virus, the receptor binding and membrane fusion functions are divided over two different envelope proteins, the hemagglutinin (H) and the fusion (F) protein, respectively. When an scFv was carboxy-terminally appended to the type II membrane glycoprotein H, the virus was successfully targeted to cells expressing the distinguishing receptor.26, 33 Obviously, appending the EGFR binding moiety of scFv 23F-425 to the amino terminus of the coronavirus S protein might also expand the targeting possibilities of these viruses.
Materials and methods
Viruses, cells, and antibodies
Recombinant fMHV15 and FIPV strain 79-114634 stocks were produced and titrated in parallel on feline FCWF-4 cells, yielding titers of 5 × 106 and 1 × 107 TCID50/ml (tissue culture infectious dose 50), respectively. Wild-type Ad5 was produced and titrated on 293 human embryonic kidney cells. The recombinant vaccinia virus vTF7-3 containing the bacteriophage T7 RNA polymerase gene was used as a T7 RNA polymerase source for the T7 promoter-driven production of bispecific scFv in OST7-1 cells.35
OST7-1 (obtained from B Moss), NIH 3T3, HeLa, OVCAR-3, HCT-8, Caco-2, WiDr, HepG2, A431 (American Type Culture Collection), and FCWF-4 cells (obtained from NC Pedersen) were grown in Dulbecco's modified Eagle's medium (DMEM) (Cambrex Bio Science, Verviers, Belgium) containing 10% fetal bovine serum (FBS), 100 IU of penicillin/ml, and 100 μg of streptomycin/ml (all from Life Technologies, Ltd, Paisley, UK). NIH-hEGFR cells (NIH 3T3-her14, obtained from PMP van Bergen en Henegouwen)22 were maintained in DMEM containing 10% FBS, 100 IU of penicillin/ml, 100 μg of streptomycin/ml, and 0.25 mg/ml G418 (Life Technologies, Ltd, Paisley, UK). HeLa-fAPN and OVCAR-fAPN were maintained in the same medium supplemented with 0.5 and 0.25 mg/ml G418, respectively. The hybridoma cell line producing the 23F8.1 monoclonal antibody (MAb) against the FIPV spike protein36 was cultured in CD Hybridoma medium supplemented with 2 mM Glutamax (all from Life Technologies, Ltd, Paisley, UK).
C428, ascitic fluid from an FIPV-infected cat (kindly provided by BJ Haijema), was used as a source of polyclonal antibodies to FIPV. The rabbit antiserum K134 raised against purified MHV37 and the MAb R-G-4 directed against the fAPN receptor have been described previously.37, 38 The culture supernatant of the hybridoma cell line 23F8.136 was used as a source of the MAb 23F8.1 directed against the FIPV and fMHV spike. For EGFR detection, the MAb 425 was used (culture supernatant from the hybridoma cell line 425, ATCC). To detect Myc-tagged scFv 23F-425, the anti-Myc antibody Myc was used (culture supernatant from the hybridoma cell line Myc 9E10, ATCC).
Production and characterization of fAPN-expressing cell lines
The expression plasmid pCR3-fAPN, containing the fAPN cDNA under the control of the cytomegalovirus promoter,5 was used to transiently express fAPN on different cancer cell lines following transfection with Lipofectamine PLUS reagent (Life Technologies, Ltd, Paisley, UK). Transfected HeLa and OVCAR-3 cells were also cultured in G418-containing cell culture medium to select for stable transfectants. HeLa and OVCAR-3 G418-resistant cells were cloned by two and four rounds of limiting dilution, respectively, and tested for their susceptibility to FIPV infection and for fAPN expression by fluorescent activated cell sorter (FACS) analysis. Furthermore, FIPV infection was blocked by the anti-fAPN antibody R-G-4, confirming that the infections occurred via fAPN.
Monitoring of virus growth
An amount of 5 × 105 cells/10 cm2 well was seeded and inoculated the next day with virus at an MOI of 5 PFU/cell for 1 h in serum-free culture medium. The cells were washed three times with PBS, and cultured for up to 48 h. At several time points p.i., the medium was harvested, centrifuged for 10 min at 3000 r.p.m., and stored at −80°C until analysis. The amount of virus produced at each time point p.i. was determined by end point titration on feline FCWF-4 cells.
Monolayer cytotoxicity analysis
An amount of 5 × 104 HeLa-fAPN or OVCAR-fAPN cells was seeded per 0.32 cm2 well and infected in triplicate with various amounts of FIPV or fMHV. At several time points after inoculation, the cells were cultured for 1 h in 10% WST-1 (Roche Diagnostics GmbH, Mannheim, Germany), after which the OD450 was measured. Viability was expressed relative to uninfected control cells, after subtraction of background values of WST-1 incubated in the absence of cells.
Spheroid cytotoxicity analysis
Three-dimensional multilayer spheroids were produced by incubating 5 × 104 OVCAR-fAPN cells in 0.32 cm2 wells coated with 100 μl 2% Multi Purpose agarose (Roche Diagnostics GmbH, Mannheim, Germany) in PBS for 24 h on a spinner-platform set at 125 r.p.m., at 37°C and 5% CO2. Subsequently, the spheroids were cultured for 7 days at 37°C and 5% CO2, reaching a diameter of approximately 600 μm, before they were used for infection experiments. Spheroids were infected in a total volume of 100 μl and cultured for 2, 5, or 7 days, after which they were incubated for 5 h in 10% WST-1. The OD450 was measured directly in spheroid-containing wells. Viability was expressed relative to uninfected control spheroids, after subtraction of background values of WST-1 incubated in the absence of spheroids. Statistical significance between different groups was determined by the t-test.
Construction of the bispecific scFv 23F-425
The hybridoma cell line 23F8.1 was used to isolate mRNA by using the Quickprep Micro mRNA Purification Kit (Amersham Pharmacia Biotech Europe GmbH, Freiburg, Germany). The Mouse ScFv Module/Recombinant Phage Antibody System (Amersham) was used to generate the scFv 23F. The mRNA was isolated from 1 × 107 23F8.1 hybridoma cells and the cDNA was produced by RT-PCR, according to the Mouse ScFv Module protocol. The variable domains of both the heavy chain (VH) and the light chain (VL) of the 23F8.1 antibody cDNA were isolated by PCR using VH and VL primers of the scFv isolation system. The fragments were cloned separately into the pGemTeasy cloning vector (Promega, Madison, USA) and sequenced. To introduce a 16-amino-acid residue middle linker, a fusion PCR was performed joining the 23F8.1-VH and 23F8.1-VL fragments in a VH–VL configuration via a 48-nucleotide linker DNA. The linker DNA consists of two primers, each comprising a part of the linker sequence of scFv 42539, 40 (underlined), and flanking sequences overlapping the 3′ 23F8.1-VH (5′-CAGAGCCACCTCCGCCTGAACCGCCTCCACCTGAGGAGACGGTGACCGT-3′) and the 5′ 23F8.1-VL (5′-CAGGCGGAGGTGGCTCTGGCGGTGGCGGATCGGACATCCAGATGACCCA-3′), resulting in a Ser-(Gly4-Ser)3 linker. In a subsequent PCR, the assembled scFv DNA was amplified and restriction sites were added by using the RS primers of the scFv isolation system. The resulting DNA fragment contained a 5′ SfiI site and a 3′ NotI site, and was subsequently cloned into pGemTeasy and sequenced. The sequence was compared to the sequences of the independent 23F8.1-VH and 23F8.1-VL fragments, and the resulting correct clone was named pGemTeasy-23FVHVL. The pGemTeasy-23FVHVL plasmid was digested by SfiI and NotI to isolate the 754 bp scFv 23F, which was ligated into the pCANTAB derivative pSTCF40 that had been digested by SfiI and NotI, resulting in pSTCF23F. The scFv 425 directed against the EGFR was isolated from pSTCFS11-42539, 40 by NotI digestion and ligated in a VH–VL configuration into the NotI site downstream of the scFv 23F in pSTCF23F creating a three Ala linker between the two scFv fragments. This resulted in the expression vector pSTCF23F-425, which contains the 1587 bp bispecific cDNA construct encoding the anti-spike scFv 23F linked to the anti-EGFR scFv 425 in fusion with an amino-terminal Igκ signal sequence and a carboxy-terminal Myc tag under the control of CMV and T7 promoters.
Metabolic labeling and immunoprecipitation
To determine whether scFv 23F-425 was produced and secreted into the culture medium, subconfluent monolayers of OST7-1 cells in 2 cm2 tissue culture dishes were inoculated with vTF7-3 at 5 PFU/cell (t=0 h) and subsequently transfected (t=1 h) without DNA, with pSTCF23F-425, or with pSTCFS11-425 plasmid DNA by using lipofectin (Life Technologies, Ltd, Paisley, UK).39, 40 At t=5 h, the cells were starved for 30 min in cysteine- and methionine-free modified Eagle's medium containing 10 mM HEPES, pH 7.2, and 5% FBS. The medium was then replaced by 200 μl of similar medium containing 100 μCi of 35S in vitro cell-labeling mixture (Amersham Pharmacia Biotech Europe GmbH, Germany). After 1 h, the cells were either lysed or the labeling medium on the cells was replaced by 240 μl normal culture medium and incubation continued for 0, 4, or 16 h. The cells were lysed in 300 μl TESV lysis buffer (20 mM Tris-HCl (pH 7.3), 1 mM EDTA, 100 mM NaCl, 1 mM PMSF, 1% Triton X-100). To the cleared medium, 60 μl of 5 × TESV lysis buffer was added. Proteins were immunoprecipitated from the medium or the lysed cells by using the anti-Myc antibody diluted 1:10. The immune complexes were adsorbed to Pansorbin cells (Calbiochem, La Jolla, USA) as described previously.29 Equal volumes of the immunoprecipitates were analyzed by SDS–PAGE containing 10% polyacrylamide.
Production of the bispecific scFv 23F-425
For the production of scFv 23F-425, subconfluent monolayers of OST7-1 cells were inoculated at an MOI of 5 with vTF7-3 (t=0 h) and transfected (t=1 h) with pSTCF23F-425, or mock transfected (no plasmid DNA), by using lipofectin (Life Technologies, Ltd, Paisley, UK). The medium was refreshed at t=4.5 h, harvested at t=20 h, and centrifuged for 10 min at 3000 r.p.m. to clear it from cell debris. The mock supernatant and the supernatant containing the bispecific scFv were loaded onto a 20% sucrose cushion, centrifuged for 30 min at 13 000 r.p.m. to clear the supernatant from vTF7-3 virus, and stored at −20°C in aliquots until use. The supernatant from OST7-1 cells infected with vTF7-3, but not transfected with plasmid DNA, was used as a control supernatant. A single batch of scFv 23F-425 and control supernatant was used for all experiments described.
Determination of optimal amount of 23F-425 to be used in targeted infections
To determine the optimal amount of scFv 23F-425 to be used in targeted infections, FIPV (MOI 5) was preincubated for 1 h with various amounts of scFv 23F-425 and inoculated in a total volume of 500 μl on A431 cells in a 2 cm2 well. After 1 h at 37°C, the inoculum was replaced by regular culture medium and incubation of the cells was continued for 15 h. An immunostaining with serum directed against FIPV proteins was performed, after which the stained cells were counted and their numbers plotted against the amount of scFv 23F-425 used. The titration results revealed the optimal amount of bispecific antibody needed for maximal targeting efficiency under these standard conditions to be 200 μl scFv 23F-425. More antibody did not increase but, rather, decreased the level of infection, presumably because excess antibodies in the inoculum may bind to EGFR on cells thereby blocking their use by the virus. Cells inoculated with FIPV preincubated with mock control supernatant remained negative (data not shown).
Immunostaining
Cells were incubated with C428 anti-FIPV ascites fluid diluted 1:500, or K134 anti-MHV serum diluted 1:300, followed by goat anti-cat peroxidase (DAKO, Glostrup, Denmark) diluted 1:400, or swine anti-rabbit peroxidase (DAKO, Glostrup, Denmark) diluted 1:300, respectively. The cells were stained by AEC (Brunschwig, Amsterdam, The Netherlands) according to the manufacturer's protocol, and analyzed by light microscopy.
Antibody blocking experiments
To determine whether scFv 23F-425 interacts with the EGFR, 2 × 105 HeLa cells per 2 cm2 well were incubated with or without 500 μl hybridoma supernatant containing MAb 425 for 30 min at 4°C in order to block the interaction of EGFR and scFv 23F-425. Next, the cells were inoculated for 15 h at 37°C with 1 × 105 PFU fMHV preincubated with 200 μl scFv 23F-425 for 1 h at 4°C in a total volume of 500 μl. Thereafter, the cells were fixed, permeabilized, and immunostained for the presence of fMHV. The number of infected cells was counted by using light microscopy.
The interaction of scFv 23F-425 and the spike protein was analyzed by incubating 1 × 105 PFU fMHV with or without 200 μl hybridoma supernatant containing anti-fMHV-S antibody 23F8.1. After 1 h of incubation at 4°C, 200 μl scFv 23F-425 was added for 1 h of incubation at 4°C in a total volume of 500 μl. Next, the infection mixes were inoculated on 2 × 105 HeLa cells per 2 cm2 well for 15 h at 37°C. The cells were fixed, permeabilized, and immunostained for the presence of fMHV. Again, the number of infected cells was determined by using light microscopy.
Analysis of scFv 23F-425-mediated fusion
An amount of 5 × 104 A431 cells per 0.32 cm2 well was inoculated at an MOI of 5 for 2 h with fMHV preincubated with 50 μl of scFv 23F-425 for 1 h at 4°C. The cells were washed three times with PBS and incubated further in the presence or absence of 50 μl scFv 23F-425. An immunostaining was performed 24 h after infection, and the number of nuclei per syncytium was determined by light microscopy.
Production of the fHR1 peptide
For the production of the fHR1 peptide corresponding to amino acids 1047–1156 of the FIPV spike protein (acc. no. VGIH79), a PCR fragment was prepared using as a template the sequence from cDNA clone B1, which contains the FIPV spike gene.41 The fHR1 peptide was expressed in Escherichia coli, purified, and quantified as described elsewhere.11, 42
Analysis of the fusion mechanisms of coronavirus infection
Sensitivity of the normal virus–cell fusion process to fHR1 was studied by inoculating feline FCWF-4 with fMHV at an MOI of 0.5 in the presence or absence of 0.5 μM fHR1 peptide for 8 h. The effect of fHR1 on the targeted fusion process was analyzed by preparing in parallel two inoculation mixtures by preincubating fMHV with scFv 23F-425 for 1 h at 4°C after which 0.5 μM fHR1 peptide was added to one mixture. Two cultures of human A431 cells were washed with PBS and inoculated at an MOI of 5 for 16 h at 37°C with the infection mixes. The cells were fixed, permeabilized, and stained for the presence of fMHV using anti-MHV rabbit antiserum K134. The number of infected cells was counted by using light microscopy.
The sensitivity of the cell–cell fusion process during normal infection of feline FCWF-4 cells was analyzed by inoculating cells with fMHV at an MOI of 0.5 for 1 h at 37°C, washing them three times with PBS, and incubating for 7 h in culture medium with or without 0.5 μM fHR1 peptide. For a similar analysis of cell–cell fusion after targeted infection, cultures of A431 cells were inoculated at an MOI of 5 with fMHV preincubated with scFv 23F-425 for 1 h at 4°C. Next, the cells were washed three times with PBS, and incubated for 22 h in culture medium with or without 0.5 μM fHR1 peptide. The cells were fixed, permeabilized, and immunostained for the presence of fMHV proteins. The number of nuclei per syncytium was counted under the light microscope.
Acknowledgements
We are grateful to Tsutomu Hohdatsu (Kitasato University, Towada, Aomori, Japan) for providing anti-fAPN antibody R-G-4, to Kathryn V Holmes (University of Colorado Health Sciences Center, Denver, USA) for supplying the fAPN expression vector pCR3-fAPN, to Ed Dubovi (Cornell University, Ithaca, USA) for providing the hybridoma cell line 23F8.1, and to Paul MP van Bergen en Henegouwen (Utrecht University, Utrecht, The Netherlands) for the gift of NIH 3T3-Her14 cells.
This work was supported by the Dutch Cancer Society (UU 2001-2430). Victor W van Beusechem is supported by a research fellowship of the Royal Netherlands Academy of Arts and Sciences (KNAW).
References
- 1.Ring CJ. Cytolytic viruses as potential anti-cancer agents. J Gen Virol. 2002;83:491–502. doi: 10.1099/0022-1317-83-3-491. [DOI] [PubMed] [Google Scholar]
- 2.Russell SJ. RNA viruses as virotherapy agents. Cancer Gene Ther. 2002;9:961–966. doi: 10.1038/sj.cgt.7700535. [DOI] [PubMed] [Google Scholar]
- 3.Holmes KV, Lai MM. Fields Virology. Philadelphia: Lippincott-Raven Publishers; 1996. The viruses and their replication; pp. 1075–1093. [Google Scholar]
- 4.Cavanagh D. The Coronaviridae. 1995. The coronavirus surface protein; pp. 73–103. [Google Scholar]
- 5.Vennema H, et al. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J Virol. 1990;64:339–346. doi: 10.1128/jvi.64.1.339-346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tresnan DB, Levis R, Holmes KV. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J Virol. 1996;70:8669–8674. doi: 10.1128/jvi.70.12.8669-8674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dveksler GS, et al. Several members of the mouse carcinoembryonic antigen-related glycoprotein family are functional receptors for the coronavirus mouse hepatitis virus-A59. J Virol. 1993;67:1–8. doi: 10.1128/jvi.67.1.1-8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taguchi F. The S2 subunit of the murine coronavirus spike protein is not involved in receptor binding. J Virol. 1995;69:7260–7263. doi: 10.1128/jvi.69.11.7260-7263.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sturman LS, Ricard CS, Holmes KV. Conformational change of the coronavirus peplomer glycoprotein at pH 8.0 and 37 degrees C correlates with virus aggregation and virus-induced cell fusion. J Virol. 1990;64:3042–3050. doi: 10.1128/jvi.64.6.3042-3050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoo DW, Parker MD, Babiuk LA. The S2 subunit of the spike glycoprotein of bovine coronavirus mediates membrane fusion in insect cells. Virology. 1991;180:395–399. doi: 10.1016/0042-6822(91)90045-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benbacer L, et al. Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J Virol. 1997;71:734–737. doi: 10.1128/jvi.71.1.734-737.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wentworth DE, Holmes KV. Molecular determinants of species specificity in the coronavirus receptor aminopeptidase N (CD13): influence of N-linked glycosylation. J Virol. 2001;75:9741–9752. doi: 10.1128/JVI.75.20.9741-9752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Compton SR, et al. Coronavirus species specificity: murine coronavirus binds to a mouse-specific epitope on its carcinoembryonic antigen-related receptor glycoprotein. J Virol. 1992;66:7420–7428. doi: 10.1128/jvi.66.12.7420-7428.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo L, et al. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J Virol. 2000;74:1393–1406. doi: 10.1128/JVI.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haijema BJ, Volders H, Rottier PJ. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J Virol. 2003;77:4528–4538. doi: 10.1128/JVI.77.8.4528-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolibaba KS, Druker BJ. Protein tyrosine kinases and cancer. Biochim Biophys Acta. 1997;1333:F217–248. doi: 10.1016/s0304-419x(97)00022-x. [DOI] [PubMed] [Google Scholar]
- 18.Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciardiello F, Tortora G. A novel approach in the treatment of cancer: targeting the epidermal growth factor receptor. Clin Cancer Res. 2001;7:2958–2970. [PubMed] [Google Scholar]
- 20.Grill J, et al. The organotypic multicellular spheroid is a relevant three-dimensional model to study adenovirus replication and penetration in human tumors in vitro. Mol Ther. 2002;6:609–614. [PubMed] [Google Scholar]
- 21.Enger PO, et al. Adeno-associated viral vectors penetrate human solid tumor tissue in vivo more effectively than adenoviral vectors. Hum Gene Ther. 2002;10:1115–1125. doi: 10.1089/104303402753812511. [DOI] [PubMed] [Google Scholar]
- 22.Schalkwijk CG, et al. Maximal epidermal growth-factor-induced cytosolic phospholipase A2 activation in vivo requires phosphorylation followed by an increased intracellular calcium concentration. Biochem J. 1996;313:91–96. doi: 10.1042/bj3130091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckert DM, Kim PS. Mechanisms of viral membrnae fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 24.Rao PV, Kumari S, Gallagher TM. Identification of a contiguous 6-residue determinant in the MHV receptor that controls the level of virion binding to cells. Virology. 1997;229:336–348. doi: 10.1006/viro.1997.8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu X, Zhang X. Potent systemic antitumor activity from an oncolytic herpes simplex virus of syncytial phenotype. Cancer Res. 2002;62:2306–2312. [PubMed] [Google Scholar]
- 26.Peng KW, et al. Oncolytic measles viruses displaying a single-chain antibody against CD38, a myeloma cell marker. Blood. 2003;101:2557–2562. doi: 10.1182/blood-2002-07-2195. [DOI] [PubMed] [Google Scholar]
- 27.Anderson BD, Nakamura T, Russell SJ, Peng KW. High CD46 receptor density determines preferential killing of tumor cells by oncolytic measles virus. Cancer Res. 2004;64:4919–4926. doi: 10.1158/0008-5472.CAN-04-0884. [DOI] [PubMed] [Google Scholar]
- 28.van Beusechem VW, et al. Conditionally replicative adenovirus expressing a targeting adapter molecule exhibits enhanced oncolytic potency on CAR-deficient tumors. Gene Therapy. 2003;10:1982–1991. doi: 10.1038/sj.gt.3302103. [DOI] [PubMed] [Google Scholar]
- 29.de Haan CA, et al. Coronaviruses as vectors: position dependence of foreign gene expression. J Virol. 2003;77:11312–11323. doi: 10.1128/JVI.77.21.11312-11323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Haan CA, et al. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haijema BJ, Volders H, Rottier PJ. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J Virol. 2004;78:3863–3871. doi: 10.1128/JVI.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Haan CA, et al. Coronaviruses maintain viability despite dramatic rearrangements of the strictly conserved genome organization. J Virol. 2002;76:12491–12502. doi: 10.1128/JVI.76.24.12491-12502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bucheit AD, et al. An oncolytic measles virus engineered to enter cells through the CD20 antigen. Mol Ther. 2003;7:62–72. doi: 10.1016/S1525-0016(02)00033-3. [DOI] [PubMed] [Google Scholar]
- 34.Evermann JF, et al. Characterization of a feline infectious peritonitis virus isolate. Vet Pathol. 1981;18:256–265. doi: 10.1177/030098588101800214. [DOI] [PubMed] [Google Scholar]
- 35.Elroy-Stein O, Moss B. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc Natl Acad Sci USA. 1990;87:6743–6747. doi: 10.1073/pnas.87.17.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corapi WV, Olsen CW, Scott FW. Monoclonal antibody analysis of neutralization and antibody-dependent enhancement of feline infectious peritonitis virus. J Virol. 1992;66:6695–6705. doi: 10.1128/jvi.66.11.6695-6705.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rottier PJ, Horzinek MC, van der Zeijst BA. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J Virol. 1981;40:350–357. doi: 10.1128/jvi.40.2.350-357.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hohdatsu T, et al. Differences in virus receptor for type I and type II feline infectious peritonitis virus. Arch Virol. 1998;143:839–850. doi: 10.1007/s007050050336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller KM, Arndt KM, Pluckthun A. A dimeric bispecific miniantibody combines two specificities with avidity. FEBS Lett. 1998;432:45–49. doi: 10.1016/S0014-5793(98)00829-1. [DOI] [PubMed] [Google Scholar]
- 40.Haisma HJ, et al. Targeting of adenoviral vectors through a bispecific single-chain antibody. Cancer Gene Ther. 2000;7:901–904. doi: 10.1038/sj.cgt.7700198. [DOI] [PubMed] [Google Scholar]
- 41.de Groot RJ, et al. Stably expressed FIPV peplomer protein induces cell fusion and elicits neutralizing antibodies in mice. Virology. 1989;171:493–502. doi: 10.1016/0042-6822(89)90619-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volkel D, Blankenfeldt W, Schomburg D. Large-scale production, purification and refolding of the full-length cellular prion protein from Syrian golden hamster in Escherichia coli using the glutathione S-transferase-fusion system. Eur J Biochem. 1998;251:462–471. doi: 10.1046/j.1432-1327.1998.2510462.x. [DOI] [PubMed] [Google Scholar]


