Abstract
Interferon (IFN) lambda (IFN-λ or type III IFN) gene polymorphisms were discovered in the year 2009 to have a strong association with spontaneous and treatment-induced clearance of hepatitis C virus (HCV) infection in human hosts. This landmark discovery also brought renewed interest in type III IFN biology. After more than half a decade since this discovery, we now have reports that show that genetic association of IFNL gene polymorphisms in humans is not limited only to HCV infections but extends beyond, to include varied diseases such as non-alcoholic fatty liver disease, allergy and several other viral diseases including that caused by the human immunodeficiency virus. Notably, all these conditions have strong involvement of host innate immune responses. After the discovery of a deletion polymorphism that leads to the expression of a functional IFN-λ4 as the prime ‘functional’ variant, the relevance of other polymorphisms regulating the expression of IFN-λ3 is in doubt. Herein, I seek to critically address these issues and review the current literature to provide a framework to help further understanding of IFN-λ biology.
Supplementary information
The online version of this article (doi:10.1038/gene.2016.24) contains supplementary material, which is available to authorized users.
Subject terms: Immunology, Genetics
Introduction
In the year 2003, two different groups reported on the presence of three novel genes closely placed to each other, on human chromosome 19 that coded for interferons (IFNs) with potent antiviral properties.1, 2 These genes due to their relatedness to the interleukin 10 (IL-10) family were initially christened IL-28A, IL-28B and IL-29, and subsequently changed to IFNL2, IFNL3 and IFNL1.3 The genes encode IFNλ-2, IFNL-λ3 and IFNL-λ1, respectively; together with the newly discovered IFNL-λ4 (or IFNL4) they constitute the type III IFNs or the lambda IFNs (IFNL-λs). IFNL-λ1, 2 and 3 activate antiviral responses through the JAK–STAT (janus kinase–signal transducer and activator of transcription) pathway by utilizing a distinct receptor complex made of a heterodimer formed between IFN-λR1 and IL-10R2.2, 3 Subsequent studies showed that unlike the receptors that bind to type I IFNs, the IFN-λ receptors were expressed on selective cell types mainly of epithelial origin, hepatocytes and some immune cells.3, 4, 5, 6 The discovery of IFN-λs was seminal in the sense that it showed the presence of an alternate system to the well-known type I IFNs (IFN-α and IFN-β) that the different nucleated cells in the body, especially the ones on epithelial surfaces, can utilize to combat viral infections. Later studies in mice have shown that type III IFNs form a strong barrier at the host–environment interface, which encompasses large regions of epithelial lining to the respiratory, gastrointestinal and urogenital tracts of mammals.7, 8, 9, 10
A major boost in the area of IFN-λ research came after another discovery in the year 2009. Three independent groups conducted genome-wide association studies (GWASs) involving treatment response to chronic hepatitis C virus (HCV) infections, in three different geographical regions of the world, and reported that single-nucleotide polymorphisms (SNPs) in the IFNL locus (Figure 1), had strong association with treatment-induced HCV clearance irrespective of ethnicity and geographical location of the hosts.11, 12, 13 The search for a ‘causative’ or a ‘functional’ SNP at the IFNL locus that could give a biological explanation for the HCV-GWAS results was taken up rigorously by several groups, but none seem to have given a better explanation to the HCV-IFN-λ ‘puzzle’ than the group from the National Institutes of Health, USA, that discovered the presence of another IFNL upstream to IFNL3, named as IFNL4 (refs 14, 15; Figure 1; in Figure 1b, the alleles for the respective SNPs are shown as beneficial (B) or non-beneficial (NB) with respect to the studies on HCV (reviewed in ref. 15; the major allele for each of the SNPs depicted in Figure 1b has been shown to be the beneficial one in HCV infections). Functional IFN-λ4 is expressed only in a subset of individuals, due to a frameshift-causing deletion polymorphism (IFNL4-ΔG; rs368234815) in the first exon of IFNL4 (Figure 1).14 The presence of the alternate allele (IFNL4-TT) renders IFNL4 a pseudogene and this allele is seen in ~50% of the European population and in most of the east Asian population, but IFNL4 is a functional gene (IFNL4-ΔG allele) in majority (~95%) of the African population,16 suggesting that human evolution has played an active role in elimination of a functional IFN-λ4 in the human species.17 In fact, the pseudogene shows strong positive selection in human evolution, whereas the functional gene is conserved in other mammalian species except in mice and rats where the gene is completely absent.17
Figure 1.

Gene structure at the IFNL locus. (a) The SNPs that will be discussed in the text are shown. Arrows indicate the direction of the open reading frames of genes. UTR, untranslated region. (b) The linkage disequilibrium (LD; r2) values and minor allele frequency (MAF) are shown for the SNPs depicted in a. The alleles for the respective SNPs are shown as beneficial (B) or non-beneficial (NB) with respect to the studies on HCV (reviewed in ref. 15). The minor allele (Mi) is the non-beneficial allele (that leads to expression of a functional IFN-λ4 and also lowers expression of IFN-λ3) and the major allele (Ma) is the beneficial allele (that does not produce a functional IFN-λ4 and is associated with higher levels of expression of IFN-λ3) for all SNPs, again with respect to HCV studies.15 The information on LD values has been obtained from 1000 Genomes Project reference panel (http://www.1000genomes.org), October 2014 release; some LD (r2) values for YRI (marked with *) were obtained from ref. 14. The MAF values were obtained from dbSNP (National Center for Biotechnology Information). In Asian population, MAFs of rs12979860, rs8099917 and rs4803217 are from JPT population; and MAFs of rs368234815, rs8103142 and rs28416813 are from CHB-JPT populations. YRI, Yoruba in Ibadan, Nigeria; CEU, residents with ancestry from northern and western Europe; CHB/JPT, Han Chinese in Beijing, China/Japanese in Tokyo, Japan; —, MAF information not available for the TA repeat polymorphism rs59702201.
In this article, I review the literature on genetic association studies that have shown the involvement of the HCV-GWAS SNPs in non-HCV disorders that involve both viral diseases and some non-infectious conditions. I also update the progress on transcriptional studies of the IFNL4 gene and examine whether the functional IFN-λ4-generating SNP is sufficient to explain the molecular mechanism of causality in the diseases it is associated with, and whether the other IFNL locus SNPs (mainly the ones regulating IFN-λ3 expression) may have any functional roles to play in the observed phenotypes.
Genetic association of IFNL locus polymorphisms is not limited to HCV infections: innate immunity is the key
IFNL-λs and innate immunity against viruses
Innate and adaptive immunity are the two indispensable arms of the mammalian immune system. Although we had a clearer understanding of the principles of functioning of the adaptive immunity arm, a lack of advanced molecular techniques and incomplete understanding of molecular mechanisms made us remain unaware of the intricacies of functioning of the innate immunity arm, for a long time.18 With the advent of superior molecular biology techniques and the discovery of the pathogen-associated molecular pattern (PAMP) or pattern recognition receptors,19 we now have better understanding of how nucleated cells can differentially recognize different classes of pathogens and propagate signals to their surroundings, in the process raising the immediate alarm in the host.19 Large strides were made in the area of molecular recognition of viral PAMPs and signal transduction that leads to raising of antiviral states within virus-infected cells.10, 20 The epithelial cells, being at the interface between the host and the environment in the respiratory, gastrointestinal and the urogenital tract, are not only prone to a variety of viral infections but are strategically located to respond and propagate alarm signals to the underlying immune cells (Figure 2). However, the primary function of the epithelium is to provide a physical barrier between the underlying lamina propria and the lumen of the cavity or the exterior. Even though they can sense and respond to PAMPs and damage-associated molecular patterns,21 the epithelial cells are not professional immune cells and due to their high level of differentiation, may lack the plasticity required to send out amplified and prolonged signals to the lamina propria. Therefore, a crucial link still remained missing about how an adaptive immune response is shaped within distantly located lymph nodes that have obligatory ‘immune-rich environments’, by taking cues from signals generated by viral infections at the epithelium (Figure 2).
Figure 2.
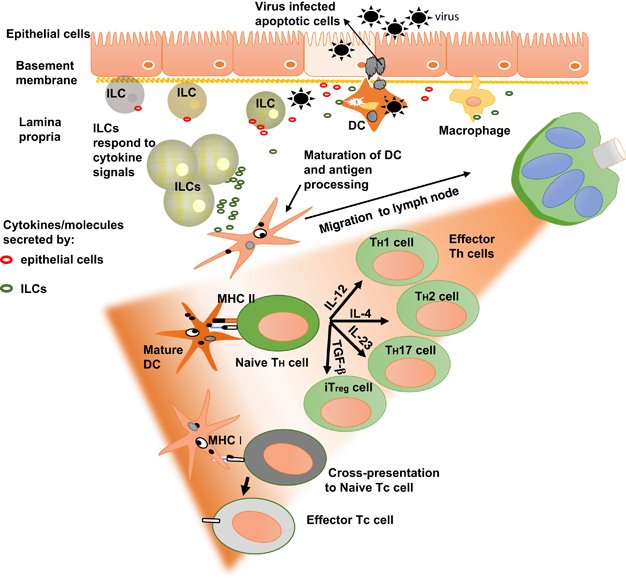
Immune responses to viral infections of the epithelia. Resident macrophages and dendritic cells (DCs) form the first line of cellular resistance to invading viruses. The newly discovered innate lymphoid cells (ILCs)22 may have important roles due to their ability to respond to signaling molecules/alarmins secreted by the epithelial cells. ILCs diversify and respond by secreting large amounts of effector cytokines and chemokines locally. These effector molecules can potentially lead to the polarization of DCs and macrophages, and therefore may be critical in shaping the adaptive immunity mediated by T-helper (Th) and T-cytotoxic (Tc) cells that develop in the nearby lymphatic tissue (green lobed structure). MHC, major histocompatability complex.
The discovery of the new class of effector immune cells called the innate lymphoid cells (ILCs) may seem to have provided the answer to this puzzle (Figure 2). ILCs are derivatives of common lymphoid precursors along with T and B cells.22 These cells are stationed near the epithelial surfaces in larger numbers and respond to signals from the surrounding cells by secreting cytokines and chemokines in large quantities, thereby acting as signal amplifiers in both health and disease.23 The ILCs are considered the innate immunity counterparts to the various Th (T helper) cell (CD4+ (cluster of differentiation)) subsets of the adaptive immune system (Figure 2; ref. 24). For example, ILC2s respond to IL-33 generated from influenza virus-infected epithelial cells by secreting IL-5 and IL-13, both known inducers of Th2 immunity.25 The natural killer cells, now classified as ILC1 cells, are considered the Tc (T cytotoxic) cell (CD8+) counterparts.24 Even though most studies so far on ILCs have been on mice, ILCs have also been characterized in a variety of human tissues26 and are deregulated in many human diseases.27 With emerging roles of type III IFNs at the epithelium,8, 10, 28 ILCs and IFN-λs now seem to be the major players in innate immunity at barrier surfaces.10, 29 How ILCs respond and interact with the epithelial cells during viral infections and how they cross-talk with other immune cells at the barrier surfaces in shaping and maintaining the optimal Th responses, will form the next exciting wave in innate immunity research.
Similarly, research on the production, regulation and functions of IFN-λs in viral infections has also been exciting in the last decade. Human IFN-λs are secreted (except IFN-λ4 that is poorly secreted30) in response to the detection of viral RNA intermediates from the cytoplasm of epithelial cells via the toll-like receptor and retinoic acid inducible gene I-like receptor) pathways.4, 7, 31 IFN-λs are also known to be secreted by macrophages, plasmacytoid dendritic cells, monocyte-derived dendritic cells and hepatocytes.4, 6, 32, 33, 34 The IFN-λR1 receptor is expressed on limited cell types including epithelial cells, hepatocytes, B cells and monocytes.5, 35 There is currently no information on whether the newly discovered ILCs secrete any of the IFN-λs and whether they express IFN-λR1. IFN-λs act in a paracrine and/or autocrine manner to raise an antiviral state in the infected and to-be-infected cells by reprogramming the target cell gene expression patterns.28, 36 The IFNL SNPs would have functional roles if they can affect the diversification of innate and adaptive immune cell subsets. Diversification of ILC subsets will lead to the polarization of dendritic cells and macrophages and will eventually influence the Th1/Th2 balance by favoring either a Th1 or a Th2 response (Figure 3).37 Although there is no evidence for this belief, it is known that the IFNL SNPs do affect the expression of IFN-λ3 (ref. 15) and that they give rise to a new IFN (IFN-λ4).14 IFN-λ1, 2 and 3 are known to deflect the Th1/Th2 balance to a Th1 predominant mode in vitro and also in vivo in humans and mice38, 39 (reviewed in ref. 37). No such studies are reported for IFN-λ4. One hypothesis is that different levels of IFN-λ3 expression dictated by the underlying genetic polymorphisms are responsible for eliciting a Th1 or a Th2 response.37 The role played by IFN-λ4 in this process is only speculative at this stage (Figure 3). How IFN-λs interact with the newly discovered ILCs is also unknown. A recent report showed that rotavirus infection in mice is controlled by IL-22 produced by ILC3s for which the presence of an intact IFN-λ signaling pathway is required.40 An even more important question in humans will be on what role does the IFNL SNPs, and therefore IFN-λ4, play in the diversification and functioning of ILCs.
Figure 3.
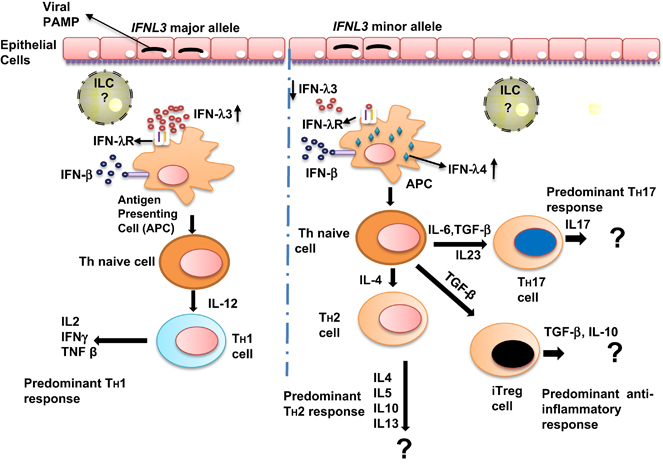
IFNL SNPs influence Th1/Th2 responses by affecting expression of IFN-λ3 and IFN-λ4. The model is based on the hypothesis proposed by Egli et al.37 Although IFN-λ3 is known to upregulate Th1 responses and simultaneously downregulate Th2 responses,86 the role played by IFN-λ4 is unknown. As the SNPs that can potentially affect IFN-λ3 expression (rs59702201, rs28416813 and rs4803217; reviewed in ref. 15) are in strong LD with rs368234815 that generates a functional IFN-λ4, lower levels of IFN-λ3 expression (caused by minor/non-beneficial alleles) will be accompanied by expression of a biologically active IFN-λ4, and therefore should not favor a predominant Th1 response. However, experimental proof is lacking for the role of IFN-λ4 in affecting the Th1/Th2 balance (denoted as ?). The role of the newly discovered innate lymphoid cells (ILCs)22, 23, 24 may be critical in the pathway. IL, interleukin; PAMP, pathogen-associated molecular patterns; TGF, transforming growth factor. The APCs in a viral infection are predominantly dendritic cells (DCs).
IFN-λ4 apart from being antiviral to HCV14 is also known to inhibit other flaviviruses such as dengue virus, yellow fever virus41 and human corona viruses.30 Some of the other IFN-λs are known to be induced by several human pathogens including M. tuberculosis,42 human papilloma virus,43 influenza virus44 and human metapneumovirus (also induces IFN-λ4),45 and have clear antiviral activities against some but not other viruses in mice (reviewed in ref. 46). In a recent finding, murine IFN-λ3 but not IFN-α/IFN-β was responsible for protecting mice against norovirus persistence in mice colon, strikingly, even in the absence of T and B lymphocytes.9 Similar results were seen in reovirus infections of mice colon.8 In conclusion, IFN-λs are potent antiviral molecules and their cross-talk with innate immune cells can potentially orchestrate innate immunity against several viruses and they may be particularly important at the barrier surfaces. IFN-λs also modulate adaptive immunity by affecting Th1/Th2 balance, tilting it to a Th1-favoring response that is required for clearing viral infections. The function of the newly discovered IFN-λ4 in these immune processes remains to be determined.
Association of IFNL locus polymorphisms with human diseases
In cognizance of the potential that IFN-λs may hold in innate immunity, researchers across the world have got intrigued with the IFNL SNPs and have started to test them in candidate gene case–control studies in both infectious and non-infectious diseases. So far, association has been reported with non-alcoholic fatty liver disease (NAFLD), allergy and infections with several viruses.
Cytomegalovirus
Cytomegalovirus (CMV) is a herpes virus that infects plasmacytoid dendritic cells and epithelial cells in humans causing chronic infections and is especially a problem in immunosuppresed individuals. Egli et al.47 tested the association of the IFNL SNP rs8099917 with CMV replication in a small number of solid organ transplant patients (n=38) who were seronegative for CMV (but received an organ transplant from a CMV-seropositive donor) and who had stopped receiving antiviral prophylaxis. They found that the minor allele at rs8099917 (G) was associated with decreased CMV replication (P=0.036) in a dominant model of inheritance. Further, their in vitro studies provided evidence for a beneficial effect of the minor allele against CMV replication.47 A study in CMV-seropositive kidney transplant patients48 also found that the minor allele (T) at rs12979860 had a dominant beneficial effect against CMV replication. Although in another study in allogenic stem cell transplant recipients,49 the minor allele T at rs12979860 protected recipients against CMV infection in a recessive model of inheritance.
Two more studies have also reported on the association of IFNL SNPs with CMV.50, 51 The first report by Bibert et al.50 tested for the occurrence of CMV retinitis among those human immunodeficiency virus (HIV)-infected patients who were at risk of developing CMV retinitis due to low CD4 counts and CMV seropositivity. They found that carriers of two copies of the minor allele (ΔG/ΔG), increased the risk of getting CMV retinitis (P=0.007) in multivariate regression models. In the second report by Manuel et al.,51 solid organ transplant patients were tested for the association of rs368234815 with cumulative incidence of CMV replication. The results showed that the minor allele homozygotes (ΔG/ΔG), only in the pre-emptive antiviral therapy group but not among those receiving antiviral prophylaxis, had higher incidence of CMV replication. These results are indeed very interesting and sound, but the paradoxical findings of the five groups in terms of the model of inheritance of IFNL SNPs raise some questions. Although two groups showed that their results fit best with a recessive model of inheritance of IFNL SNPs50, 51 in affecting CMV replication/retinitis wherein minor homozygosity was non-beneficial, the other three studies show an opposite trend where the minor allele had a beneficial effect against CMV replication in both patients and cell culture experiments, involving either dominant47, 48 or recessive models of inheritance.49 In stark contrast, a dominant model of inheritance (of the non-beneficial IFNL SNP minor allele) has consistently given the best explanation on the observed phenotypes in association studies with both spontaneous clearance and IFN-based treatment response in chronic HCV infections.11, 12, 13, 15 This discrepancy within the CMV studies may be partly due to statistical fluctuations owing to low sample sizes,47, 50 and high heterogeneity among patient groups, end points and antiviral regimens that are followed in the different studies. The discrepancy needs be resolved by well-designed replication studies to gain a proper understanding of the role of IFNL SNPs in CMV replication and disease.
Human T-lymphotrophic virus
Human T-lymphotrophic virus (HTLV)-1 is an ancient retrovirus causing chronic infections in humans and is associated with adult T-cell leukemia/lymphoma. It is also associated with inflammatory disorders such as HTLV-1-associated myelopathy/tropical spastic papaparesis (HAM/TSP), HTLV-associated arthropathy and several other related disorders including rheumatoid arthritis. A Spanish study reported for the first time that an IFNL SNP (rs12979860) was associated with HAM/TSP in a small number of patients (n=41) who had HTLV-1 proviral DNA in their blood cells.52 They showed that the presence of the minor allele made 12/41 patients to have a sixfold higher risk (P=0.03) of having symptoms of HAM/TSP compared with asymptomatic carriers (n=29) in a dominant model of inheritance. However, proviral DNA load was a confounder in this association; covariate analysis suggested that both the SNP and proviral DNA load were linked in their association with HAM/TSP. The minor allele-carrying genotypes (CT and TT) were indeed having higher proviral DNA in their blood cells compared with the major homozygotes (P=0.01). The major drawback of this study seems to be the low sample number; although the strength of the study was that there was significant effect of the minor allele on proviral DNA copy number, a functional association that could be directly linked with the antiviral role of IFN-λ3. It is known that the IFNL SNP minor alleles are associated with decreased expression of IFN-λ3 (reviewed in ref. 15, although an opposite effect is evident in chronic HCV infections, where the minor allele carriers have low baseline virus levels11). In a later study from Brazil,53 both rs8099917 and rs12979860 were tested in a cohort consisting of 229 HTLV-positive subjects (93 HAM/TSP patient and 136 asymptomatic carriers). The minor allele G of rs8099917 was significantly associated with HAM/TSP in both univariate and multivariate analysis with a recessive model of inheritance (P<0.001). In this study, the proviral DNA load as a covariate did not seem to interfere with the association of rs8099917 with HAM/TSP unlike the previous Spanish study.52 With respect to rs12979860, the minor allele T was associated with HAM/TSP only as a heterozygote in univariate analysis (P=0.01) and weakly in multivariate analysis (P=0.06).
However, a series of studies have also reported conflicting results to the above two reports on the association of IFNL SNPs with HTLV-1-associated diseases. First, Sanabani et al.54 show clearly a lack of association of IFNL SNP rs12979860 with HAM/TSP and/or adult T-cell leukemia/lymphoma. This Brazilian study had more number of samples (n=112) than the Spanish study of Trevino et al. (n=41).52 They also did not find any correlation of proviral DNA load with rs12979860 genotypes. Another report from Brazil with a sample size of 79 also reflected similar findings, where they found no association of rs12979860 with HAM/TSP and proviral DNA load.55 This study also compared 300 healthy controls with 79 HTLV-1-positive subjects and found no association with rs12979860 and HTLV-1 infection. Yet another recent study from Brazil analyzed the genotypes at rs8099917, rs12979860 and rs8103142 in 300 healthy controls and 96 HTLV-1-infected individuals, and found no association with HTLV-associated arthropathy and any of the three SNPs when tested individually.56 When they carried out haplotype analysis, they found some association (P=0.01) with HTLV-1 infection and one of the seven haplotypes (CCT) involving the three SNPs (rs8099917, rs12979860 and rs8103142); with HTLV-associated arthropathy and another haplotype (TTG; P=0.05). They also found an association of the three SNPs individually with levels of some cytokines (such as IFN-γ) and proviral DNA load (P<0.05).56 However, no multiple testing corrections seem to have been carried out in their analysis, thus severely undermining the results.56 Further, a Japanese study also failed to see any association with adult T-cell leukemia/lymphoma and rs8099917 and also with HTLV-1 and HCV mono- or co-infections.57 Last, a study from France on 95 HTLV-1-positive subjects of Afro-Carribean lineage compared the distribution of genotypes of rs12979860 and the IFN-λ4-generating SNP rs368234815, and found no association with HAM/TSP.58 In summary, the results so far are not entirely convincing on a true association of the IFNL SNPs with HTLV infection-related diseases. Further, in the two reports52, 53 that did see an association, the models of inheritance used to fit the phenotype data do not agree with each other, raising doubts on the underlying functionality of the observed genetic associations. Therefore, more functional characterization of the observed association may be needed to rule out false positivity.
Hepatitis B virus
If at all there is another human disease where IFNL SNPs were expected to have associations as strong as that of HCV infections, it was the case of hepatitis B virus (HBV). This expectation is because both viruses are hepatotropic and cause chronic infections; both diseases can be effectively treated using IFN-α even though they differ in their PAMP ligands recognized by innate immune receptors.59, 60 Mixed results have been obtained about the role of IFNL SNPs in both spontaneous clearance and IFN-α-induced clearance of HBV and the literature until the year 2013–2014 has been reviewed elsewhere.61, 62, 63 More studies have also been conducted since then (results from 10 of them are summarized in Supplementary Table 1), but have largely failed to resolve the conflict.
Two studies have also reported on a lack of association of IFNL SNPs on spontaneous as well as IFN-induced clearance of hepatitis D virus, a co-infecting satellite virus that requires HBV for replication and assembly.64, 65 Although consistent results were obtained across numerous studies with HCV–IFNL SNP association mainly because they had only virological end point phenotypes15 that correlated well with serological and biochemical parameters of the infection, several drawbacks in case of HBV studies may be responsible for the inconsistent results. Some of them are: presence of different HBV genotypes as mixed infections, with some genotypes (genotype D62) showing better association than others; variation in treatment regimens (IFN alone or with nucleotide analogs); end point phenotypes varying from serology to viral load, to biochemical parameters without a common quantitative parameter to assess the phenotype; prolonged clinical course of the disease with fluctuating virological, serological and biochemical markers; and prevalence rates of disease differing in different ethnicities/populations, to name a few. The reports that have shown an association of IFNL SNPs with HBV spontaneous or treatment-induced clearance are largely in agreement with the results from HCV studies in terms of the model of inheritance (Supplementary Table 1). Although the conflicting data so far on HBV-IFNL SNP association do lead to doubts about its true positive nature, the data are also not fully supportive of the notion that the association may be false positive. In fact it has been argued that if properly assessed, the association between HBV disease progression and IFNL SNPs could have clinical value.62 But it appears that the effect of the IFNL SNPs on HBV persistence and/or progression within the human host is highly variable; it may involve more complex interactions with other variables and genes than was observed in chronic HCV infections.
Human immunodeficiency virus
Another important viral disease that has been tested for the influence of IFNL SNPs is acquired immunodeficiency syndrome (AIDS) caused by HIV. Particularly, it was of interest to see how the IFNL SNP beneficial alleles are distributed in a unique group of HIV-infected individuals that can suppress the virus from replicating to high levels (defined as elite controllers/suppressors or natural viral suppressors) and defer the progression to AIDS (called as long-term non-progressors) without any antiretroviral therapy; and another unique group that remains HIV-seronegative despite being at high risk for infection due to intravenous drug use (highly exposed seronegative or exposed seronegative)). More interestingly, the natural viral suppressor patients have also been known to efficiently clear HCV in HCV-HIV co-infections, compared with controls in both African Americans66 and Caucasians.67
A few reports have come up in this area, some showing no correlation of HIV infection/disease progression to the IFNL SNPs, whereas others see clear association.68, 69, 70, 71, 72, 73, 74 One of the earlier reports tested the association of rs12979860 in 291 high-risk seronegative and 1221 HIV-positive subjects comprising both white and black subjects in the USA.68 No association was evident for either HIV positivity or for disease progression within the infected subjects. In another study reported by Rallon et al.,69 the association of rs12979860 was tested in two different cohorts. The first comprised of 30 long-term non-progressors and 38 typical progressors to AIDS; the second included 29 exposed seronegative and 29 HIV-positive partners. Thus, the study tested both HIV progression and protection, although in a small number of patients. No significant difference in distribution of genotypes between cases and controls was evident in both cohorts, although the beneficial CC genotype was more frequent in the exposed seronegative patients compared with HIV-positive patients (62% vs 45%), suggesting that there may be a protective role for the CC genotype against HIV that was not detected in the study, likely due to inadequate power. Another study from the USA tested whether the beneficial CC genotype at rs12979860 was over-represented in 25 African-American elite controllers/suppressors compared with HIV-infected patients with high viral loads, and found no statistically significant difference.70 Confirming this report, Sajadi et al.71 also found that CC genotype (for rs12979860) was not significantly over-represented in 48 natural viral suppressors of African-American origin compared with HIV-positive (n=124) and -negative controls (n=173).
Three reports published subsequently have seen association with rs12979860 and rs368234815, and spontaneous control of HIV and/or AIDS progression.72, 73, 74 Interestingly, all three studies were carried out on whites/Caucasians, whereas the reports that failed to see an association described above were mostly carried out on African Americans (except that by Rallon et al69 that used white subjects) or mixed group of blacks and whites.68 First, Machmach et al.72 found an association of rs12979860 with spontaneous HIV control when tested on 53 white natural viral suppressors and 389 matched non-controllers at P=0.02 after correcting for occurrence of HLA-B57 (human leukocyte antigen) protective alleles (which also have independent association with HIV and HCV spontaneous clearance) and gender, in multivariate analysis. Second, Machmach et al.73 confirmed and extended their results in another report, where they found the association of rs368234815 with AIDS progression. Last, a recent study carried out on a well-characterized Spanish white cohort of HCV-seropositive men exposed to HIV infection through shared needles shows a clear association of IFN-λ4-generating rs368234815 with HIV positivity.74 The study had 213 men who were HIV seropositive and 188 were highly exposed seronegative. They found that the protective TT/TT genotype was over-represented in the highly exposed seronegative group (0.49 vs 0.41; P=0.006). Further, this association had no interaction with the HIV-protective CCR5 (C–C chemokine receptor type 5) deletion (P=0.7). Interestingly, all three studies that found an association show data that the non-beneficial minor alleles are following a recessive model of inheritance,72, 73, 74 suggesting a common underlying functionality in the observed genetic association between the three studies. This is however different from the dominant model of inheritance seen in HCV studies,11, 12, 13, 15 suggesting that the two viruses may have different interactions with IFN-λ-driven immune responses. However, unlike in the case of CMV studies discussed above,47, 48, 49, 50, 51 all three HIV studies72, 73, 74 show that the minor alleles are non-beneficial, similar to the observations in chronic HCV infections.15 In summary, it is evident that IFNL SNPs do associate with HIV replication and disease progression, even though in an ethnicity-specific manner. It appears that the beneficial alleles of IFNL SNPs have protective roles in whites/Caucasians. In African Americans, more studies with the functional IFN-λ4-generating SNP rs368234815 rather than rs12979860 will reveal any true association. This is especially true as the two SNPs are not in strong linkage disequilibrium (LD) in this population (Figure 1b).
Herpes simplex virus
Herpes simplex virus resides inside the human host latently in sensory neurons, but gets reactivated due to the altered host immunity and replicates efficiently in epithelial cells causing genital or oral lesions (the latter is referred to as ‘cold sores’). An association of rs12979860 with the recurrence and severity of herpes simplex virus-1-induced ‘cold sores’ was reported from a study on a small number of individuals (n=57).75 A recent report carried out on a large number of individuals (n=2192 for genital herpes; n=1511 for oral herpes) clearly found no association of rs368234815 with recurrence of genital or oral herpes episodes.76 Among other phenotypes, the latter report ruled out an effect of the SNP on ‘frequency of recurrence’ of oral herpes, it however did not rule out an effect of the SNP on severity of infection.
Non-infectious diseases and miscellaneous conditions
IFNL SNPs have also been tried as candidate gene SNPs in some non-infectious inflammatory diseases. The association of IFNL SNPs with NAFLD was earlier reported by Petta et al.77 This study with 160 subjects identified an independent association with the CC genotype at rs12979860 and the severity of lobular inflammation (CC genotype positively correlated with severity of inflammation) in a cohort of patients with NAFLD (P=<0.001). The study was conducted taking lead from previous reports of increased hepatic necroinflammation in HCV patients with the CC genotype at rs12979860.78 Another report showed no such association in 195 Caucasian biopsy-confirmed NAFLD patients.79 However, all the patients in this cohort were obese (with body mass index >30), whereas in the previous study only 40% were obese. After the latter report, Petta et al.80 revisited their data and indeed found an association with the IFNL SNP only with their non-obese patients (n=94; P=0<0.001) but not with the obese patients (n=66; P=0.13). These initial doubts seem to have been resolved with a recent report by Eslam et al.81 who worked on a relatively large number of NAFLD patients (n=488) and confirmed the association of the CC genotype at rs12979860 with increased severity of liver inflammation and fibrosis (P=<0.0001). They also found a similar strong association with hepatic inflammation and fibrosis in separate cohorts of viral hepatitis C (n=3129) and B (n=555), strongly suggesting that even though the major allele genotype CC is beneficial against HCV and HBV, it is also responsible for excess inflammation of the liver in viral and non-viral hepatitis. All four studies77, 78, 79, 80, 81 report on dominant models of inheritance for the minor alleles; but unlike in the context of chronic HCV infections,11, 12, 13 the minor alleles in NAFLD are beneficial rather than being non-beneficial, as they are responsible for less severe hepatic inflammation.
Extending the potential role for IFNL SNPs in inflammatory diseases, there is also a recent report that showed association of rs12979860 and allergy in children aged <5 years.82 Although the study involved a small sample size (non-allergic, n=35; allergic cohort 1, n=35; food allergy cohort 2, n=30), a large effect size (odds ratio=4.56, P=0.004, for cohort 1) was seen wherein the non-beneficial T allele at rs12979860 was over-represented in the allergy group in a dominant model of inheritance. Another interesting finding in this study was that the effect of the IFNL SNP was more pronounced in females than males. This report suggests that the non-beneficial IFNL SNP minor allele-carrying genotypes may predispose children to an allergic phenotype by skewing the Th1/Th2 balance to a Th2 predominant one. This extrapolation is also based on the findings from a recent study on mice model of allergic asthma, which showed that IFN-λ2 was able to rescue mice from allergy by suppressing Th2 cytokines.39 These conclusions would also suggest a role for IFNL SNPs in immune response to respiratory viral infections, which account for disease exacerbations in conditions such as asthma.83 A study carried out on infants with respiratory syncytial virus-induced bronchiolitis shows no association of r12979860 and rs8099917 with viral load or other clinical features.84 Interestingly, the non-beneficial TT genotype of rs12979860 was associated with early age of hospitalization in the infants (P=0.005). More studies are awaited on the association of IFNL SNPs with allergy and allergy-related diseases. In the autoimmune disorder multiple sclerosis, rs8099917 and rs12979860 did not show any association with IFN-β treatment.85
As evidence for a potential role of IFN-λs and IFNL SNPs in adaptive immune responses, a study has linked IFNL SNPs with development of effective vaccine response against human influenza virus in immunosuppressed individuals.86 The study shows that minor allele (G)-carrying individuals at rs8099917 show better vaccine responses (odds ratio=1.99; P=0.038) in a dominant model of inheritance, and that T cells from minor allele-carrying individuals produce more IL-4 (a Th2 cytokine). This study also showed that recombinant IFN-λ3 increased Th1 responses from human peripheral blood mononuclear cells while inhibiting Th2 responses (Th2 cytokines are needed for effective seroconversion).86
Transcription studies on the IFNL4 gene
Studies have accumulated evidence on the different transcription factors (TFs) that bind and drive transcription from all four IFNL genes. Roles for several virus-inducible TFs such as NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells), IFN regulatory factor-3 and IFN regulatory factor-7 have been defined for IFNL1, 2 and 3 genes, and have been reviewed elsewhere.28, 87, 88 We recently showed that apart from these three TFs, specificity protein 1 (a GC-rich DNA-binding TF) also has a role in driving transcription from the IFNL4 promoter in A549 cells89 (Figure 4a). A CpG island of ~1.5 kb is located upstream of the IFNL3 gene and overlapping the IFNL4 gene, and also includes two of the most important IFNL SNPs rs12979860 and rs368234815. Epigenetic regulation of the transcriptional activity at the IFNL locus is likely to involve this CpG island and needs further exploration.90, 91
Figure 4.

Transcription studies on IFNL4 gene expression. (a) A schematic of the IFNL4 promoter showing the different transcription factors (TFs) that were recently discovered89 that are important in driving the expression of IFNL4 gene. The gene structure of IFNL4 is shown as having five exons with the positions of the two SNPs also indicated. Tall arrow indicates translation start site. The transparent triangle (green) shows the CpG island (position: chr19:39737690-39739288; band: 19q13.2; CpG count-137, UCSC genome browser, https://genome.ucsc.edu) that covers most of the exonic region and a part of the 5′-UTR (untranslated region) over a length of ~1.5 kb. IRF, IFN regulatory factor; P, phosphorylated TFs; TSS, transcription start site. (b) Schematic representation of primer-binding sites used in several recent studies to measure the expression of IFNL4. A schematic of the IFNL4 gene locus in chromosome 19q13 is depicted. Position of SNP rs368234815, which creates functional IFN-λ4, is shown. Black standing arrow represents the translational start site (ATG). Primer-binding sites of six different studies (eight primer pairs in total) are shown (a, refs 45, 89; b, ref. 94; c, ref. 41; d–g, ref. 93; h, refs 14, 96; i, ref. 92). Expected amplicon size of each primer pair is shown as dotted line between two small black arrows. The primer sequences are listed in Supplementary Table 2. TGA, translational stop site.
Even though IFNL4 messenger RNA (mRNA) was shown to be highly expressed from stimulated primary hepatocytes in the original report that described the discovery of IFNL4,14 subsequent studies have not seen robust expression levels of IFNL4 gene in human samples. For example, Amanzada and others found IFNL4 transcripts expressed at 4.3–5.6-fold lower levels than IFNL2/3 mRNA in liver biopsies of HCV-infected patients.92 In another report, IFNL4 mRNA was detectable in only in 7/23 and 8/23 patient-derived peripheral blood mononuclear cells that were stimulated or not with IFN-poly(I:C), respectively.93 All 23 patients carried at least one copy of the functional IFN-λ4-generating allele, ΔG. In the report by Liang and colleagues also, IFNL4 transcripts were seen in only 33/70 liver biopsies from HCV-infected patients.94 Furthermore, Lu et al.41 found that only 2% of the transcripts generated from poly(I:C)-stimulated primary hepatocytes derived from two heterozygous (TT/ΔG at rs368234815) individuals, represented functional IFN-λ4 transcripts that originated from the ΔG allele. Similarly, they also found no expression of the full-length functional IFN-λ4 mRNA transcripts from poly(I:C)-stimulated A549 cells. It is possible that the qPCR assays utilized in these studies may be suffering from technical difficulties in dealing with several different mRNA isoforms of IFNL4 gene.14 All the four studies described above have used primer sets where at least one of the primers binds to the exon/intron/exon–intron junctions (Figure 4b). Using a primer set that uniquely binds to the 3′-untranslated region of IFNL4 mRNA, IFN-λ4 induction was seen at levels similar to that of IFN-λ2/3, upon human metapneumovirus infection of A549 cells by Banos-Lara et al.45 We used the same primer set and found similar high expression levels upon poly(I:C)/HCV RNA transfection in A549 cells.89 Two other studies have reported no such problems in amplifying IFNL4 transcripts from liver biopsies.95, 96 Although Honda et al.96 used the allele-specific Taqman assay described earlier,14 Konishi et al. do not provide the primer sequence information that they used in their Taqman assays.95 The inconsistency in the above reports in detection of IFNL4 transcripts (either the full-length functional IFN-λ4 isoform or the other isoforms) suggests that RNA-sequencing may be the optimal approach to measure IFNL4 transcripts. Furthermore, the pre-mRNA splicing mechanism that leads to the expression of different IFN-λ4 isoforms14 under different stimulation and cell-type conditions needs to be examined.
The relevance of other ‘IFN-λ3 functional SNPs’ after the discovery of functional IFN-λ4-generating SNP
Till date, three functional SNPs have been identified that regulate IFNL3 gene transcription/translation (referred to as ‘IFN-λ3 functional SNPs’ in this section). Although rs28416813 is a SNP present at ~37 nucleotides upstream of the start codon of IFNL3 and is known to affect downstream gene expression by differentially binding to NF-κB,97 rs4803217 was identified in the 3′-untranslated region region of IFNL3 (Figure 1a) that affects stability of the mRNA by interfering with AU-rich element decay (AMD).98 Both these SNPs are in high LD with each other and with rs12979860 and were predicted to be two of the four potential causal SNPs from among the HCV-GWAS hits.15, 99 A recent study defines another mechanism by which rs4803217 affects the stability of RNA by remodeling its secondary structure.100 Furthermore, a third functional SNP is the TA repeat polymorphism rs59702201, originally reported by the Mizokami group.101 This SNP located within the proximal promoter affects transcription of IFNL3, depending on the number of repeats present.101 Recent reports have confirmed the significance of this SNP in association studies involving chronic HCV infections.102, 103, 104 Apart from several reports that showed genotype-dependent differences in expression levels of IFN-λ3 (reviewed in ref. 15) recent reports also have confirmed this finding in ex vivo and in vivo conditions.94, 105 These results suggest that IFN-λ3 expression levels dictated by the alleles present at the three functional SNPs may have a role in the observed phenotype in health and disease.
However, the discovery of the IFN-λ4-generating SNP (rs368234815, referred to as ‘IFN-λ4 functional SNP’ in this section) has overshadowed the importance of the ‘IFN-λ3 functional SNPs’, as most of the new reports have chosen to test for the ‘IFN-λ4 functional SNP’ rather than rs12979860, which is the tag-SNP for the ‘IFN-λ3 functional SNPs’.15 Another SNP within the coding region of IFNL4 that leads to a non-synonymous mutation in the functional IFN-λ4 protein is thought to be a better marker of association along with rs368234815, in chronic HCV infections.106 So the question arises: is the IFN-λ4-generating SNP the sole functional SNP or the other SNPs regulating the expression of IFN-λ3 also have independent roles? The answer to this question will be difficult to obtain under in vivo conditions due to high LD between these SNPs in most populations that precludes any attempts to assess their independent effects (Figure 1b). A recent report compared the strength of association of rs4803217 and rs368234815 in IFN treatment response in HCV-infected African Americans, who have the lowest reported LD values among all ethnicities between rs368234815 and rs12979860 SNPs (Figure 1b).107 They found that rs368234815 was more strongly associated with the outcome than rs4803217.107 In another independent report, Lu et al. applied multivariate regression to test the independence of rs368234815 from rs4803217 in predicting response to IFN treatment in HCV patients of African-American decent.100 They found that correcting for the effect of s368234815 on rs4803217 abolished the latter SNP’s association with treatment response, whereas correcting for the effect of the latter SNP reduced the strength of association of the former SNP (from P=0.004 to 0.065).100 This could have resulted due to the low sample size (n=169); nevertheless, more results are awaited to conclude that IFN-λ4-generating SNP is the sole functional SNP and that the observations giving credence to the functionality of the other ‘IFN-λ3 functional SNPs may only be artifacts.
Studies so far point to the functional IFN-λ4-generating SNP rs368234815 as the prime causal variant at the human IFNL locus. Therefore, a detailed investigation is needed on the function of IFN-λ4 as both an antiviral cytokine in different viral diseases and as a potential player in diversification and maintenance of innate and adaptive immune cell subsets. In case of IFN treatment response in chronic HCV infections, even though the IFN-λ4 -generating SNP can explain a large amount of variance in the observed phenotypes,107 questions still remain on its role in spontaneous clearance of HCV. It is known that expression of functional IFN-λ4 is associated with high levels of IFN-stimulated gene (ISG) expression in non-responders who further fail to upregulate their ISG expression upon IFN-α treatment.15 However, such an elegant explanation is lacking in the case of spontaneous clearance of HCV (Figure 5). Although the majority of studies on spontaneous HCV clearance are with rs12979860 and not rs368234815, the fact that these two SNPs are in high LD in most populations14 allows us to extrapolate the results. So, how does the expression of a functional IFN-λ4 in patients who get an acute HCV infection will lead them to become chronically infected? One explanation could be that similar to the IFN treatment non-responders group, the subjects expressing functional IFN-λ4 also have higher baseline levels of ISGs, which do not get further upregulated upon acute HCV infection. Having a higher basal innate immune response may have helped those populations with higher frequency of the functional IFN-λ4-generating allele in dealing with prevalent viral infections. Higher baseline levels of ISGs in people who can express a functional IFN-λ4 may be offering protection against excess inflammation of the liver in hepatitis of non-viral origin such as NAFLD.78, 79, 80, 81 However, as is evident from studies on spontaneous and treatment-induced clearance of HCV, such a pre-emptive state may become non-beneficial in dealing with hepatitis of viral origin. Further, there may be other conditions that we do not yet know in which expression of a functional IFN-λ4 may be beneficial. Therefore, an epidemiological investigation to assess basal ISG expression levels, in humans both in health and disease, and their correlation with expression of IFN-λ4, is needed. Further, studies are also needed to examine why hepatic IFN-λ3 levels are lower in the beneficial allele/genotype-carrying individuals during chronic HCV infection and how this phenomenon is reversed once treatment is initiated (Figure 5c; ref. 94).
Figure 5.

IFN-λ3 and IFN-λ4 expression, and spontaneous and IFN-based therapy-induced clearance of HCV infections in humans. The schematic is a summary and interpretation of data available so far from different studies (reviewed in ref. 15) dealing with HCV infections. All patients who get infected by HCV can be divided in to the three groups shown in the schematic. (a) Patients who spontaneously clear HCV are known to have higher levels of IFN-λ3 protein in their serum compared with patients who carry on with the infection108 and it is likely that their IFN-stimulated gene (ISG) levels are also influenced by the IFN-λ3 levels, thus helping to clear the virus infection. Once patients get chronically infected with HCV, they undergo IFN-α-ribavirin treatment for 24–48 weeks. (b, c) Those who do not respond to this therapy have high frequency of the functional IFN-λ4-generating allele ΔG at rs368234815 (ref. 14) and therefore are expected to express IFN-λ4.94 A recent study has documented that type III IFNs including IFN-λ2/3 and IFN-λ4 expression levels correlate with ISG expression in the liver of patients undergoing anti-HCV therapy.94 The IFN-λ2/3 and IFN-λ4 levels and their associated ISG levels are higher in the liver of patients who will eventually not respond to the therapy94 compared with those who will respond, suggesting that an unknown effect inhibits the expression of IFN-λ3 in the latter group of patients (shown as an ellipse in c). It seems that IFN-α-ribavirin treatment induces the expression of IFN-λ3 once treatment begins (depicted as a circle in c) preferably in those patients who will eventually respond to the treatment94 (These patients have lower frequency of the functional IFN-λ4-generating alleles14 and hence are shown without IFN-λ4 expression (c)). This induction is further dependent on whether the patients carry beneficial alleles at rs12979860. The patients who have the beneficial alleles show higher increase in their hepatic IFN-λ3 levels than those who do not, once treatment is initiated.94 A previous report also had seen similar changes in serum IFN-λ3 levels.109 The ISG levels are shown in correlation with IFN-λ3 expression.
Although IFN-λ1, 2 and 3 are known to associate with a Th1 response,38, 39, 86 no information is available in this regard for IFN-λ4. If the hypothesis that higher levels of IFN-λ3 expression (due to presence of major alleles) will lead to a Th1 predominant response is to be believed, then by extrapolation, IFN-λ4 should promote Th2 responses (Figure 3; as ‘IFN-λ3 functional SNPs’ and ‘IFN-λ4 functional SNP’ are in strong LD and the minor allele will give rise to IFN-λ4). This explanation is difficult to reconcile with the observations that IFN-λ4, except for being a poorly secreted IFN, requires the same dimeric receptor for signaling and can stimulate not only similar set of ISGs but also at similar levels when compared with IFN-λ3.14, 30, 36 Besides, both IFN-λ4 and IFN-λ3 have similar antiviral potencies in vitro.14, 41 Therefore, expression of IFN-λ4 and its associated ISGs in a subset of individuals carrying minor alleles is less likely to favor a completely opposite phenotype to that seen in those carrying major alleles that induce higher levels of IFN-λ3 expression.
Conclusions
Human IFNL SNPs came to the limelight after genetic studies showed their relevance in chronic HCV infections in the year 2009. Since then, case–control studies in humans have been reported involving IFNL SNPs and several other viral diseases, NAFLD, allergy and even in vaccine responses. Although it appears that the associations are well replicated (such as in NAFLD and AIDS) and strong in some diseases (such as in pediatric allergy), conflicting reports weaken the association in some (such as those involving HTLV and HBV), whereas further studies are needed in others (such as those involving CMV and herpes simplex virus) to clear discrepancies. The IFNL-λs may have critical roles in shaping innate and adaptive immunity in general and in viral infections in particular. However, unlike their effect in HCV infections, the IFNL SNPs may involve interactions with variables other than just standard end point phenotypes in many of the reported diseases/conditions. Therefore, future studies should aim at dissecting these interactions to arrive at meaningful results. Well-designed replication studies and functional studies to strengthen the genetic association results are required to arrive at firm conclusions. Importantly, IFN-λ4 has emerged as the key causal link to the genetic associations, but many questions remain on its functional role in the diversification and shaping of innate and adaptive immune responses in viral infections and other inflammatory conditions.
Supplementary information
Acknowledgements
The author dedicates this article to Professor Partha P. Majumder, Director, National Institute of Biomedical Genomics, Kalyani, India, on a successful journey in nurturing human genetics research in India. The author was (partially) supported as a visiting scientist by the Healthy Ageing Research Centre (HARC) project (REGPOT-2012-2013-1, 7FP). The author thanks Professor Marek L. Kowalski, Chair, Department of Immunology, Rheumatology and Allergy and Director, HARC, Medical University of Lodz, Poland, for his support and critical comments on the manuscript. The author also thanks Anand Bhushan and Sumona Ghosh for help in preparing the Figures and Supplementary Table 2. The funding for this work was provided by NIBMG, India and HARC, Poland.
Competing interests
The author declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Genes and Immunity website
References
- 1.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2002;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 2.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE. IL-28, IL-29 and their class II cytokine receptor IL-28 R. Nat Immunol. 2002;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 3.Witte K, Witte E, Sabat R, Wolk K. IL-28A, IL-28B, and IL-29: promising cytokines with type I interferon-like properties. Cytokine Growth Factor Rev. 2010;21:237–251. doi: 10.1016/j.cytogfr.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Coccia EM, Severa M, Giacomini E, E, Monneron D, Remoli ME, Julkunen I. Viral infection and Toll like receptor agonists induce a differential expression of type I andλ interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol. 2004;34:796–805. doi: 10.1002/eji.200324610. [DOI] [PubMed] [Google Scholar]
- 5.Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-λ) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mihm S, Frese M, Meier V, Wietzke-Braun P, Scharf J-G, Bartenschlager R. Interferon type I gene expression in chronic hepatitis C. Lab Invest. 2004;84:1148–1159. doi: 10.1038/labinvest.3700135. [DOI] [PubMed] [Google Scholar]
- 7.Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
- 8.Mahlakoiv T, Hernandez P, Gronke K, Diefenbach A, Staeheli P. Leukocyte-derived IFN-α/β and epithelial IFN-λ constitute a compartmentalized mucosal defense system that restricts enteric virus infections. PLoS Pathog. 2015;11:e1004782. doi: 10.1371/journal.ppat.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M. Interferon-λ cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wack A, Terczyńska-Dyla E, Hartmann R. Guarding the frontiers: the biology of type III interferons. Nat Immunol. 2015;16:802–809. doi: 10.1038/ni.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 12.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 14.Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H. Avariant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinnaswamy S. Genetic variants at the IFNL3 locus and their association with hepatitis C virus infections reveal novel insights into host-virus interactions. J Interferon Cytokine Res. 2014;34:479–497. doi: 10.1089/jir.2013.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Brien TR, Prokunina-Olsson L, Donnelly RP. IFN-λ4: the paradoxical new member of the interferon lambda family. J Interferon Cytokine Res. 2014;34:829–838. doi: 10.1089/jir.2013.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Key FM, Peter B, Dennis MY, Huerta-Sánchez E, Tang W, Prokunina-Olsson L. Selection on a variant associated with improved viral clearance drives local, adaptive pseudogenization of interferon lambda 4 (IFNL4) PLoS Genet. 2014;10:e1004681. doi: 10.1371/journal.pgen.1004681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beutler B. Innate immunity: an overview. Mol Immunol. 2004;40:845–859. doi: 10.1016/j.molimm.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 22.Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med. 2015;21:698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 24.Annunziato F, Romagnani C, Romagnani S. The 3 major types of innate and adaptive cell-mediated effector immunity. J Allergy Clin Immunol. 2015;135:626–635. doi: 10.1016/j.jaci.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Jackson DJ, Makrinioti H, Rana BM, Shamji BW, Trujillo-Torralbo MB, Footitt J. IL-33-dependent type 2 inflammation during rhinovirus-induced asthma exacerbations in vivo. Am J Respir Crit Care Med. 2014;190:1373–1382. doi: 10.1164/rccm.201406-1039OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazenberg MD, Spits H. Human innate lymphoid cells. Blood. 2014;124:700–709. doi: 10.1182/blood-2013-11-427781. [DOI] [PubMed] [Google Scholar]
- 27.Hepworth MR, Sonnenberg GF. Regulation of the adaptive immune system by innate lymphoid cells. Curr Opin Immunol. 2014;27:75–82. doi: 10.1016/j.coi.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazear HM, Nice TJ, Diamond MS. Interferon-λ: Immune functions at barrier surfaces and beyond. Immunity. 2015;43:15–28. doi: 10.1016/j.immuni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wojno EDT, Artis D. Innate lymphoid cells: balancing immunity, inflammation and tissue repair in the intestine. Cell Host Microbe. 2012;12:445–457. doi: 10.1016/j.chom.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamming OJ, Terczyńska-Dyla E, Vieyres G, Dijkman R, Jørgensen SE, Akhtar H. Interferon lambda 4 signals via the IFNλ receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J. 2013;32:3055–3065. doi: 10.1038/emboj.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siren J, Pirhonen J, Jullkunen I, Matikainen S. IFN-alpha regulates TLR-dependent gene expression of IFN-alpha, IFN-beta, IL-28 and IL-29. J Immunol. 2005;174:1932–1937. doi: 10.4049/jimmunol.174.4.1932. [DOI] [PubMed] [Google Scholar]
- 33.Yin Z, Dai J, Deng J, Sheikh F, Natalia M, Shih T. Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. J Immunol. 2012;189:2735–2745. doi: 10.4049/jimmunol.1102038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hillyer P, Mane VP, Schramm LM, Puig M, Verthelyi D, Chen A. Expression profiles of human interferon-alpha and interferon-lambda subtypes are ligand- and cell-dependent. Immunol Cell Biol. 2012;90:774–783. doi: 10.1038/icb.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Witte K, Gruetz G, Volk HD, Looman AC, Asadullah K, Sterry W. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes Immun. 2009;10:702–714. doi: 10.1038/gene.2009.72. [DOI] [PubMed] [Google Scholar]
- 36.Lauber C, Vieyres G, Terczyńska-Dyla E, Anggakusuma, Dijkman R, Gad HH. Transcriptome analysis reveals a classical interferon signature induced by IFNλ4 in human primary cells. Genes Immun. 2015;16:414–421. doi: 10.1038/gene.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egli A, Santer DM, O'Shea D, Tyrell DL, Houghton M. The impact of the interferon-lambda family on the innate and adaptive immune response to viral infections. Emerg Microbes Infect. 2014;3:e51. doi: 10.1038/emi.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jordan WJ, Eskdale J, Srinivas S, Pekarek V, Kelner D, Rodia M. Human interferon lambda-1 (IFN-lambda1/IL-29) modulates the Th1/Th2 response. Genes Immun. 2007;8:254–261. doi: 10.1038/sj.gene.6364382. [DOI] [PubMed] [Google Scholar]
- 39.Koltsida O, Hausding M, Stavropoulos A, Koch S, Tzelepis G, Ubel C. IL-28 A (IFN-λ2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO Mol Med. 2011;3:348–361. doi: 10.1002/emmm.201100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernandez PP, Mahlakoiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F. Interferon-l and interleukin 22act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol. 2015;16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu YF, Goldstein DB, Urban TJ, Bradrick SS. Interferon-λ4 is a cell-autonomous type III interferon associated with pre-treatment hepatitis C virus burden. Virology. 2015;476:334–340. doi: 10.1016/j.virol.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Travar M, Vucic M, Petkovic M. Interferon lambda-2 levels in sputum of patients with pulmonary Mycobacterium tuberculosis infection. Scand J Immunol. 2014;80:43–49. doi: 10.1111/sji.12178. [DOI] [PubMed] [Google Scholar]
- 43.Cannella F, Scagnolari C, Selvaggi C, Stentella P, Recine N, Antonelli G. Interferon lambda 1 expression in cervical cells differs between low-risk and high-risk human papillomavirus-positive women. Med Microbiol Immunol. 2014;203:177–184. doi: 10.1007/s00430-014-0330-9. [DOI] [PubMed] [Google Scholar]
- 44.Killip MJ, Fodor E, Randall RE. Influenza virus activation of the interferon system. Virus Res. 2015;209:11–22. doi: 10.1016/j.virusres.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baños-Lara Mdel R, Harvey L, Mendoza A, Simms D, Chouljenko VN, Wakamatsu N. Impact and regulation of lambda interferon response in human metapneumovirus infection. J Virol. 2015;89:730–742. doi: 10.1128/JVI.02897-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hermant P, Michiels T. Interferon-λ in the context of viral infections: production, response and therapeutic implications. J Innate Immun. 2014;6:563–574. doi: 10.1159/000360084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Egli A, Levin A, Santer DM, Joyce M, O'Shea D, Thomas BS. Immunomodulatory function of interleukin 28B during primary infection with cytomegalovirus. J Infect Dis. 2014;210:717–727. doi: 10.1093/infdis/jiu144. [DOI] [PubMed] [Google Scholar]
- 48.Fernández-Ruiz M, Corrales I, Arias M, Campistol JM, Giménez E, Crespo J. Association between individual and combined SNPs in genes related to innate immunity and incidence of CMV infection in seropositive kidney transplant patients. Am J Transplant. 2015;15:1323–1335. doi: 10.1111/ajt.13107. [DOI] [PubMed] [Google Scholar]
- 49.Bravo D, Solano C, Giménez E, Remigia MJ, Corrales I, Amat P. Effect of the IL28B Rs12979860 C/T polymorphism on the incidence and features of active cytomegalovirus infection in allogeneic stem cell transplant patients. J Med Virol. 2014;86:838–844. doi: 10.1002/jmv.23865. [DOI] [PubMed] [Google Scholar]
- 50.Bibert S, Wojtowicz A, Taffé P, Manuel O, Bernasconi E, Furrer H. The IFNL3/4 ΔG variant increases susceptibility to cytomegalovirus retinitis among HIV-infected patients. AIDS. 2014;28:1885–1889. doi: 10.1097/QAD.0000000000000379. [DOI] [PubMed] [Google Scholar]
- 51.Manuel O, Wójtowicz A, Bibert S, Mueller NJ, van Delden C, Hirsch HH. Influence of IFNL3/4 polymorphisms on the incidence of cytomegalovirus infection after solid-organ transplantation. J Infect Dis. 2015;211:906–914. doi: 10.1093/infdis/jiu557. [DOI] [PubMed] [Google Scholar]
- 52.Treviño A, Lopez M, Vispo E, Aquilera A, Ramos JM, Benito R. Development of tropical spastic paraparesis in human T-lymphotropic virus type 1 carriers is influenced by interleukin 28B gene polymorphisms. Clin Infect Dis. 2012;55:e1–e4. doi: 10.1093/cid/cis343. [DOI] [PubMed] [Google Scholar]
- 53.Assone T, de Souza FV, Gaester KO, Fonseca LA, Luiz Odo C, Malta F. IL28B gene polymorphism SNP rs8099917 genotype GG is associated with HTLV-1 carriers. PLoS Negl Trop Dis. 2014;8:e3199. doi: 10.1371/journal.pntd.0003199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanabani SS, Nukui Y, Pereira J, de Costa AC, de Oliveira AC, Pessôa R. Lack of evidence to support the association of a single IL28B genotype SNP rs12979860 with the HTLV-1 clinical outcomes and proviral load. BMC Infect Dis. 2012;12:374. doi: 10.1186/1471-2334-12-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vallinoto AC, Santana BB, Sá KS, Ferreira TC, Sousa RC, Azevedo VN. HTLV-1- associated myelopathy/ tropical spastic paraparesis is not associated with SNP rs12979860 of the IL-28B gene. Mediators Inflamm. 2015;2015:804167. doi: 10.1155/2015/804167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Sá KS, Santana BB, de Souza Ferreira TC, Sousa RC, Caldas CA, Azevedo VN. IL28B gene polymorphisms and Th1/Th2 cytokine levels might be associated with HTLV-associated arthropathy. Cytokine. 2016;77:79–87. doi: 10.1016/j.cyto.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Kamihira S, Usui T, Ichikawa T, Ono N, Morinaga Y, Mori S. Paradoxical expression of IL-28B mRNA in peripheral blood in human T-cell leukemia virus type-1 mono-infection and co-infection with hepatitis C virus. Virol J. 2012;9:40. doi: 10.1186/1743-422X-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jeannin S, Costa JM, Poveda JD, Belrose G, Lezin A, Cabie A. Absence of association of IFNL3/IL28B rs 12979860 and IFNL4 ss 469415590 polymorphisms with the neurological status of HTLV-1 Afro-Caribbean subjects in Martinique. Retrovirology. 2015;2(Suppl 1):P61. [Google Scholar]
- 59.Leong CR, Oshiumi H, Suzuki T, Matsumoto M, Seya T. Nucleic acid sensors involved in the recognition of HBV in the liver-specific in vivo transfection mouse models-Pattern recognition receptors and sensors for HBV. Med Sci. 2015;3:16–24. doi: 10.3390/medsci3020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horner Jr, SM, Gale M. Regulation of hepatic innate immunity by hepatitis C virus. Nat Med. 2013;19:879–888. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galmozzi E, Viganó M, Lampertico P. Systematic review with meta-analysis: do interferon lambda 3 polymorphisms predict the outcome of interferon-therapy in hepatitis B infection? Aliment. Pharmacol Ther. 2014;39:569–578. doi: 10.1111/apt.12631. [DOI] [PubMed] [Google Scholar]
- 62.Jilg N, Chung RT. One more piece in the interleukin 28B gene puzzle? The case of hepatitis B. Hepatology. 2013;57:870–872. doi: 10.1002/hep.26026. [DOI] [PubMed] [Google Scholar]
- 63.Takahashi T. Interleukin 28B genetic polymorphism and hepatitis B virus infection. World J Gastroenterol. 2014;20:12026–12030. doi: 10.3748/wjg.v20.i34.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Visco-Comandini U, Lapa D, Taibi C, Angeletti C, Capobianchi MR, Garbuglia AR. No impact of interleukin-28B polymorphisms on spontaneous or drug induced hepatitis delta virus clearance. Dig Liver Dis. 2014;46:348–352. doi: 10.1016/j.dld.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 65.Yilmaz E, Baran B, Soyer OM, Onel M, Onel D, Ormeci AC. Effects of polymorphisms in interferon λ3 (interleukin 28B) on sustained virologic response to therapy in patients with chronic hepatitis D virus infection. Clin Gastroenterol Hepatol. 2014;12:1753–1758. doi: 10.1016/j.cgh.2014.01.043. [DOI] [PubMed] [Google Scholar]
- 66.Sajadi MM, Shakeri N, Talwani R, Redfield RR. Hepatitis C infection in HIV-1 viral suppressors. AIDS. 2010;24:1689–1695. doi: 10.1097/QAD.0b013e32833a2a32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruiz-Mateos E, Machmach K, Romero-Sanchez MC, Ferrando-Martinez S, Viciana P, Del Val M. Hepatitis C virus replication in Caucasian HIV controllers. J Viral Hepat. 2011;18:e350–e357. doi: 10.1111/j.1365-2893.2010.01431.x. [DOI] [PubMed] [Google Scholar]
- 68.Martin MP, Qi Y, Goedert JJ, Hussain SK, Kirk GD, Hoots WK. IL28B polymorphism does not determine outcomes of hepatitis B virus or HIV infection. J Infect Dis. 2010;202:1749–1753. doi: 10.1086/657146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rallon NI, Restrepo C, Naggie S, Lopez M, Del Romero J, Goldstein D. Interleukin-28B gene polymorphisms do not influence the susceptibility to HIV-infection or CD4 cell decline. AIDS. 2011;25:269–271. doi: 10.1097/QAD.0b013e328341b84e. [DOI] [PubMed] [Google Scholar]
- 70.Salgado M, Kirk GD, Cox A, Rutebemberwa A, Huggins Y, Astemborski J. Protective interleukin-28B genotype affects hepatitis C virus clearance, but does not contribute to HIV-1 control in a cohort of African-American elite controllers/suppressors. AIDS. 2011;25:385–387. doi: 10.1097/QAD.0b013e328341b86a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sajadi MM, Shakeri N, Talwani R, Howell CD, Pakyz R, Redfield RR. IL28B genotype does not correlate with HIV control in African Americans. Clin Transl Sci. 2011;4:282–284. doi: 10.1111/j.1752-8062.2011.00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Machmach K, Abad-Molina C, Romero-Sánchez MC, Abad MA, Ferrando-Martínez S, Genebat M. IL28B single-nucleotide polymorphism rs12979860 is associated with spontaneous HIV control in white subjects. J Infect Dis. 2013;207:651–655. doi: 10.1093/infdis/jis717. [DOI] [PubMed] [Google Scholar]
- 73.Machmach K, Abad-Molina C, Romero-Sánchez MC, Dominquez-Molina B, Moyano M, Rodriquez MM. IFNL4 ss469415590 polymorphism is associated with unfavourable clinical and immunological status inHIV-infected individuals. Clin Microbiol Infect. 2015;21:e1–e4. doi: 10.1016/j.cmi.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 74.Real LM, Herrero R, Rivero-Juárez A, Camacho A, Macías J, Vic S. IFNL4 rs368234815 polymorphism is associated with innate resistance to HIV-1 infection. AIDS. 2015;29:1895–1897. doi: 10.1097/QAD.0000000000000773. [DOI] [PubMed] [Google Scholar]
- 75.Griffiths SJ, Koegl M, Boutell C, Zenner HL, Crump CM, Pica F. A systematic analysis of host factors reveals a Med23-interferon-λ regulatory axis against herpes simplex virus type 1 replication. PLoS Pathog. 2013;9:e1003514. doi: 10.1371/journal.ppat.1003514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lang Kuhs KA, Kuniholm MH, Pfeiffer RM, Chen S, Desai S, Edlin BR. Interferon lambda 4 genotype is not associated with recurrence of oral or genital herpes. PLoS One. 2015;10:e0138827. doi: 10.1371/journal.pone.0138827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petta S, Grimaudo S, Cammá C, Cabibi D, Di Marco V, Licata G. IL28B and PNPLA3 polymorphisms affect histological liver damage in patients with non-alcoholic fatty liver disease. J Hepatol. 2012;56:1356–1362. doi: 10.1016/j.jhep.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 78.Thompson AJ, Clark PJ, Zhu M, Zhu Q, Ge D, Sulkowski MS. Genome wide-association study identifies IL28B polymorphism to be associated with baseline Alt and hepatic necro-inflammatory activity in chronic hepatitis C patients enrolled in the ideal study. Hepatology. 2010;52:1220 A–1221 A. [Google Scholar]
- 79.Garrett ME, Abdelmalek MF, Ashley-Koch A, Hauser MA, Moylan CA, Pang H. IL28B rs12979860 is not associated with histologic features of NAFLD in a cohort of Caucasian north American patients. J Hepatol. 2013;58:402–403. doi: 10.1016/j.jhep.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Petta S, Craxi A. Reply to: "IL28B rs12979860 is not associated with histologic features of NAFLD in a cohort of Caucasian north American patients". J Hepatol. 2013;58:403–404. doi: 10.1016/j.jhep.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 81.Eslam M, Hashem AM, Leung R, Romero-Gomez M, Berg T, Dore GJ. Interferon-λ rs12979860 genotype and liver fibrosis in viral and non-viral chronic liver disease. Nat Commun. 2015;6:6422. doi: 10.1038/ncomms7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gaudieri S, Lucas M, Lucas A, McKinnon E, Albloushi H, Rauch A. Genetic variations in IL28B and allergic disease in children. PLoS One. 2012;7:e30607. doi: 10.1371/journal.pone.0030607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dhariwal J, Edwards MR, Johnston SL. Anti-viral agents: potential utility in exacerbations of asthma. Curr Opin Pharmacol. 2013;13:331–336. doi: 10.1016/j.coph.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scagnolari C, Midulla F, Riva E, Monteleone K, Solimini A, Bonci E. Evaluation of interleukin 28B single nucleotide polymorphisms in infants suffering from bronchiolitis. Virus Res. 2012;165:236–240. doi: 10.1016/j.virusres.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Malhotra S, Morcillo-Suárez C, Brassat D, Goertsches R, Lechner-Scott J, Urcelay E. IL28B polymorphisms are not associated with the response to interferon-β in multiple sclerosis. J Neuroimmunol. 2011;239:101–104. doi: 10.1016/j.jneuroim.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 86.Egli A, Santer DM, O'Shea D, Barakat K, Syedbasha M, Vollmer M. IL-28B is a regulator of B- and T- cell vaccine responses against influenza. PLoS Pathog. 2014;10:e1004556. doi: 10.1371/journal.ppat.1004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iversen MB, Paludan SR. Mechanisms of type III interferon expression. J Interferon Cytokine Res. 2010;30:573–578. doi: 10.1089/jir.2010.0063. [DOI] [PubMed] [Google Scholar]
- 88.Durbin RK, Kotenko SV, Durbin JE. Interferon induction and function at the mucosal surface. Immunol Rev. 2013;255:25–39. doi: 10.1111/imr.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chinnaswamy S, Bhushan A, Behera AK, Ghosh S, Rampurkar V, Chandra V. Roles for transcription factors Sp1, NF-ĸB, IRF3, and IRF7 in expression of the human IFNL4 gene. Viral Immunol. 2016;29:49–63. doi: 10.1089/vim.2015.0076. [DOI] [PubMed] [Google Scholar]
- 90.Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med. 2013;210:1109–1116. doi: 10.1084/jem.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smith KR, Suppiah V, O’Connor K, Berg T, Weltman M, Abate ML. Identification of improved IL28B SNPs and haplotypes for prediction of drug response inn treatment of hepatitis C using massively parallel sequencing in a cross-sectional European cohort. Genome Med. 2011;3:57. doi: 10.1186/gm273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Amanzada A, Kopp W, Spengler U, Ramadori G, Mihm S. Interferon-λ4 (IFNL4) transcript expression in human liver tissue samples. PLoS One. 2013;8:e84026. doi: 10.1371/journal.pone.0084026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Murakawa M, Asahina Y, Nakagawa M, Sakamoto N, Nitta S, Kusano-Kitazume A. Impaired induction of interleukin 28B and expression of interferon λ4 associated with nonresponse to interferon-based therapy in chronic hepatitis C. J Gastroenterol Hepatol. 2015;30:1075–1084. doi: 10.1111/jgh.12902. [DOI] [PubMed] [Google Scholar]
- 94.Noureddin M, Rotman Y, Zhang F, Park H, Rehermann B, Thomas E. Hepatic expression levels of interferons and interferon-stimulated genes in patients with chronic hepatitis C: a phenotype-genotype correlation study. Genes Immun. 2015;16:321–329. doi: 10.1038/gene.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Konishi H, Motomura T, Matsumoto Y, Harimoto N, Ikegami T, Yoshizumi T. Interferon-lambda4 genetic polymorphism is associated with the therapy response for hepatitis C virus recurrence after a living donor liver transplant. J Viral Hepat. 2014;21:397–404. doi: 10.1111/jvh.12154. [DOI] [PubMed] [Google Scholar]
- 96.Honda M, Shirasaki T, Shimakami T, Sakai A, Horii R, Arai K. Hepatic interferon-stimulated genes are differentially regulated in the liver of chronic hepatitis C patients with different interleukin-28B genotypes. Hepatology. 2014;59:828–838. doi: 10.1002/hep.26788. [DOI] [PubMed] [Google Scholar]
- 97.Chinnaswamy S, Chatterjee S, Boopathi R, Mukherjee S, Bhattacharjee S, Kundu SK. A single nucleotide polymorphism associated with hepatitis C virus infections located in the distal region of the IL28B promoter influences NF-ĸB mediated gene transcription. PLoS One. 2013;8:e75495. doi: 10.1371/journal.pone.0075495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McFarland AP, Horner SM, Jarret A, Joslyn RC, Bindewald E, Shapiro BA. The favourable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nat Immunol. 2014;15:72–79. doi: 10.1038/ni.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.du lulio J, Ciuffi A, Fitzmaurice K, Kelleher D, Rotger M, Fellay J. Estimating the net contribution of interleukin-28B variation to spontaneous hepatitis C virus clearance. Hepatology. 2011;53:1446–1454. doi: 10.1002/hep.24263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu YF, Mauger DM, Goldstein DB, Urban TJ, Weeks KM, Bradrick SS. IFNL3 mRNA structure is remodelled by a functional non-coding polymorphism associated with hepatitis C virus clearance. Sci Rep. 2015;5:16037. doi: 10.1038/srep16037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sugiyama M, Tanaka Y, Wakita T, Nakanishi M, Mizokami M. Genetic variation of the IL-28B promoter affecting gene expression. PLoS One. 2011;6:e26620. doi: 10.1371/journal.pone.0026620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thong VD, Wasitthankasem R, Tangkijvanich P, Vongpunsawad S, Poovorawan Y. Prevalence of thymine—adenine dinucleotide repeat, IL28B and IFNL4 in Thai population and correlation with spontaneous clearance and treatment outcome of hepatitis C infection. PLoS One. 2015;10:e0125400. doi: 10.1371/journal.pone.0125400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Masaki N, Sugiyama M, Shimada N, Tanaka Y, Nakamuta M, Izumi N. Pretreatment prediction of the outcome of response-guided peginterferon-α and ribavirin therapy for chronic hepatitis C. J Gastroenterol Hepatol. 2014;29:1996–2005. doi: 10.1111/jgh.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hiramine S, Sugiyama M, Furusyo N, Uto H, Ido A, Tsubouchi H. A thymine-adenine dinucleotide repeat polymorphism near IL28B is associated with spontaneous clearance of hepatitis C virus. J Gastroenterol. 2015;50:1069–1077. doi: 10.1007/s00535-015-1056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kurbanov F, Kim Y, Latanich R, Chaudhari P, El-Diwany R, Knabel M. IFNL3 genotype is associated with differential induction of IFNL3 in primary human hepatocytes. Antivir Ther. 2015;20:805–814. doi: 10.3851/IMP2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Terczyńska-Dyla E, Bibert S, Duong FH, Krol I, Jørgensen S, Collinet E. Reduced IFNλ4 activity is associated with improved HCV clearance and reduced expression of interferon-stimulated genes. Nat Commun. 2014;5:5699. doi: 10.1038/ncomms6699. [DOI] [PubMed] [Google Scholar]
- 107.O'Brien TR, Pfeiffer RM, Paquin A, Lang kuhs KA, Chen S, Bonkovsky HL. comparison of functional variants in IFNL4 and IFNL3 for association with HCV clearance. J Hepatol. 2015;63:1103–1110. doi: 10.1016/j.jhep.2015.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shi X, Pan Y, Wang M, Wang D, Li W, Jiang T. IL-28B genetic variation is associated with spontaneous clearance of hepatitis C virus, treatment response, serum IL-28B levels in Chinese population. PLoS One. 2012;7:e37054. doi: 10.1371/journal.pone.0037054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rallón NI, Soriano V, Naggie S, Restrepo C, Hutchison J, Vispo E. Impact of IL28B gene polymorphisms on interferon-λ3 plasma levels during pegylated interferon-α/ribavirin therapy for chronic hepatitis C in patients coinfected with HIV. J Antimicrob Chemother. 2012;67:1246–1249. doi: 10.1093/jac/dkr598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


