Abstract
The prevalence of adeno-associated virus (AAV) has been investigated in bat populations, but little is known about the biological properties of this virus. In this study, four full-length bat AAV capsid genes were isolated in China, with their amino acid sequences sharing 61% identity with those of AAV2 on average. These capsid genes could package AAV particles in combination with AAV2 rep and ITRs, albeit at a lower efficiency. Bat AAVs could only slightly infect mouse liver but could transduce mouse muscle to some extent after systemic administration with a higher muscle/liver ratio than that of primate AAVs. Bat AAV 10HB showed moderate muscle transduction, similar to that of AAV2, during direct intramuscular injection and, compared with other AAV serotypes, was also relatively efficient in resisting human antibody neutralization after intramuscular injection. Evolutionary analysis revealed a number of codons in bat AAV capsid genes subject to positive selection, with sites corresponding to V259 and N691 in 10HB capsids being localized on the surface of the AAV2 capsid. Mutagenesis studies indicated that the positive selection in bat AAV capsids is driven by their tropism evolution in host species. Taken together, the results of this study indicate that bat AAV 10HB vector has the possible applications for muscular gene therapy, especially in the presence of human AAV neutralizing antibodies.
Subject terms: Virology, Gene therapy, Genetic transduction, Adaptive immunity, Microbial genetics
Introduction
Adeno-associated virus (AAV) is a Dependovirus genus parvovirus that was originally identified as a contaminant in a simian adenovirus preparation [1]. AAV requires the assistance of other viruses, such as adenovirus and herpes simplex virus, for competent replication. In the absence of a helper virus, wild-type AAV2 can integrate its genome into the human chromosome 19. The AAV genome encodes at least three open-reading frames, with the rep gene encoding the replication proteins, the cap gene encoding the capsid proteins, and the recently identified assembly-activating protein (AAP) being related to virion assembly [2].
AAVs have been universally employed as gene therapy vectors because of their stable transduction of numerous tissues and relative scarcity of the immune response. Thirteen AAV serotypes and over 100 AAV variants have been discovered from non-human primate and human tissue samples [3, 4], with variable tissue tropism and immune profiles. Some representative AAV serotypes have been utilized in clinical trials for gene therapy of genetic diseases, such as AAV1 for lipoprotein lipase deficiency [5], AAV2 for Leber’s congenital amaurosis [6], AAV8 for hemophilia B [7], and AAV9 for spinal muscular atrophy [8].
Despite the universal application of AAVs in gene therapy, an important concern for the efficacy of AAV is the high prevalence of neutralizing antibodies within human populations. A prevalence of up to 59% was observed for neutralizing antibodies against AAV2 in an investigated French population, and even the AAV8 originally isolated from primates had a prevalence of 19% [9]. Furthermore, different AAV serotypes showed some common footprints for antibody binding, suggesting cross-reactivity among primate AAV serotypes and their neutralizing antibodies [10].
The discovery of novel AAV serotypes and directed evolution remain two important genetic approaches to enhance AAV in its capacity to evade preexisting human neutralizing antibodies. For instance, numerous AAV serotypes have been successively isolated from cattle, goats, and pigs [11–13]. The bovine and caprine AAVs showed potential to evade neutralization of human antibodies, while the porcine AAV demonstrated distinct tissue tropism from primate AAVs. These results suggest the potential application of nonprimate mammalian AAVs as gene therapy vectors to overcome the human immunological barrier. At the same time, some notable studies have been performed to evolve AAV for optimized immunological properties [14, 15], which further expanded the application of natural AAV serotypes.
Bats are a large reservoir for different viruses, including the notorious SARS coronavirus [16]. In our previous work, we sequenced the complete rep and cap sequences of a bat AAV Yunnan strain and investigated primarily the epidemiology of bat AAVs in several Chinese provinces [17]. However, the potential of bat AAV to be used as a gene therapy vector has not been thoroughly elucidated to date. In this study, a number of bat AAV capsid genes will be cloned and characterized with a focus on their tissue tropism and immunological properties, which will establish a foundation for their further application in gene therapy, especially in the presence of human AAV antibodies.
Materials and methods
Cell cultures
For this study, 293 (human embryo kidney cells), A549 (human lung carcinoma cells), Huh7 (human hepatoma cells), HT29 (human colon cancer cells), and U87 (human glioma cells) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, and 0.1 mg of streptomycin/ml. Cells were maintained at 37 °C in a 5% CO2 humidified atmosphere.
Cloning and sequence determination of bat AAV capsid genes
The bat total fecal samples were collected in 2007 and 2009 from Yunnan Province and in 2010 from Hubei Province in China. The bat intestinal tissues were collected from Yunnan in 2007. Viral nucleic acid was extracted from the fecal samples with a QIAamp RNA Mini kit (Qiagen) following the manufacturer’s instructions. These RNA samples have been used for epidemiological studies on RNA viruses. However, since bat genomic DNA was detected from them, they were also used for amplification of AAV capsid genes in this study. Viral DNA was also directly isolated from the intestinal samples using the DNeasy Blood and Tissue kit (Qiagen). A set of primers was selected from the bat AAV Yunnan strain genomic sequence (GenBank no. GU226971) published previously: Bt-CAP5′ (5′-CCCAAGCTTCGGGGGAATCTGACTCCGTGAACTTCGCCGAGA-3′) and Bt-CAP3′ (5′-GGAAGATCTCGCAGAGACCAAAGTTCAACTGAAACGA-3′). These primers were used to amplify the complete capsid gene from different bat AAV fecal samples. A 2.5-kb PCR product was obtained, treated successively with BglII, Klenow fragment, and HindIII and inserted into the UF1-AAV8 vector treated successively with NotI, Klenow fragment, and HindIII. The UF1-AAV8 vector contains the AAV2 rep gene and ITR sequences and was used for cloning shuffled AAV capsid genes [18]. This bat AAV capsid gene was designated 07YN considering its sampling from Yunan in 2007. To adapt the PCR to more bat DNA samples, a degenerate primer dCAP3′ was designed based on alignment of the 3′ untranslated region of mammalian AAV genomes (5′-GGAAGATCTACTGAMACGAAT(H/-)AMMCGGTTTATTG-3′, modified from Bello et al. [19]. By the use of the Bt-CAP5′/dCAP3′ primers, two additional bat AAV capsid gene sequences were amplified from bat fecal samples and cloned into the UF1-AAV8 vector. The sequences were designated 09YN and 10HB according to their sampling time and regions. Furthermore, a Bt-CAP3′-N1 primer was selected from the downstream sequence of the 07YN capsid gene obtained by 3′ rapid amplification of genomic DNA ends (RAGE; data not shown): (5′-GGAAGATCTACGCGAGAGACCGGACTTGACTGAAACGAA-3′). A fourth bat AAV capsid gene was amplified from the Myotis ricketti intestinal DNA sample using Bt-CAP5′/Bt-CAP3′-N1 primers and designated as 1285 according to the bat code. The 09YN, 10HB, and 1285 capsid genes were cloned into the UF1-07YN plasmid using HindIII/BglII digestion to generate UF1-09YN, UF1-10HB, and UF1-1285, respectively. All bat AAV capsid genes were sequenced using the ABI automated sequencer.
Construction of recombinant bat AAV packaging plasmids
A series of cloning experiments were performed to prepare for the production of recombinant bat AAVs. The 07YN bat AAV capsid gene was released from the UF1-07YN plasmid by treatment by successive treatment with BglII/Klenow/HindIII and ligated into the AAV2/9 vector treated successively with NotI/Klenow/HindIII to obtain AAV2-07YN. The 09YN, 10HB, and 1285 capsid genes were similarly cloned into the AAV2-07YN vector via HindIII/BglII digestion to obtain AAV2-09YN, AAV2-10HB, and AAV2-1285, respectively. With respect to AAV2-10HB, due to its lower packaging efficiency, we attempted to mutate the start codon of its AAV2 rep gene from ATG to ACG by transferring the rep gene from UF1-10HB to this plasmid via SpeI/HindIII digestion.
Production and purification of AAV vectors
The UF1-serial plasmids contained the AAV2 rep gene, bat AAV cap genes and AAV2 ITR. These plasmids were used together with the human adenovirus 5 helper plasmid pHelper for the generation of wild-type bat AAV vectors. Moreover, the AAV2-serial plasmids contained the AAV2 rep gene and bat AAV cap genes. These plasmids were used together with pHelper and a plasmid encompassing a reporter gene expression cassette and AAV2 ITR, AAV-CMV-luc1, or AAV2.1-CMV-lacZ (kind gifts from Dr. Xiao Xiao) for the generation of recombinant bat AAV vectors. Thus, two or three plasmids were used for Ca3PO4 transfection of 293 cells [20]. In addition, recombinant AAVs, including AAV2, AAV8, and AAV1, were generated for use as controls in bat AAV transduction experiments. AAV vectors were purified by fractionation with CsCl gradient centrifugation. DNase-resistant genome copy titers of the vector preparations were determined by dot blots with probes specific to the AAV2 rep gene or the CMV promoter.
In vitro transduction assay
Titration of recombinant AAVs was performed in Huh7, U87, A549, 293, and HT29 cells. Cells were maintained in DMEM complete medium and plated into a 24-well plate at a 1:3 ratio. Twenty-four hours later, AAV2-, AAV8-, 1285-, 09YN- and 10HB-CMV-LacZ were used for infection in duplicate at a multiplicity of infection (MOI) of 5 × 103, 5 × 104, and 5 × 105, respectively. Forty-eight hours after infection, the cells were resuspended in 250 µl of lysis buffer (100 mM K3PO4, pH 7.8; 0.2% Triton X-100) and centrifuged at 10,000 × g for 3 min. The supernatants were used for detection of β-galactosidase activity using the Tropix Galacto-Light kit (Applied Biosystems). The protein quantitation was accomplished using the BCA protein assay kit (Beyotime).
In vivo transduction in mice
Animal use in this study was approved and monitored by the Sichuan University Institutional Animal Care and Use Committee. To investigate the transduction efficiency of AAV vectors in systemic administration, 5 × 1011 vector genomes (v.g.) of AAV2-, AAV8-, 1285-, 09YN-, or 10HB-CMV-Luc vectors were injected into male 7-week-old C57B6 mice by tail vein with four mice per group. Two weeks later, mouse tissues, including the heart, liver, brain, lung, kidney, spleen, muscle and testis, were collected for the luciferase assay. One hundred milligrams of the tissue samples was homogenized using a Tissue-Tearor device (BioSpec) in 0.8 ml of lysis buffer on ice, and the lysates were centrifuged at 10,000 × g for 3 min. The supernatants were used for detection of luciferase activity using the luciferase assay system (Promega). The protein concentration was determined using the Bradfold assay (Beyotime). Genomic DNA was extracted from mouse tissues for quantitative detection of vector genome copies by TaqMan probes (Applied Biosystems) with a single-copy endogenous gene (glucagon gene) as the diploid cell number reference.
To study the direct muscle transduction of AAV vectors, 1010 v.g. of AAV2-, AAV1-, 07YN-, 1285-, 09YN-, or 10HB-CMV-Luc vectors were directly injected into the gastrocenemius muscles of male 7-week-old C57B6 mice with bilateral muscles of three mice treated as a group. Two weeks later, the mouse muscles were collected for luciferase assays and real-time PCR analysis as described above.
In vivo neutralization assays
Male 7-week-old CB17 SCID mice (Vital River) were injected with 294 µl of 50 mg/ml intravenous human immunoglobulin (IVIG; Shuanglin Biopharmaceutics, S20003024) by tail vein injection. Twenty-four hours later the amount of IVIG circulating in the mouse blood was expected to decrease by half to an approximate antibody concentration of 5 mg/ml, which is close to the physiological level of the normal human body [21]. Next, 5 × 1011 v.g. of AAV2-, 09YN-, or 10HB-CMV-Luc vectors were injected into the SCID mice by tail vein with four mice per group. Furthermore, for each AAV vector, an identical number of mice were injected with the same vector dose but without IVIG pretreatment. Two weeks later, gastrocenemius muscles were collected from the mouse groups for detection of luciferase activity.
To investigate the effects of human antibody on direct intramuscular injection, 602 µl of 50 mg/ml IVIG was injected into male 7-week-old CB17 SCID mice to yield an approximate antibody concentration of 10 mg/ml 24 h after treatment. 1010 v.g. of AAV2-, 09YN- and 10HB-CMV-Luc vectors were directly injected into the gastrocenemius muscles with four mice per group. Furthermore, for each AAV vector, an identical number of mice were injected with the same vector dose but without antibody treatment. Two weeks later, gastrocenemius muscles were collected from each mouse group for the luciferase assay as described above.
Evolutionary analysis on bat AAV capsid genes
Twenty-eight complete coding sequences of dependovirus capsids [22] were downloaded from GenBank in April 2014 (http://www.ncbi.nlm.nih.gov/; Table S1). In addition to the four bat AAV capsid genes listed in this study, a total of 32 dependovirus sequences were aligned using ClustalX2.1 [23]. The resulting fasta file was further aligned using the codon algorithm implemented in PRANSTER to improve the quality of alignment [24]. GENECONV [25], MaxChi [26], and GARD [27] were used to detect recombination breakpoints among dependovirus capsid gene sequences. The sequence alignment was therefore partitioned into 10 larger segments that contained no recombination breakpoints to exclude the disturbance of recombination on detection of positive selection [28]. Tree topologies were inferred from these sequence segments by the maximum-likelihood method implemented in DNAML of PHYLIP3.69 [29].
The CODEML program of PAML4.6 [30] was used for the detection of positive selection across dependovirus gene lineages. The nested evolution models M1a/M2a and M7/M8 were compared using likelihood ratio tests to assess the significance of positive selection, and empirical Bayes methods enabled the identification of individual positively selected codon sites a posteriori. Furthermore, positive selection could occur episodically in the specific dependovirus capsid gene lineage. The branch-site test implemented in CODEML and mixed effects model of evolution implemented in MEME [31] allow selection pressure to vary both among protein sites and across tree branches. These methods were used for detection of episodic diversifying selection across bat AAV lineages. Conditions existed in which an identical serine residue was observed through all of the gene lineages and was detected as positively selected by the programs. However, this finding was attributed to toggling selection or doublet mutation [32] but not strict positive selection, because eventually, no replacement could be observed.
Mutagenesis experiment of bat AAV capsid gene
T250 and E694 were inferred to be subject to positive selection in bat AAV capsid gene lineages and to be exposed on the exterior capsid surface. The bat AAV 10HB was selected as a prototype for evolutionary studies, and the corresponding V259 and N691 in its capsid were mutated by PCR methods. The mutagenesis primers for the V259 site were as follows: V259T-1: 5′-CCAAAGTACGTCGCGTCCCCGGTTCCGCTGCCGTT-3′; V259H-1: 5′-CCAAAGTAGTGCGCGTCCCCGGTTCCGCTGCCGTT-3′; V259-2: 5′-TTACAGCACGCCTTGGGGATACTT-3′. The mutagenesis primers for the N691 site were as follows: N691-1: 5′-CCGTGTACTGGATCTCGGGGTTCC-3′; N691E-2: 5′-CCAACTTCGAGAACAGCGCCAACGCGCAGTTCT-3′; N691A-2: 5′-CCAACTTCGCCAACAGCGCCAACGCGCAGTTCT-3′; N691K-2: 5′-CCAACTTCAAAAACAGCGCCAACGCGCAGTTCT-3′; N691S-2: 5′-CCAACTTCAGTAACAGCGCCAACGCGCAGTTCT-3′. These six mutated 10HB capsid genes were used for the production of recombinant bat AAV encompassing the luciferase reporter gene. These modified vectors were compared to the parental 10HB and control AAV2 vectors on their capacity to transduce mouse skeletal muscle after intramuscular injection using the methods described above.
Results
Isolation of four bat AAV capsid genes
After a series of PCR experiments, four full-length bat AAV capsid genes were amplified and sequenced (Fig. 1). The bat AAV capsid genes shared an amino acid sequence identity of 60.7–61.4% with that of the AAV 2 capsid gene (Table 1). Moreover, different sequence homology was shown within the bat AAV group. The 07YN and 1285 capsids shared 99.6% amino acid sequence identity, with replacement of only three residues. The 09YN capsid had a weaker relationship with 07YN with 99.0% of sequence identity and seven replacements. The close relationships of 07YN, 1285, and 09YN capsids inferred them as a single serotype. By contrast, the 10HB capsid had a lower (82%) sequence identity with the other three bat AAV variants, inferring it as a distinct serotype. Generally, the ~60% sequence identity between bat AAVs and AAV2 capsid VP1 proteins suggested their markedly different transduction and immunological properties.
Fig. 1.
Amino acid sequences of the bat AAV capsid genes. Coding sequences of four bat AAV capsid genes were obtained by PCR, and their amino acid sequences were aligned with those of AAV2 using the PRANK program [24]. Dots represented the same residues as those of bat AAV 07YN capsid. Dashes represented deletions. The putative N-termini of the bat AAV VP proteins are shown by arrows
Table 1.
Percentage of VP1 amino acid sequences identity of bat AAVs and AAV2
| 07YN | 1285 | 09YN | 10HB | AAV2 | |
|---|---|---|---|---|---|
| 07YN | |||||
| 1285 | 99.6 a | ||||
| 09YN | 99.0 | 99.2 | |||
| 10HB | 81.9 | 82.1 | 81.9 | ||
| AAV2 | 61.4 | 61.4 | 61.1 | 60.7 b |
a, bThe underlined numbers represent the highest and lowest similarity of AAV capsids
Generation of recombinant bat AAV vectors
To produce bat AAV vectors for additional studies, we employed the pseudotyping strategy. The bat AAV capsid genes were cloned into the wild-type AAV plasmid UF1-AAV8 with the AAV2 rep gene and ITRs, or the recombinant AAV plasmid AAV2/9 with the AAV2 rep gene. The UF1-series plasmids were used in combination with the adenovirus helper plasmid to generate of the wild-type bat AAVs, and the AAV2-series plasmids were used in combination with the pHelper and the plasmid encompassing the AAV2ITRs and a reporter gene, i.e., luciferase or β-galactosidase. The generated recombinant AAV particles were then detected by dot blotting (Table 2). All bat AAV capsid genes were competent in packaging wild-type and recombinant AAV vectors. However, these genes displayed different efficiencies for vector production. Specifically, the 07YN, 1285, and 09YN had similar capsid coding sequences, but the 07YN vector showed a lower production titer; thus, only 1285 and 09YN were selected representatively for comparison of their tissue tropism and immunological properties with 10HB and primate AAVs in most experiments.
Table 2.
Yields of AAV serotype and bat AAV production on 293 cells
| AAV variants | Vector genomes per 15 cm cell disha |
|---|---|
| AAV2 | 1.6 × 1012 |
| AAV1 | 2.5 × 1010 |
| AAV8 | 4 × 1011 |
| 07YN | 1.25 × 1010 |
| 1285 | 2 × 1011 |
| 09YN | 4 × 1011 |
| 10HB | 5 × 1010 |
aThe AAV variants were purified by CsCl ultracentrifugation and titrated by dot-blot
In vitro transduction by bat AAV vectors
A panel of the following human cell lines was transduced with serial dilutions of AAV2, AAV8, or bat AAVs: hepatoma (Huh7), glioma (U87), lung carcinoma (A549), embryo kidney (293), and colon cancer (HT29) cells. Generally, AAV2 exhibited the highest transduction for most of the cells (data not shown). By contrast, AAV8 could only efficiently transduce Huh7 cells with similar β-galactosidase activity as that of AAV2. Furthermore, none of the three bat AAV variants could efficiently transduce any of the five human cell lines tested. Interestingly, the bat AAVs were not capable of infecting the bat kidney cell line BK from Myotis davidii (data not shown). Thus, the nondetection of bat AAV infection on human cell lines might infer a potential host species barrier for bat AAVs to infect human cells. However, the different properties of human cells in vitro and in vivo could also be the reason for their nonpermissiveness for bat AAV infection.
In vivo tissue tropism of bat AAV vectors
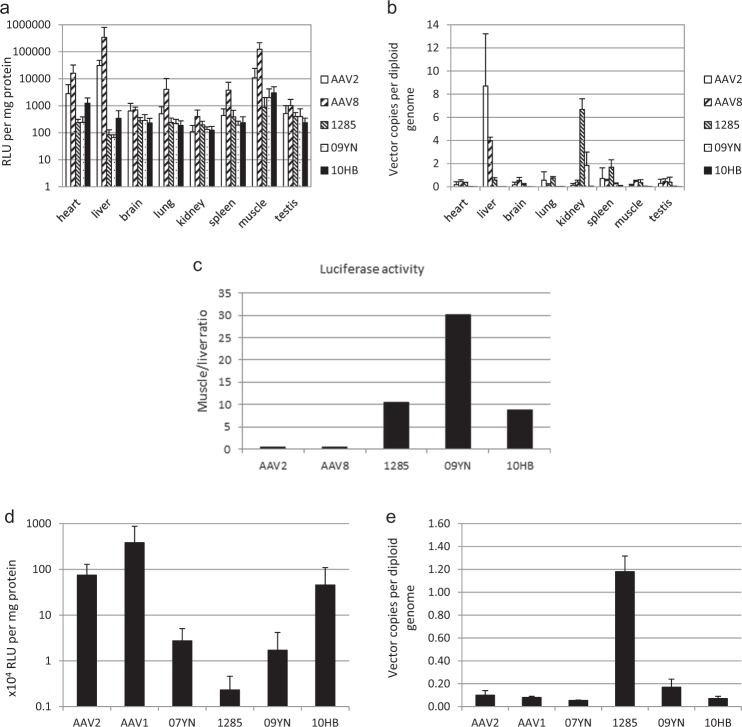
We next investigated bat AAVs to determine their systemic gene delivery efficiency and tissue tropism. The AAV luciferase reporter gene cassette was packaged in the viral particles of AAV2, AAV8, and bat AAVs for a side-by-side comparison. At 2 weeks after tail vein injection of AAV vectors in 7-week-old C57B/6 mice, luciferase activities and vector DNA copy numbers in various tissues were analyzed (Fig. 2). Consistent with a previous report [33], the AAV8 vector transduced mouse liver as well as heart and muscle efficiently (Fig. 2a). AAV2 predominantly infected the liver with considerably lower transduction in other organs. In contrast, the bat AAVs only slightly infected mouse liver but could transduce the heart and muscle to some extent. In keeping with the transgene expression, bat AAV vector copy numbers in the liver were considerably lower than those of AAV2 and AAV8, and considerably lower than those of AAV8 in the heart and muscle (Fig. 2b). However, in the kidney and spleen, the bat AAV 1285 vector showed a significantly higher vector copy number than AAV2 and AAV8, although no obvious transgene expression was observed in these organs. Furthermore, the muscle versus liver transduction ratio was calculated for the five AAV vectors examined (Fig. 2c). Interestingly, in contrast to the muscle/liver ratio of 0.4 observed for AAV2 and AAV8, the ratios of 11.0, 30.2, and 8.8 were observed for bat AAV 1285, 09YN, and 10HB, respectively. These data showed the distinct in vivo tissue distribution of bat AAVs from that of the primate AAVs given their weak transduction in mouse liver and lower transduction in other tissues.
Fig. 2.
Tissue tropism of bat AAVs in C57B6 mice after intravenous or intramuscular injection. a, b Luciferase activity (a) and rAAV genome copy numbers (b) in mouse tissues 14 days after intravenous injection of 5 × 1011 vector genomes of rAAV2-, rAAV8- or bat AAV-CMV-luciferase. Bars represent the luciferase activity of relative light units per microgram protein (a) or vector genome copies per diploid genome (b). The data are mean values ± s.d. (c) Relative transduction efficiency in muscle versus liver among AAV2, AAV8, and bat AAV vectors inferred by luciferase activity. d, e Luciferase activity (d) and rAAV vector genome copies (e) in mouse gastrocnemius muscle 14 days after intramuscular injection of 1010 vector genomes of AAV2, AAV1, or bat AAV vectors. The data are mean values ± s.d
Since bat AAVs showed muscle transduction to some extent in systemic administration, their performance in direct intramuscular injection was further investigated. The AAV2, AAV1, and four bat AAV vectors expressing luciferase were used for direct intramuscular injection of mouse gastrocnemius muscle (Fig. 2d, e). At 2 weeks post injection, the highest transgene expression was observed in AAV1-treated mouse muscles, moderate luciferase expression was observed in AAV2-transduced and 10HB-transduced muscles, while transgene expression was only slightly detected in 07YN-, 1285-, and 09YN-injected mouse tissues (Fig. 2d). Intriguingly, no association was observed between the vector genome copies and transgene expression levels. Most vector genome copies were detected in mouse muscles injected with the 1285 vector, while the other five vectors showed the muscle gene copy numbers at the same level (Fig. 2e). These data reiterated the results from systemic administration of AAV vectors in that bat AAV 1285 might be persistent in mouse tissues, such as kidney, spleen, or muscle for a long time, but the rate-limiting factors after cellular entry might hinder their efficient transduction. Notably, the vector genome copy number and transgene expression in skeletal muscle of the 10HB vector were similar to those of AAV2, which inferred its potential application as a vector for muscular gene therapy.
Neutralization assay of bat AAV by human sera
We considered utilizing the in vitro neutralization assay to study the immunological properties of the novel bat AAVs. However, none of the available cell lines, including those from humans or bats, was permissive for bat AAVs. In contrast, bat AAVs could transduce mouse muscle in vivo both by systemic administration and by direct intramuscular injection. Furthermore, the SCID mice infused with IVIG, the pooled human antibodies, have been demonstrated as an excellent model to assess the immune evasion of antibody neutralization by AAVs [12, 21]. We thus sought to inject the bat AAVs into the SCID mice to evaluate their capacity to transduce muscle in the presence of IVIG.
To evaluate the neutralization properties of the AAV vector in systemic administration, 294 µl of 50 mg/ml IVIG was injected into the male SCID mice, which should result in an ~5 mg/ml antibody concentration after 24 h mimicking the lower antibody level in human plasma. SCID mice were then injected intravascularly with 5 × 1011 v.g. of AAV2, bat AAV 09YN or 10HB-CMV-luciferase vectors. Without IVIG treatment, the AAV2 transduction of the mouse muscle was 4.2-fold of that of the 09YN and 1.7-fold of that of 10HB (Fig. 3a). However, after IVIG treatment, the AAV2 transduction in the mouse muscle decreased to approximately one-sixth of the untreated control, and the transduction of 09YN and 10HB, respectively, decreased to approximately one second and one-third of the untreated controls. The 10HB subsequently had the muscle transduction efficiency similar to that of the AAV2. Thus, the capacity to resist human antibody neutralization for the three vectors in the case of intravenous injection appeared to be 09YN > 10HB > AAV2.
Fig. 3.
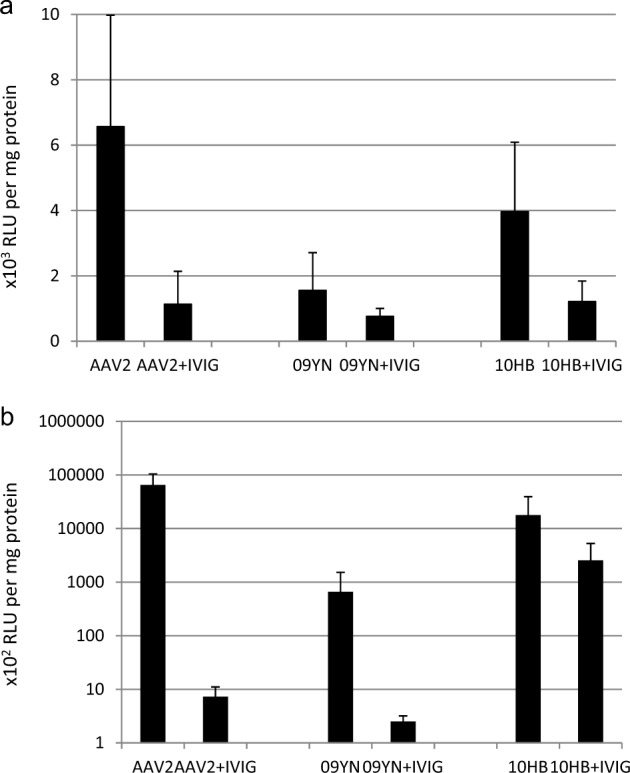
rAAV transduction of muscle of SCID mice after IVIG pretreatment. a Adult SCID mice were injected with 294 µl of 50 mg/ml IVIG via the tail vein or without IVIG treatment as a control group. 24 h later, 5 × 1011 v.g. of recombinant AAV2, bat AAV 09YN or 10HB vectors were injected into the mice via tail vein. b Adult SCID mice were injected with 602 µl of 50 mg/ml IVIG via the tail vein or without IVIG treatment as a control group. 24 h later, 1010 v.g. of recombinant AAV2, bat AAV 09YN or 10HB vectors were intramuscularly injected into the mouse muscles. Mouse gastrocnemius muscle was collected for the luciferase assay 14 days after vector treatment. The data are mean values ± s.e
To assess the three AAV vectors on their neutralization properties in intramuscular injection, 602 µl of 50 mg/ml of IVIG was injected into the SCID mice, corresponding to an ~10 mg/ml antibody concentration 24 h later, mimicking the higher antibody level in human blood. In the absence of IVIG pretreatment, the bat AAV09YN and 10HB vectors produced luciferase activity that was 98.6-fold and 3.7-fold lower than that produced by the AAV2 vector, respectively. In the presence of IVIG, the luciferase activity produced by the AAV2 and 09YN vectors was completely inhibited close to the baseline (Fig. 3b). By contrast, the transgene expression from the 10HB vector was reduced to 14.4% of the level produced in the absence of IVIG, with absolute luciferase activity over 2.5 × 105 RLU/mg protein. Therefore, the muscle transduction by bat AAV 10HB was considerably more resistant to IVIG neutralization than the 09YN and AAV2. The significant transgene expression in the presence of human antibodies suggested the application potential of the 10HB vector for muscular gene therapy in the case of preexisting AAV immunity.
Positive selection and mutagenesis analysis of bat AAVs
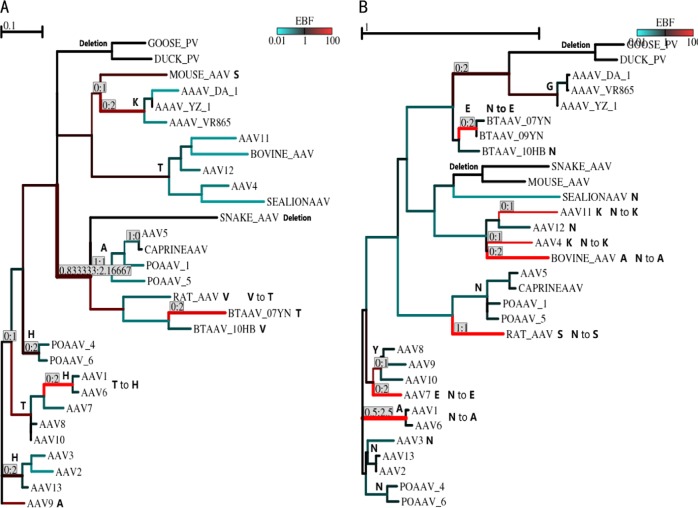
The evolution of bat AAVs has been primarily investigated in our previous report [17]. We were interested in the evolutionary mechanism of bat AAVs with respect to their unique capsid sequences, tissue tropism, and immunological properties. Using the maximum-likelihood method, we constructed a phylogenetic tree for bat AAVs and other representative dependovirus taxa based on their complete capsid coding sequences (Fig. 4). The four bat AAV variants formed a monophyletic group distinct from other dependovirus taxa, which inferred their independent evolutionary origin. A branch composed of rat AAV/PoAAV1/PoAAV5/caprine AAV/AAV5 was directly clustered with the bat AAVs in the molecular phylogeny which demonstrated their closer phylogenetic relationship with bat AAVs.
Fig. 4.
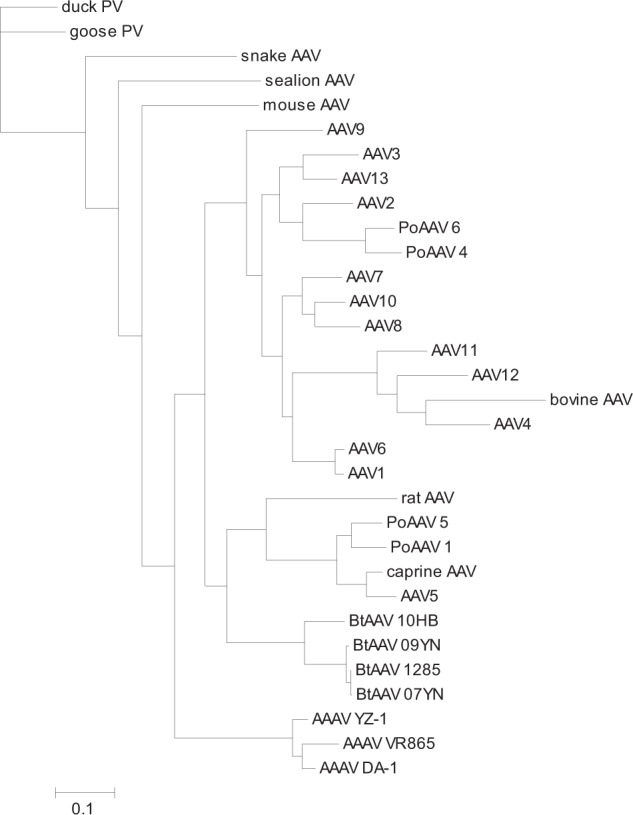
Maximum-likelihood phylogeny based on coding sequences of vertebrate dependovirus capsid genes. The tree topology was estimated using the DNAML program in the PHYLIP package [29]
In accordance with the condition in primate AAVs [3], a number of recombination events were detected among dependovirus capsid genes by RDP and GARD analysis (data not shown). However, no recombination event was detected during the evolution of the four bat AAV capsid genes. Since recombination could interfere with the detection of positive selection [28], the dependovirus capsid sequence alignment was partitioned into 10 segments according to the detected recombination breakpoints. These sequence segments were then subjected to selection analysis using the branch-site test in the PAML program [30] or the MEME method in the HYPHY program [31]. With respect to the branch site test, two sites corresponding to S238 and M420 in the 09YN capsid were subject to positive selection in the whole bat AAV lineage; one site corresponding to N344 in the 09YN capsid was selected within the 07YN/09YN/1285 lineage; and four sites corresponding to Q213, R297, A422, and M664 sites in the 10HB capsid were selected in the 10HB branch (Table 3). Interestingly, the results from the MEME analysis differed partially from those obtained from the branch-site test. The site corresponding to S238 in the 09YN capsid was positively selected for the whole bat AAV lineage, and the three sites corresponding to T259, N344, and E694 in the 09YN capsid were selected for the 07YN/09YN/1285 lineage. The site corresponding to S214 in the 10HB capsid was selected for the 10HB branch. These results suggest that positive selection but not genetic recombination play the crucial role in the speciation of bat AAV variants.
Table 3.
Detection of positive selection during evolution of bat AAV capsid genes
| Bat AAV lineages | Positively selected sites in bat AAV capsids | Correspondent residues in AAV2 capsid |
|---|---|---|
| Branch-site test | ||
| The whole bat AAV lineage | S238 and M420 in 09YN capsid; or S238 and M418 in 10HB capsid | A248 and A425 |
| 07YN/09YN/1285 lineage | N344 in 09YN capsid | S356 |
| 10HB branch | Q213, R297, A422 and M664 in 10HB capsid | N223, K309, S429 and V678 |
| MEME | ||
| The whole bat AAV lineage | S238 in 09YN capsid; or S238 in 10HB capsid | A248 |
| 07YN/09YN/1285 lineage | T259, N344 and E694 in 09YN capsid | H271, S356 and N705 |
| 10HB branch | S214 in 10HB capsid | S224 |
Note: The correspondent sites in AAV2 capsid were determined by checking the related sequence alignment of dependovirus capsid genes generated by PRANKSTER
For a better comprehension of their functional roles, the positively selected sites of bat AAVs were projected onto the AAV2 structure [34]. All three bat AAV sites from the 10HB branch were projected onto the interior surface of the AAV2 capsids (data not shown). The sites corresponded to Q213, S214, and R297 in the 10HB capsid and N223, S224 and K309 in the AAV2 capsid. By contrast, both bat AAV sites from 07YN/09YN/1285 lineage were projected onto the exterior surface of the AAV2 capsids (Fig. 5). The sites corresponded to T259 and E694 in the 09YN capsid, V259 and N691 in the 10HB capsid, and H271 and N705 in the AAV2 capsid. Unfortunately, no functional information is currently available for the three bat AAV sites localized on the interior surface of AAV2 capsids. However, mutations of H271 and N705, two projected sites on the exterior surface of AAV2 capsids, were associated with the altered transduction or neutralization properties of AAV2 [35]. Thus, it was plausible that positive selection within the corresponding sites in bat AAVs was related to their adaptation to uncharacterized bat species. To test this hypothesis, it would be interesting to perform mutagenesis experiments on the bat AAV 10HB capsid gene to examine their effects in muscle transduction in mice. The 10HB variant was selected because of its moderate muscle transduction and effective evasion of antibody neutralization (Figs. 2and 3).
Fig. 5.
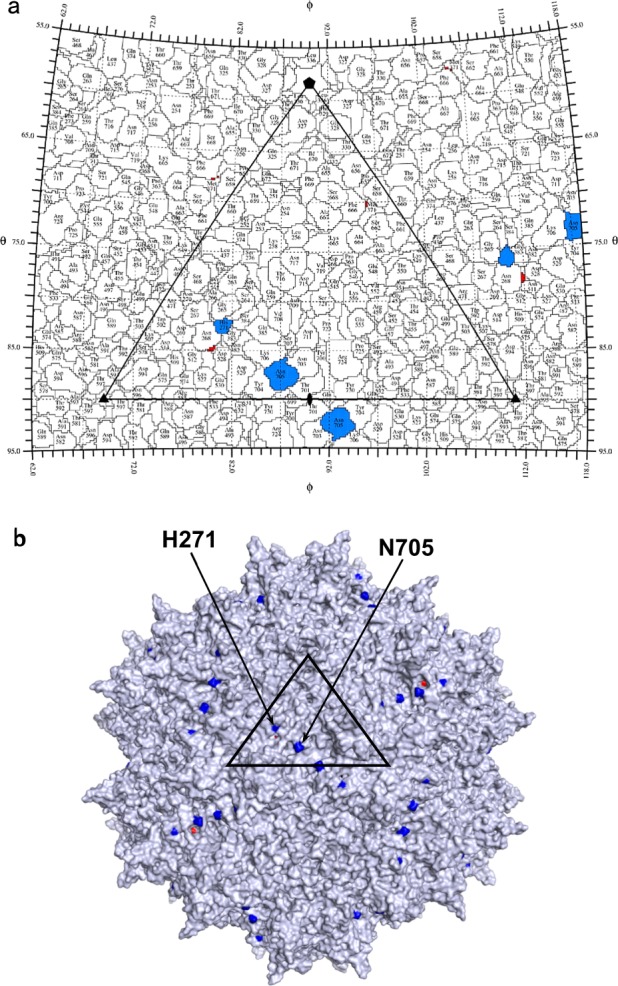
Roadmap (a) and spatial localization (b) of positively selected sites in bat AAV capsid genes on the exterior surface of AAV2 capsids. The two selected residues are displayed in blue, corresponding to V259 and N691 in bat AAV 10HB capsid, or H271 and N705 in AAV2 capsid. The axes of symmetry are shown by solid pentagon (five-fold), triangle (three-fold), and oval (two-fold) on the AAV2 capsid structure
The evolutionary pathway of the V259 site in the 10HB capsid (corresponding to T259 in the 09YN capsid and H271 in the AAV2 capsid) is demonstrated in Fig. 6a. Positive selection was detected in two dependovirus lineages with this codon being mutated from T to H in the AAV1/AAV6 lineage and from V to T in the bat AAV 07YN/1285/09YN lineage. Moreover, the evolutionary pathway of the N691 site in the 10HB capsid (corresponding to the E694 site in the 09YN capsid and the N705 site in the AAV2 capsid) is demonstrated in Fig. 6b. This codon was replaced from the ancestral N to E in bat AAV 07YN/1285/09YN lineage and AAV7, to K in AAV4, to A in bovine AAV and AAV1/AAV6 lineage, and to S in rat AAV.
Fig. 6.
Selection pressure and evolutionary pathway for sites 259 (a) and 691 (b) of the bat AAV 10HB capsid gene. The full-length dependovirus capsid coding sequences were subjected to recombination analysis to be split into segments without recombination. Segments 2290–2535 (a) and 6766–7074 (b) were used for MEME analysis [31]. Branches are colored according to the magnitude of the empirical Bayes factor (EBF) for the event of positive selection: red—evidence for positive selection, teal—evidence for neutral evolution or negative selection, black—no information. The amino acid residue for each branch was demonstrated at the root, and the replacements inferred as positive selection were displayed on top of or adjacent to the branches
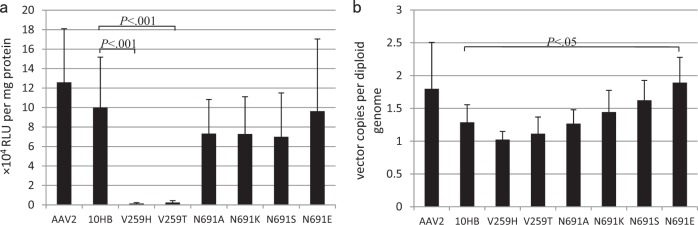
Based on these data, the V259 site in the 10HB capsid gene was mutated to T or H, and the N691 site was mutated to E, K, A, or S to evaluate the effects on AAV transduction (Fig. 7). Moreover, when the six bat AAV 10HB mutants were compared side-by-side with the wild-type 10HB virus, they showed variable transduction properties in mouse muscle. The transduction of 10HB V259T and V259H fell to approximately one-seventieth and one-fortieth of that of wild-type 10HB, respectively (Fig. 7a). This result was consistent with previous observations of AAV2 vector with mutations at the orthologous H271 site [35]. However, mutations at the N691 site within the 10HB capsid only slightly reduced the muscle transduction of the 10HB vector, which was also in keeping with the results in the AAV2 vector with mutations at the orthologous N705 site. Intriguingly, the 10HB V259H and V259T mutants showed only slightly reduced AAV genome copy numbers in mouse muscle (Fig. 7b). Furthermore, the 10HB mutants N691A, N691K, and N691S exhibited highly similar vector genome copies to those of the wild-type 10HB vector. The 10HB N691E mutant was distinct in its significantly improved genome copy number in mouse muscle. The V259H and V259T mutations dramatically altered the intracellular trafficking of the 10HB vector in mouse muscle, which determined their severely reduced transgene expression. Although the cellular entry of the 10HB mutants N691A, N691K, and N691S was not affected, their intracellular trafficking was slightly reduced. With respect to the 10HB N691E mutant, it was unique in that the increased vector genome copies balanced its reduced intracellular trafficking such that no alteration in transgene expression was observed in muscle for this vector.
Fig. 7.
Muscle transduction by bat AAV 10HB mutants at the selected sites 259 and 691. A total of 1010 vector genomes of AAV2, wild-type or mutated bat AAV 10HB vectors as indicated were intramuscularly injected into the gastrocnemius muscle of adult C57B/6 mice, and luciferase activity (a) and rAAV vector genome copies (b) were detected 14 days after treatment. The data are mean values ± s.e.P-values from the unpaired two-tailed t-test are shown above the bars
Discussion
In this study, four AAV variants were isolated from bat fecal or intestinal samples from two regions of China. The 07YN, 1285, and 09YN were sampled from the same location in Yunnan Province at different times. Consequently, the variants showed only a limited number of sequence variations among one another. Specifically, 1285 capsid differed from the 07YN capsid by three amino acid residues, while 09YN capsid differed from 07YN by seven amino acid replacements (Fig. 1). By contrast, the 10HB capsid from Hubei Province differed from 07YN by 132 residues. These results inferred that the genetic diversity of the bat AAV capsid gene was dictated by the geographic distribution of the original bat populations. Furthermore, the bats might be an important source for the isolation of novel AAV serotypes because the four AAV variants were isolated from a limited number of bat samples and because AAV sequences were highly prevalent in bat samples from different regions of China [17].
The bat AAV capsids showed ~60% sequence identity with that of AAV2 (Table 1), which demonstrated the distinct biological properties of bat and primate AAVs. In another aspect, the bat AAV capsid genes could be used in combination with the AAV2 rep gene and ITR sequences to generate recombinant AAV particles, albeit with the lower titers, which suggested a structural and functional conservation of AAV capsid genes across a large number of dependovirus species.
The pseudotyping strategy was successfully applied to the production of bat AAV vectors to investigate their transduction and immunological properties. Intriguingly, the bat AAVs could not efficiently infect the human cell lines (data not shown), which might be attributable to their high sequence variation from human AAVs and a potential interspecies barrier for transmission of bat AAV to human populations. Furthermore, when used for systemic administration, the bat AAVs could not transduce most of the organs including the liver in mice, but the 1285 vector could persist in the kidney and spleen for at least 14 days without significant transduction (Fig. 2a, b). This unique property of 1285 inferred that its capsids retained high stability in mouse tissues without efficient capsid uncoating and transgene expression. Furthermore, the bat AAV 10 HB vector could moderately transduce the mouse muscle after intramuscular injection, exhibiting the efficiency comparable to that of AAV2 (Fig. 2d, e). Notably, the muscle versus liver ratio of transgene expression for bat AAVs was over 24 times higher than that of AAV2 and AAV8. This characteristic of bat AAVs might favor their further genetic modification for gene vehicles specifically targeting to mouse skeletal muscle.
In accordance with their distinct capsid sequences from primate AAVs, the bat AAVs also appeared more resistant to neutralization by human AAV antibodies after both systemic and local administration (Fig. 3). Significantly, the bat AAV10HB vector could still efficiently transduce the skeletal muscle in a high dose of IVIG after intramuscular injection in mice, which was in contrast to the complete loss of transgene expression from the AAV2 vector at the same antibody titer. Considering both its moderate transduction of mouse muscle and efficient evasion of human antibody neutralization, the bat AAV 10HB vector exhibited the potential to be used for muscular gene therapy in humans.
Numerous genome sequences have been reported for dependoviruses, a parvovirus genus with the unique life cycle requiring helper viruses [22]. However, little is known about their evolutionary mechanism. Bioinformatics analysis revealed a limited number of recombination events among dependovirus species (data not shown), distinct from the frequent occurrence of homologous recombination in primate AAVs [3]. Furthermore, a number of positively selected sites were detected during the speciation of bat AAVs (Table 3; Fig. 5). Mutagenesis experiments of two of these selected sites in the 10HB capsid gene supported their functional roles in mouse muscle transduction (Fig. 7). These mutations appeared to affect muscle transduction of the 10HB vector primarily by lowering their intracellular trafficking or virion uncoating with no significant alteration of the tissue distribution of the vector genome. Interestingly, mutation of the N705 site in the AAV2 capsid, which is orthologous to N691 in the 10HB capsid altered its neutralization property [35]. Thus, positive selection in bat AAV capsid genes might be driven spontaneously by selection pressure from infection and immunity similar to that occurring in canine parvovirus [36].
The bats remain an important reservoir for virus transmission and spreading. Some of these viruses could be pathogens representing potential threatening to human health [16]. However, from another perspective, the bats could also be a potential source for virus isolation and vector development. Novel AAV vectors have also been isolated from other nonprimate mammals including cattle, goats, and pigs [11–13]. A common attribute of these AAV vectors was their efficient evasion of preexisting immunity from human AAV antibodies. Unfortunately, these vectors might not be superior to primate AAVs in their transduction properties, and to the best of our knowledge, no reports have described their further genetic engineering. Ideally, these vectors could be further tailored to optimize their transduction and immunological properties [37], which could ultimately benefit their application to human gene therapy.
Supplementary information
Acknowledgements
This work was funded by grants from the National Natural Science Foundation of China (31170157 and 81471776) and the National Science and Technology Major Project of the Ministry of Science and Technology of China (2018ZX09201018-013) to LY and from the National Natural Science Foundation of China (81290341) to ZS. We are grateful to Dr. Xiao Xiao for his constructive advice in the construction and production of bat AAV vectors. We extend our appreciation to Dr. Juan Li for her technical support in intramuscular injection in mice. We are grateful for the localization of selected sites of bat AAV capsid genes on the AAV2 structure by Drs. Lin-Ya Huang and Mavis Agbandje-McKenna, and for the helpful comments on our manuscript provided by Dr. Mario Mietzsch.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41434-019-0081-8) contains supplementary material, which is available to authorized users.
References
- 1.Atchison RW, Casto BC, Hammon WMCD. Adenovirus-associated defective virus particles. Science. 1965;194:754–6. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 2.Sonntag F, Schmidt K, Kleinschmidt JA. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci USA. 2010;107:10220–5. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao G, Alvira MR, Somanathan S, Lu Y, Vandenberghe LH, Rux JJ, et al. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc Natl Acad Sci USA. 2003;100:6081–6. doi: 10.1073/pnas.0937739100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, et al. Clades of Adeno-associated viruses are widely disseminated in human tissues. J Virol. 2004;78:6381–8. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira V, Twisk J, Kwikkers K, Aronica E, Brisson D, Methot J, et al. Immune responses to intramuscular administration of alipogene tiparvovec (AAV1-LPL(S447X)) in a phase II clinical trial of lipoprotein lipase deficiency gene therapy. Hum Gene Ther. 2014;25:180–8. doi: 10.1089/hum.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maguire AM, Simonelli F, Pierce EA, Pugh EN, Jr, Mingozzi F, Bennicelli J, et al. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N Engl J Med. 2008;358:2240–8. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nathwani AC, Reiss UM, Tuddenham EG, Rosales C, Chowdary P, McIntosh J, et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendell JR, Al-Zaidy S, Shell R, Arnold WD, ROdino-Klapac LR, Prior TW, et al. Single-dose gene replacement therapy for spinal muscular atrophy. N Engl J Med. 2017;377:1713–22. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 9.Boutin S, Monteilhet V, Veron P, Leborgne C, Benveniste O, Montus MF, et al. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther. 2010;21:704–12. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 10.Gurda BL, DiMattia MA, Miller EB, Bennett A, McKenna R, Weichert WS, et al. Capsid antibodies to different adeno-associated virus serotypes bind common regions. J Virol. 2013;87:9111–24. doi: 10.1128/JVI.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt M, Katano H, Bossis I, Chiorini JA. Cloning and characterization of a bovine adeno-associated virus. J Virol. 2004;78:6509–16. doi: 10.1128/JVI.78.12.6509-6516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arbetman AE, Lochrie M, Zhou S, Wellman J, Scallan C, Doroudchi MM, et al. Novel caprine adeno-associated virus (AAV) capsid (AAV-Go.1) is closely related to the primate AAV-5 and has unique tropism and neutralization properties. J Virol. 2005;79:15238–45. doi: 10.1128/JVI.79.24.15238-15245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bello A, Chand A, Aviles J, Soule G, Auricchio A, Kobinger GP. Novel adeno-associated viruses derived from pig tissues transduce most major organs in mice. Sci Rep. 2014;4:6644. doi: 10.1038/srep06644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maheshri N, Koerber JT, Kaspar BK, Schaffer DV. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- 15.Tse LV, Klinc KA, Madigan VJ, Castellanos Rivera RM, Wells LF, Havlik LP, et al. Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion. Proc Natl Acad Sci USA. 2017;114:E4812–E4821. doi: 10.1073/pnas.1704766114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–8. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Ge X, Hon CC, Zhang H, Zhou P, Zhang Y, et al. Prevalence and genetic diversity of adeno-associated viruses in bats from China. J Gen Virol. 2010;91:2601–9. doi: 10.1099/vir.0.020032-0. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Jiang J, Drouin LM, Agbandje-McKenna M, Chen C, Qiao C, et al. A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc Natl Acad Sci USA. 2009;106:3946–51. doi: 10.1073/pnas.0813207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bello A, Tran K, Chand A, Doria M, Allocca M, Hildinger M, et al. Isolation and evaluation of novel adeno-associated virus sequences from porcine tissues. Gene Ther. 2009;16:1320–8. doi: 10.1038/gt.2009.82. [DOI] [PubMed] [Google Scholar]
- 20.Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol. 1998;72:2224–32. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scallan CD, Jiang H, Liu T, Patarroyo-White S, Sommer JM, Zhou S, et al. Human immunoglobulin inhibits liver transduction by AAV vectors at low AAV2 neutralizing titers in SCID mice. Blood. 2006;107:1810–7. doi: 10.1182/blood-2005-08-3229. [DOI] [PubMed] [Google Scholar]
- 22.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, et al. The family Parvoviridae. Arch Virol. 2014;159:1239–47. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 24.Loytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA. 2005;102:10557–62. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawyer S. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989;6:526–38. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- 26.Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–9. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 27.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–8. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 28.Scheffler K, Martin DP, Seoighe C. Robust inference of positive selection from recombining coding sequences. Bioinformatics. 2006;22:2493–9. doi: 10.1093/bioinformatics/btl427. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Seattle: Department of Genome Sciences, University of Washington; 2005.
- 30.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 31.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delport W, Scheffler K, Seoighe C. Frequent toggling between alternative amino acids is driven by selection in HIV-1. PLoS Pathog. 2008;4:e1000242. doi: 10.1371/journal.ppat.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Zhu T, Qiao C, Zhou L, Wang B, Zhang J, et al. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat Biotechnol. 2005;23:321–8. doi: 10.1038/nbt1073. [DOI] [PubMed] [Google Scholar]
- 34.Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, et al. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci USA. 2002;99:10405–10. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lochrie MA, Tatsuno GP, Christie B, McDonnell JW, Zhou S, Surosky R, et al. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J Virol. 2006;80:821–34. doi: 10.1128/JVI.80.2.821-834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shackelton LA, Parrish CR, Truyen U, Holmes EC. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc Natl Acad Sci USA. 2005;102:379–84. doi: 10.1073/pnas.0406765102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet. 2014;15:445–51. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.