Abstract
Quaternary distance restraints are essential to define the three-dimensional structures of protein assemblies. These distances often fall within a range of 10–18 Å, which challenges the high and low measurement limits of conventional nuclear magnetic resonance (NMR) and double electron-electron resonance electron spin resonance spectroscopies. Here, we report the use of 19F paramagnetic relaxation enhancement (PRE) NMR in combination with 19F/paramagnetic labeling to equivalent sites in different subunits of a protein complex in micelles to determine inter-subunit distances. Feasibility of this strategy was evaluated on a pentameric ligand-gated ion channel, for which we found excellent agreement of the 19F PRE NMR results with previous structural information. The study suggests that 19F PRE NMR is a viable tool in extracting distance restraints to define quaternary structures.
Despite the great success in the use of x-ray and cryogenic electron microscopy (cryo-EM) to determine the structure of various ion channels, the capacity of these techniques to solve the structure of flexible protein regions is often challenged. Double electron–electron resonance (DEER) electron spin resonance (ESR) spectroscopy has proven useful for measuring quaternary structural restraints without restrictions from the local dynamic properties of ion channels1–5. However, for membrane proteins DEER ESR can measure distances typically in the range of 18–60 Å and is unreliable for measuring shorter distances6.
Paramagnetic relaxation enhancement (PRE) in solution nuclear magnetic resonance (NMR) has been developed for extracting distance restraints of 13–25 Å between an NMR observable nucleus and a paramagnetic probe7–9, which is often introduced by nitroxide spin labeling of a single cysteine that exists either naturally or is introduced by mutagenesis10. The paramagnetic MTSL [(1-oxyl-2,2,5,5-tetramethyl-D3-pyrroline-3-methyl) methanethiosulfonate], commonly used for ESR studies, has been adopted for PRE NMR measurements. The unpaired electron spins of MTSL enhance nuclear longitudinal (R1) and transverse (R2) relaxation rates in a distance-dependent manner. The paramagnetic enhancement of R2 in the r−6 distance dependency for NMR nuclei within the range of 13–25 Å7 can be quantified to extract distance information. The distances resulting from PRE measurements complement short interproton distance restraints (≤ 5 Å) derived from the nuclear Overhauser effect (NOE), as well as longer distance restraints measured by DEER ESR. Another benefit of PRE NMR is that it can be used to gather structural information not only for well-folded proteins, but also for disordered proteins11. Additionally, paramagnetic probes decrease the spin-lattice relaxation time and speed up NMR data acquisition9. Thus, PRE NMR has become an invaluable tool in structure biology.
PRE experiments are commonly performed by monitoring 1H signal changes in 1H-15N NMR spectra due to the MTSL-induced R2 enhancement7, 8, 12, 13. 19F PRE NMR14, 15, however, has received recent attention, especially when larger proteins and protein complexes are under investigation. In general, 19F NMR is a valued addition to other structural approaches used for characterizing structures and dynamics of proteins and protein complexes, including ion channels3,16–18. A null 19F signal background in native biological systems prevents signal overlap, which occurs in 1H-15N spectra of large proteins and often compromises accurate measurements of PRE from individual sites. The excellent sensitivity of 19F resulting from its 100% natural abundance and high gyromagnetic ratio adds another advantage for using 19F PRE NMR in structure determinations.
In the present work, we have created a 19F/MTSL labeling scheme for pentameric ligand-gated ion channels (pLGICs) that allows us to determine inter-subunit distances by solution 19F PRE NMR. This new strategy for gaining quaternary structural information can be easily extended to other proteins and protein complexes beyond pLGICs. A key step in acquiring this quaternary structural information via 19F PRE NMR is to label both the 19F and paramagnetic probes to selected equivalent residues in a channel complex. In our experiments, the 19F probe TET [2,2,2-trifluoroethanethiol] that provides a trifluoromethyl (−CH2CF3) was tagged to a selected cysteine in a channel protein as reported previously3. The paramagnetic probe MTSL was also labeled to cysteine sites equivalent to that tagged by TET. For pLGICs, a labeling molar ratio of 1 TET:4 MTSL (where one of the five subunits is labeled with TET and the remaining four subunits are labeled with MTSL) is ideal for 19F PRE NMR to extract distances between adjacent subunits. To achieve a proper TET:MTSL labeling ratio, we tested various conditions, including the order of labeling and labeling times for each species. TET has a much lower labeling efficiency than MTSL3. Thus, it was crucial to use an excess amount of TET as compared to MTSL. It is also important to control the total labeling time (see additional details in the Methods section). The final labeling efficiencies of TET and MTSL were confirmed with respective 19F NMR and ESR3, showing ~15% TET and ~67% MTSL labeling (approximately 1:4 molar ratio) in each sample. Such labeling efficiencies assure a sufficiently high probability of each 19F TET-labeled residue to meet at least one paramagnetic center at the equivalent residue labeled with MTSL in an adjacent subunit so that a quaternary distance restraint can be measured from 19F PRE NMR experiments.
We first examined the feasibility of this new strategy using ELIC (Fig. 1a), a homomeric prokaryotic pLGIC with known x-ray structures19, 20. In addition to the x-ray structures, DEER ESR and 19F NMR experiments were also previously performed on the ELIC L253C mutant3. The variety of available structural information makes the ELIC L253C construct an ideal candidate to evaluate the 19F PRE NMR strategy. Moreover, neither the TET nor the MTSL tags at L253C affect the ion channel function of ELIC3. Residue 253 in each ELIC subunit is located at the interface of the extracellular and transmembrane domains (Fig. 1a, PDB code: 3RQU20). The distance between two L253 Cβ atoms in the adjacent subunits is 15.3 ± 0.06 Å (mean ± standard deviation) (Fig. 1b), which falls into the measurable distance range of 19F PRE NMR14, 15. In order to know how well the distances measured through the TET and/or MTSL tags in 19F PRE NMR or DEER ESR match with the Cβ distances in the x-ray structure, we modeled conformational ensembles of MTSL labels at residue 253 in the ELIC x-ray structure using MTSSLWizard software21. The calculated distances between the paramagnetic centers of MTSL in the adjacent subunits (18.2 ± 5.6 Å) were in excellent agreement with the experimental distance (18.5 ± 3.9 Å) measured by DEER ESR3. Similar MTSSLWizard calculations for TET-MTSL pairs of the adjacent residues 253 in the ELIC x-ray structure show a distance distribution (17.4 ± 4.4 Å) that also matches well with the distance derived from 19F PRE NMR described as follows.
Fig. 1.
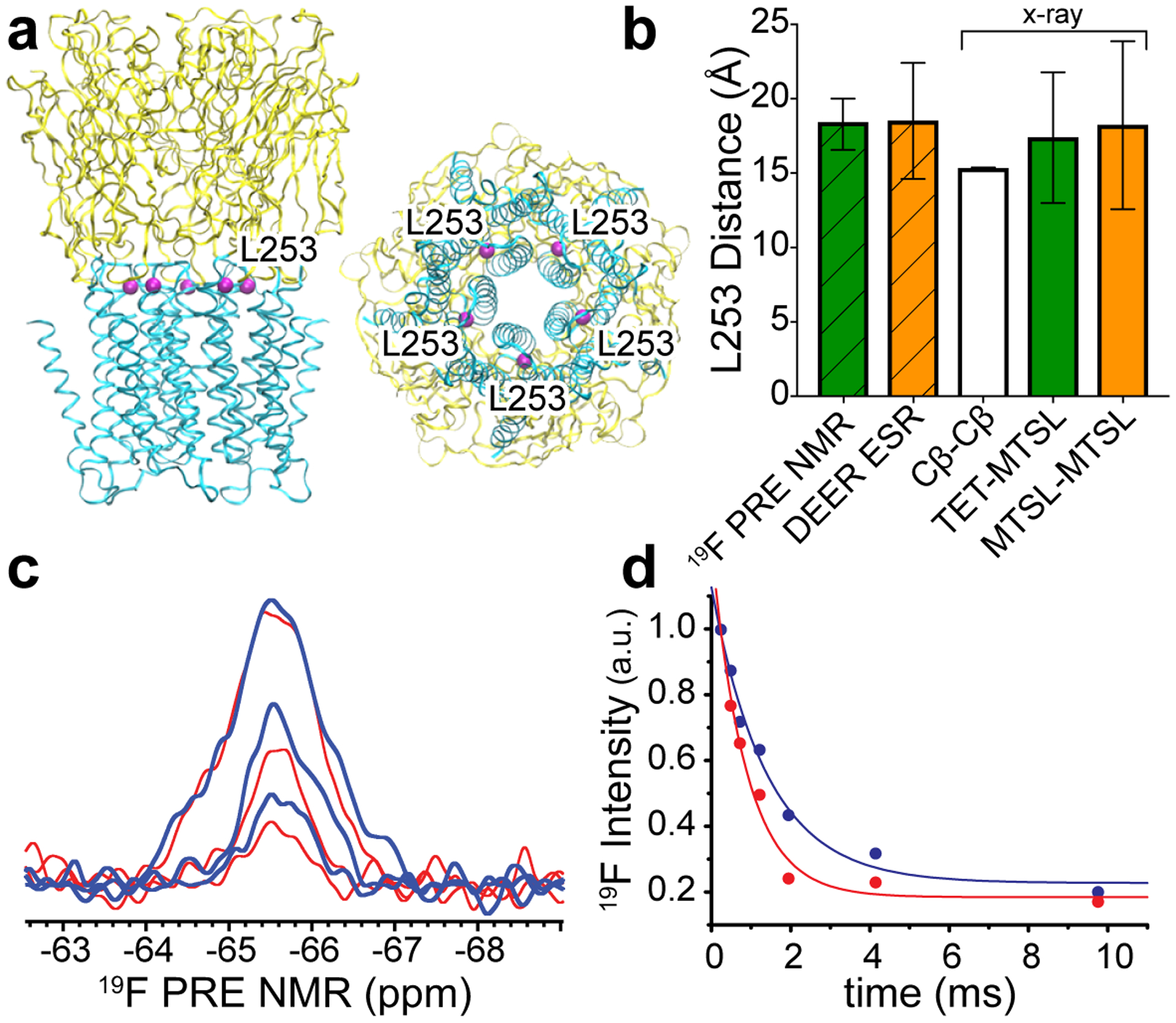
(a) Side (left) and bottom (right) views of the pentameric apo ELIC x-ray structure (PDB ID: 3RQU)20. Five equivalent L253 residues (purple) at the interface of the extracellular domain (yellow) and the transmembrane domain (cyan) are highlighted. (b) Distances obtained from 19F PRE NMR and DEER ESR experiments are compared to distances between L253 Cβ atoms (Cβ-Cβ) in adjacent subunits of the structure shown in (a). Additional comparisons include the distances between the paramagnetic center of MTSL tags (MTSL-MTSL) or the average TET fluorine positions (TET-MTSL), based on the modeled conformational ensembles of MTSL-MTSL and TET-MTSL labels in adjacent subunits of the ELIC x-ray structure using MTSSLWizard software21. Error bars represent standard deviation for all measured distances. (c) Representative 19F PRE NMR spectra of ELIC L253C labeled with TET and MTSL. The spectra collected under paramagnetic (red) and diamagnetic (blue) conditions with relaxation delays of 0.244 (top), 1.22 (middle), and 4.148 (bottom) ms are superimposed. (d) Normalized 19F NMR resonance intensity as a function of relaxation delay time under the paramagnetic (red) and diamagnetic (blue) conditions were fit to single exponential decay functions, resulting in transverse relaxation rates of R2para = 1153 ± 194 Hz and R2dia = 714 ± 123 Hz that were used to derive a distance of 18.4 ± 1.7 A between residues 253 in the adjacent ELIC subunits.
19F PRE NMR spectra of ELIC L253C labeled with TET and MTSL were collected with varied relaxation delays under paramagnetic (para) and diamagnetic (dia, after addition of ascorbic acid to the same sample) conditions (Fig. 1c). The protein was solubilized in n-dodecyl-β-D-maltoside (DDM), which was used previously for ELIC crystal structures19, 20. The corresponding resonance intensities (Ipara, Idia) as a function of relaxation times were fit to exponential decay functions to derive their respective transverse relaxation rates. The data collected in the paramagnetic state were fit to single, double, and triple exponential decay functions to test whether more than one R2para relaxation component existed in the sample. However, like R2dia, only the single exponential decay function could fit data to generate R2para (R2para = 1153 ± 194 Hz, R2dia = 714 ± 123 Hz) (Fig. 1d). A distance between the paramagnetic center of MTSL on one L253C and the fluorine atoms of TET on another L253C in the adjacent subunit of ELIC was derived based on the PRE, Γ2F = R2para - R2dia, using the Solomon-Bloembergen equation22:
where r is the distance between the 19F nucleus and the paramagnetic center, ωF is the 19F Larmor frequency (564.68 MHz) times 2π, τc is the correlation time for the nuclear–electron interaction that can be assumed to be equal to the global correlation time of the protein8, which was estimated as 218 ns at 10 °C (see Supporting Information) using Stokes’ law23. The constants in the above equation include the permeability constant μ0, the fluorine gyromagnetic ratio γF, the electron g-factor g, the Bohr magneton μB, and the electron spin quantum number S (S=1/2) of a nitroxide radical. N is the number of the paramagnetic centers adjacent to the 19F nucleus. Typically, only one paramagnetic center (N =1) is present for a chosen nucleus7, 8, 12–15. However, because of the five-fold symmetry of homo-pentameric channels and the 1 TET:4 MTSL labeling scheme, the probability to have two equivalent paramagnetic centers (N =2) in the two adjacent subunits for each 19F nucleus is extremely high. Thus, we used the equation above to obtain an adjacent inter-subunit distance of 18.4 ± 1.7 Å for the case of N =2. This distance is close to the predicted distance for modeled MTSL-TET pairs in adjacent ELIC subunits in the x-ray structure (Fig. 1b). Small distance discrepancies from three experimental methods are expected because the inter-subunit distance was measured using different reference points: Cβ atoms of two adjacent L253 residues in the x-ray structure, between two adjacent MTSL paramagnetic centers in DEER ESR, or between 19F nucleus of the labeled TET and the MTSL paramagnetic center of the adjacent subunit in 19F PRE NMR.
The 1 TET:4 MTSL labeling scheme ensures a uniform 19F PRE signal from the adjacent paramagnetic MTSL labels. However, to what degree does a non-adjacent MTSL interfere with the intended measurement for distances between adjacent subunits? The distances shown in crystal structures19, 20 and the DEER ESR results (18.5 ± 3.9 Å and 31.0 ± 5.6 Å for adjacent and non-adjacent residues 253, respectively)3 are consistent with the geometric arrangement of a pentamer, which has a distance ratio of 1.62 between equivalent residues in non-adjacent vs. adjacent subunits. A steep decay of PRE with increasing distance (r−6) makes the PRE contribution from non-adjacent MTSL almost negligible ((1.62)−6 < 6%). Thus, the non-adjacent subunit distance in a pentameric channel is too far to be measured by PRE and the distances extracted from the 19F PRE NMR in conjunction with our 19F/MTSL labeling scheme should predominantly reflect only the distances between adjacent subunits.
The same 19F PRE NMR strategy was applied to the human α7nAChR, a pentameric neurotransmitter-gated ion channel whose structures are still under investigation24, especially the structure of its intracellular domain. 19F PRE NMR experiments along with the 1 TET:4 MTSL labeling scheme were performed on two separate single-cysteine mutants (C435 and C427) of α7nAChR that are both located in the intracellular domain (Fig. 2a). 19F PRE NMR spectra of C435 and C427 in micelles collected under paramagnetic (red) and diamagnetic (blue) conditions (Fig. 2b) provided data to calculate the corresponding transverse relaxation rates R2para and R2dia (Fig. 2c), which allow for subsequent calculations of inter-subunit distances at each residue (C435 = 18.3 ± 1.7 Å; C427 = 17.2 ± 1.6 Å). Inter-subunit distances at the C435 and C427 positions are similar to distances at equivalent positions (A423 and K415, respectively) between two adjacent subunits in the cryo-EM structure (PDB code: 6BE1)25 of the resting-state 5-HT3A receptor, a pentameric ligand-gated ion channel homologous to α7nAChR (Table 1S, Supporting Information). The distances for both sites in α7nAChR are shorter than or close to the borderline of the low distance limit of DEER ESR measurements6, demonstrating the value of 19F PRE NMR as a complementary tool in quaternary structure determination.
Fig. 2.
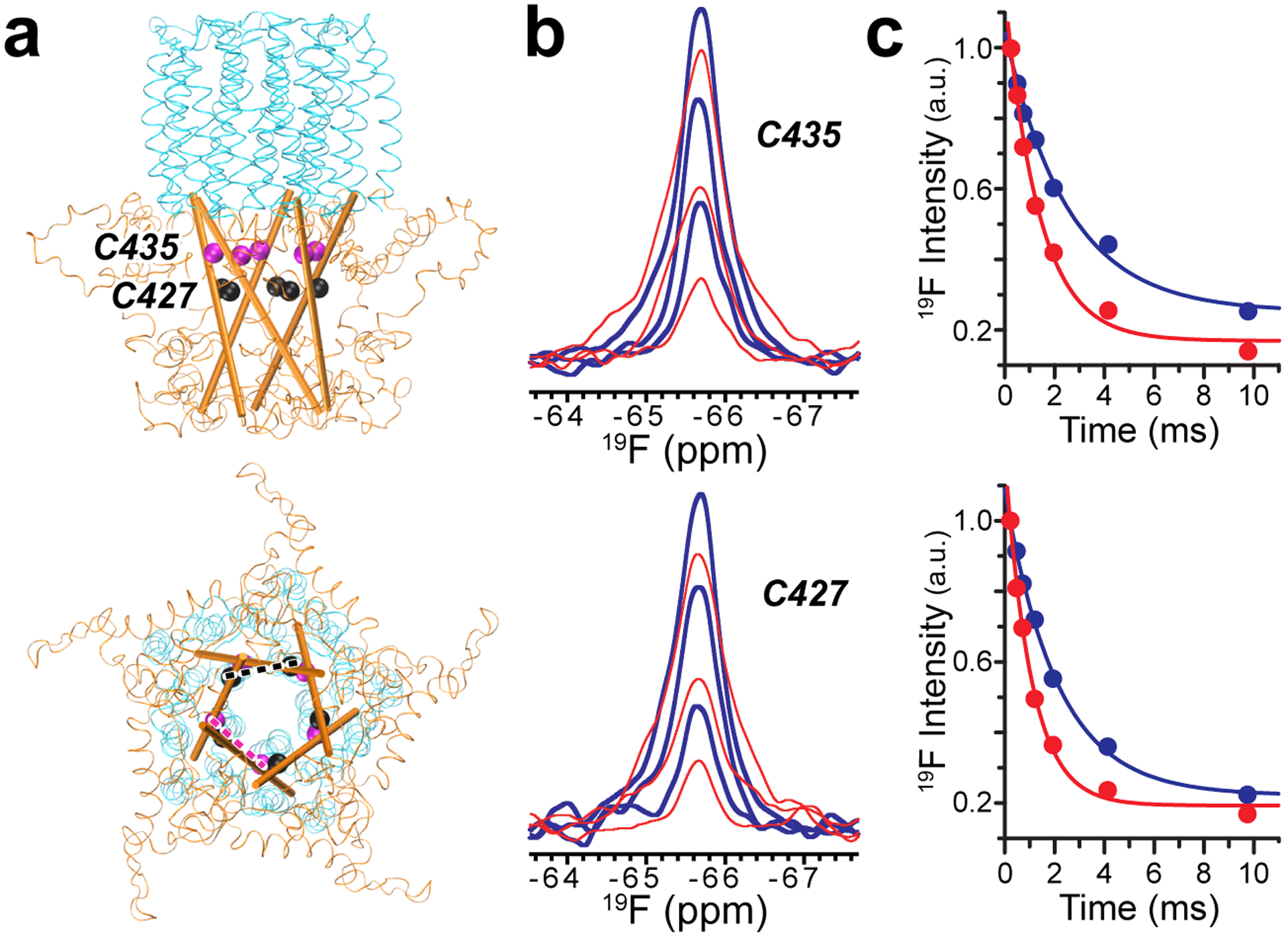
(a) Side (top) and cytoplasmic (bottom) views of the α7nAChR transmembrane domain (cyan) and intracellular domain (orange) showing selected residues along each intracellular MA helix (cartoon representation) for 19F PRE NMR experiments. Dashed lines highlight inter-subunit distances. (b) Representative 19F PRE NMR spectra for residues C435 and C427 labeled with TET and MTSL under paramagnetic (red) and diamagnetic (blue) conditions with relaxation delays of 0.244 (top), 1.22 (middle), and 4.148 (bottom) ms. (c) Normalized resonance intensities of paramagnetic (red) and diamagnetic (blue) 19F PRE NMR spectra for residues C435 and C427 as a function of relaxation delay time. Data fitting to a single exponential decay function results in transverse relaxation rates for individual sites (C435: R2para = 728 ± 88 Hz and R2dia = 393 ± 64 Hz; C427: R2para = 950 ± 80 Hz and R2dia = 473 ± 33 Hz). Corresponding inter-subunit distances were 18.3 ± 1.7 A and 17.2 ± 1.6 A for C435 and C427, respectively.
Although the disulfide-linked labels, such as MTSL and TET, have been widely used in ESR and NMR experiments, it is reasonable to question whether these labels introduce errors to the derived distances. Indeed, one should be cautious when choosing a labeling site to avoid structural disturbance to proteins. If permitted, a functionality assessment should be arranged after labeling3, 18. Battiste and Wagner previously showed good agreements between PRE-derived distances with an error bound of ±4 Å and the corresponding distances in a known protein structure7. Gottstein et al. also investigated the effect of the error margin for PRE-derived distances and found that the final structure quality was largely insensitive to the size of the error bound26. Structures with a backbone RMSD of 1.0–1.6 Å to the reference structure were obtained even with PRE error bounds up to 10 Å26. Thus, an error bound of ±4 Å for PRE-based distance restraints should ensure the structural accuracy, especially when a large number of restraints are collected from sites evenly distributed throughout the protein.
Although proteins in micelles were used in the current study, the reported method can be applied to proteins in other membrane mimics, such as nanodiscs and bicelles. The choice of membrane mimics is often determined by the protein stability and quality of NMR spectra. In most cases, membrane proteins are purified in detergent. Thus, one can complete the labeling procedures in detergent and then move the labeled protein into another mimetic membrane if it is more suitable for the protein.
Orthogonal spin labels with different spectroscopic properties have created new platforms in ESR and NMR studies of biomacromolecules with the benefit of increasing information content of experimental results27, 28. Exploiting paramagnetic probes other than nitroxide (such as chelators of Gd(III) and other lanthanide ions) in combination with labeling to non-cysteine residues (i.e. unnatural amino acids incorporated into proteins) have demonstrated great potential in various applications27, 28. All of these options can be integrated into our reported method for extracting structural information of ion channels. For example, a 19F probe can be introduced biosynthetically in protein expression16 instead of chemical modification as shown in the current study. This may become more relevant if labeling of membrane-embedded cysteine is problematic. Click chemistry, which offers a fast and highly selective biocompatible reaction between azide and alkyne groups, is a good option to tag paramagnetic probes, for which unnatural amino acids can be introduced to desired sites in the protein28. Furthermore, one has the freedom to choose whether 19F probe and paramagnetic tags are in equivalent or non-equivalent positions among different channel subunits. The final choice will be determined by protein performance in structural and functional experiments.
In conclusion, 19F PRE NMR in combination with the TET/MTSL labeling scheme presented here is a realistic alternative approach for generating quaternary distance restraints for ion channels and other protein complexes that may be difficult to be defined by a different structural tool.
Experimental Methods
Sample Preparations
ELIC was expressed and purified as reported previously3, 20, 29. The single cysteine ELIC L253C3 was constructed after replacing native C300 and C313 to alanine and serine, respectively, using the QuickChange Lightning Kit for single or multi-site mutagenesis (Agilent Technologies). Single-cysteine α7nAChR constructs (C427 and C435) containing the transmembrane domain (TMD) and intracellular domain (ICD) were prepared on the basis of the full-length WT α7nAChR construct24 by replacing native cysteines in the TMD and ICD with alanine or serine. Each construct was transformed to Rosetta (DE3) pLysS (Novagen) cells for expression in Luria-Bertani media or in the 15NH4Cl-containing M9 media. The expression was induced with 0.2 mM isopropyl β-D-1-thiogalactopyranoside when OD reached ~0.8. The expression at 15 °C lasted ~24 hours for ELIC or ~72 hours for the α7nAChR TMD+ICD. Harvested cells were re-suspended in a buffer (50 mM sodium phosphate at pH 8, 150 mM NaCl, and protease inhibitors for ELIC and 50 mM Tris at pH 8, 150 mM NaCl for α7) and lysed using a M-110Y microfluidizer processor (Microfluidics). Cell membrane was pelleted by ultracentrifugation. ELIC fused with maltose binding protein was extracted with 2% (w/v) DDM (Anatrace) and purified with a 5-mL HisTrap HP column (GE Healthcare). Maltose binding protein was cleaved off overnight using protease HRV3C (GE Healthcare) and separated from ELIC using HisTrap HP columns. The pentameric ELIC was collected in a buffer containing 25 mM sodium phosphate at pH 8, 125 mM NaCl, 0.05% (w/v) DDM by size exclusion chromatography using a Superdex 200 10/300 GL column (GE Healthcare). The single-cysteine α7nAChR TMD+ICD was extracted with 2.5% (w/v) LDAO (N,N-dimethyldodecylamine N-oxide, Sigma) and purified with 0.4% (w/v) LDAO using a HisTrap HP column and subsequently a Superdex 200 10/300 GL column as used in ELIC purifications.
Several steps are involved in the labeling of α7nAChR and ELIC with TET/MTSL (Toronto Research Chemicals). A given purified protein was first treated briefly (~1 hour) with the reduce reagent DTT (Invitrogen) (~15x the protein concentration) at room temperature to prepare all available cysteines for labeling. After removing DTT with HiTrap Desalting columns (GE Healthcare), 25-fold molar excess of MTSL to the protein was added and mixed with the sample for ~30 s. Immediately after, we added 100-fold molar excess of TET to the protein, considering that TET is more difficult to be labeled than MTSL3. A faster leaving group (the sulfinic acid, CH3SO2H) in the MTSL labeling process and suppressed sulfhydryl ionization due to a hydrophobic environment in the TET labeling sites may have contributed to their different labeling efficiencies in the channel proteins. The sample was placed on an inversion mixer and incubated for three hours at room temperature and then overnight at 4 °C. Free MTSL and TET were removed by dialysis with three changes of buffer and then subjected to size exclusion chromatography on a Superdex 200 10/300 GL column. The labeling efficiencies of TET and MTSL were assessed by 19F NMR and ESR, respectively3.
A typical sample for 19F PRE NMR contained ~100 μM protein, 20 mM sodium phosphate buffer at pH 7.7, 120 mM NaCl, and 0.5 % (w/v) DDM for ELIC or 0.5 – 1.0 % (w/v) LDAO for α7nAChR, equivalent to a molar ratio (detergent to protein) of ~100 for ELIC and ~200 for α7 TMD-ICD. 5% D2O was added for deuterium lock. The diamagnetic condition for TET/MTSL-labeled samples in 19F PRE NMR was achieved by adding a 10-fold molar excess of ascorbic acid. To determine the global rotational correlation time (τc) of the α7nAChR TMD+ICD by 1D [15N-1H]-TRACT NMR experiment30, a sample containing 15N-labeled α7nAChR TMD+ICD, 5 mM sodium acetate buffer at pH 5.0, 25 mM NaCl, and 1.0 % LDAO was used.
NMR Data Collection and Analysis
19F PRE NMR was performed at 10 °C on a Bruker Avance 600-MHz spectrometer (19F frequency: 564.68 MHz) equipped with a triple-resonance 19F-detection TXO cryoprobe (Bruker Instruments). Spectra to measure the 19F transverse relaxation rates (R2) were collected using the Carr-Purcell-Meiboom-Gill pulse sequence (CPMG) with 8192 data points, a 30-ppm spectral width and a carrier frequency at −70 ppm. For each sample, spectra were collected in the absence and presence of ascorbic acid, corresponding to paramagnetic and diamagnetic conditions, respectively, with varied relaxation delays of 0.244, 0.488, 0.732, 1.22, 1.952, 4.148, and 9.76 ms and a recycle delay of 1 second. Each sample requires 24 to 30 hours for NMR data collection and 9600 to 12000 scans for each spectrum at a given relaxation delay time. The 19F chemical shift was externally referenced to the trichlorofluoromethane resonance at 0.0 ppm.
The NMR spectra were acquired, processed and analyzed with TopSpin 3.5 (Bruker Instruments). 19F transverse relaxation rates of R2para and R2dia were obtained in the absence and presence of ascorbic acid, respectively, from fitting the 19F peak intensity (I) as a function of the relaxation delay in a single exponential decay function. The 19F PRE, Γ2F = R2para - R2dia, was calculated and used in the Solomon-Bloembergen equation22 to obtain the distance between the 19F nucleus of TET in one subunit and the paramagnetic center of MTSL in the adjacent subunit.
To determine a rotational correlation time (τc) for the α7nAChR TMD+ICD, a series of 1D [15N-1H]-TRACT NMR spectra30 with varied relaxation periods of 0.1, 0.5, 1, 2, 4, 8, 16, 32, and 64 ms were acquired with a recycle time of 1 s at 45 °C on a Bruker Avance 700 MHz spectrometer equipped with a triple-resonance inverse-detection cryoprobe TCI (Bruker Instruments). More details for τc data collection and analysis are provided in Supporting Information.
Supplementary Material
Acknowledgements
The authors thank other members of the Tang laboratory and R. Ishima for helpful discussion. The research was supported by funding from NIH (R01DA046939) and NSF (MCB 1613007 and MRI 1725678). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supporting Information
The Supporting Information is available free of charge is available free of charge via the internet at http://pubs.acs.org. Table 1S, Comparison of 19F PRE NMR measured inter-subunit distances at the selected residues in α7nAChR with the corresponding distances measured from homologous residues in the cryo-EM structure of the resting-state 5-HT3A receptor (PDB code: 6BE1). The methods used to determine rotational correlation time τc (PDF).
Competing Interests
The authors declare no competing interests.
References
- 1.Endeward B, Butterwick JA, MacKinnon R, and Prisner TF (2009) Pulsed electron-electron double-resonance determination of spin-label distances and orientations on the tetrameric potassium ion channel KcsA, J. Am. Chem. Soc 131, 15246–15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalmas O, Hyde HC, Hulse RE, and Perozo E (2012) Symmetry-constrained analysis of pulsed double electron-electron resonance (DEER) spectroscopy reveals the dynamic nature of the KcsA activation gate, J. Am. Chem. Soc 134, 16360–16369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kinde MN, Chen Q, Lawless MJ, Mowrey DD, Xu J, Saxena S, Xu Y, and Tang P (2015) Conformational Changes Underlying Desensitization of the Pentameric Ligand-Gated Ion Channel ELIC, Structure 23, 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pliotas C (2017) Ion Channel Conformation and Oligomerization Assessment by Site-Directed Spin Labeling and Pulsed-EPR, Methods Enzymol. 594, 203–242. [DOI] [PubMed] [Google Scholar]
- 5.Sahu ID, and Lorigan GA (2018) Site-Directed Spin Labeling EPR for Studying Membrane Proteins, Biomed Res Int 2018, 3248289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeschke G (2012) DEER distance measurements on proteins, Annu. Rev. Phys. Chem 63, 419–446. [DOI] [PubMed] [Google Scholar]
- 7.Battiste JL, and Wagner G (2000) Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data, Biochemistry (Mosc.) 39, 5355–5365. [DOI] [PubMed] [Google Scholar]
- 8.Clore GM, and Iwahara J (2009) Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes, Chem. Rev 109, 4108–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kocman V, Di Mauro GM, Veglia G, and Ramamoorthy A (2019) Use of paramagnetic systems to speed-up NMR data acquisition and for structural and dynamic studies, Solid State Nucl. Magn. Reson 102, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubbell WL, Lopez CJ, Altenbach C, and Yang Z (2013) Technological advances in site-directed spin labeling of proteins, Curr. Opin. Struct. Biol 23, 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eliezer D (2012) Distance information for disordered proteins from NMR and ESR measurements using paramagnetic spin labels, Methods Mol. Biol 895, 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang B, Bushweller JH, and Tamm LK (2006) Site-directed parallel spin-labeling and paramagnetic relaxation enhancement in structure determination of membrane proteins by solution NMR spectroscopy, J. Am. Chem. Soc 128, 4389–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clore GM, Tang C, and Iwahara J (2007) Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement, Curr. Opin. Struct. Biol 17, 603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi P, Li D, Li J, Chen HW, Wu FM, Xiong Y, and Tian CL (2012) Application of Site-Specific F-19 Paramagnetic Relaxation Enhancement to Distinguish two Different Conformations of a Multidomain Protein, Journal of Physical Chemistry Letters 3, 34–37. [Google Scholar]
- 15.Matei E, and Gronenborn AM (2016) (19)F Paramagnetic Relaxation Enhancement: A Valuable Tool for Distance Measurements in Proteins, Angew. Chem. Int. Ed. Engl 55, 150–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitevski-LeBlanc JL, and Prosser RS (2012) Current applications of 19F NMR to studies of protein structure and dynamics, Prog. Nucl. Magn. Reson. Spectrosc 62, 1–33. [DOI] [PubMed] [Google Scholar]
- 17.Larda ST, Simonetti K, Al-Abdul-Wahid MS, Sharpe S, and Prosser RS (2013) Dynamic equilibria between monomeric and oligomeric misfolded states of the mammalian prion protein measured by 19F NMR, J. Am. Chem. Soc 135, 10533–10541. [DOI] [PubMed] [Google Scholar]
- 18.Kinde MN, Bondarenko V, Granata D, Bu W, Grasty KC, Loll PJ, Carnevale V, Klein ML, Eckenhoff RG, Tang P, and Xu Y (2016) Fluorine-19 NMR and computational quantification of isoflurane binding to the voltage-gated sodium channel NaChBac, Proc. Natl. Acad. Sci. U. S. A 113, 13762–13767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilf RJ, and Dutzler R (2008) X-ray structure of a prokaryotic pentameric ligand-gated ion channel, Nature 452, 375–379. [DOI] [PubMed] [Google Scholar]
- 20.Pan J, Chen Q, Willenbring D, Yoshida K, Tillman T, Kashlan OB, Cohen A, Kong XP, Xu Y, and Tang P (2012) Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine, Nat. Commun 3, 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagelueken G, Ward R, Naismith JH, and Schiemann O (2012) MtsslWizard: In Silico Spin-Labeling and Generation of Distance Distributions in PyMOL, Appl Magn Reson 42, 377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solomon I, and Bloembergen N (1956) Nuclear Magnetic Interactions in the Hf Molecule, J. Chem. Phys 25, 261–266. [Google Scholar]
- 23.Cavanagh J, Fairbrother W, Palmer AI, Rance M, and Skelton N (1996) Protein NMR Spectroscopy: Principles and Practice, Academic Press, San Diego. [Google Scholar]
- 24.Tillman TS, Alvarez FJ, Reinert NJ, Liu C, Wang D, Xu Y, Xiao K, Zhang P, and Tang P (2016) Functional Human alpha7 Nicotinic Acetylcholine Receptor (nAChR) Generated from Escherichia coli, J. Biol. Chem 291, 18276–18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basak S, Gicheru Y, Rao S, Sansom MSP, and Chakrapani S (2018) Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3A receptor, Nature 563, 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottstein D, Reckel S, Dotsch V, and Guntert P (2012) Requirements on paramagnetic relaxation enhancement data for membrane protein structure determination by NMR, Structure 20, 1019–1027. [DOI] [PubMed] [Google Scholar]
- 27.Garbuio L, Bordignon E, Brooks EK, Hubbell WL, Jeschke G, and Yulikov M (2013) Orthogonal spin labeling and Gd(III)-nitroxide distance measurements on bacteriophage T4-lysozyme, J. Phys. Chem. B 117, 3145–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kucher S, Korneev S, Tyagi S, Apfelbaum R, Grohmann D, Lemke EA, Klare JP, Steinhoff HJ, and Klose D (2017) Orthogonal spin labeling using click chemistry for in vitro and in vivo applications, J. Magn. Reson 275, 38–45. [DOI] [PubMed] [Google Scholar]
- 29.Chen Q, Kinde MN, Arjunan P, Wells MM, Cohen AE, Xu Y, and Tang P (2015) Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC, Scientific reports 5, 13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee D, Hilty C, Wider G, and Wuthrich K (2006) Effective rotational correlation times of proteins from NMR relaxation interference, J. Magn. Reson 178, 72–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


