Abstract
Background
The function of 4-coumarate-CoA ligases (4CL) under abiotic stresses has been studied in plants, however, limited is known about the 4CL genes in cotton (G. hirsutum L.) and their roles in response to drought stress.
Results
We performed genome-wide identification of the 4CL genes in G. hirsutum and investigated the expression profiles of the identified genes in various cotton tissues and in response to stress conditions with an aim to identify 4CL gene(s) associated with drought tolerance. We identified 34 putative 4CL genes in G. hirsutum that were clustered into three classes. Genes of the same class usually share a similar gene structure and motif composition. Many cis-elements related to stress and phytohormone responses were found in the promoters of the Gh4CL genes. Of the 34 Gh4CL genes, 26 were induced by at least one abiotic stress and 10 (including Gh4CL7) were up-regulated under the polyethylene glycol (PEG) simulated drought stress conditions. Virus-induced gene silencing (VIGS) in cotton and overexpression (OE) in Arabidopsis thaliana were applied to investigate the biological function of Gh4CL7 in drought tolerance. The Gh4CL7-silencing cotton plants showed more sensitive to drought stress, probably due to decreased relative water content (RWC), chlorophyll content and antioxidative enzyme activity, increased stomatal aperture, and the contents of malondialdehyde (MDA) and hydrogen peroxide (H2O2). Arabidopsis lines overexpressing Gh4CL7, however, were more tolerant to drought treatment, which was associated with improved antioxidative enzyme activity, reduced accumulation of MDA and H2O2 and up-regulated stress-related genes under the drought stress conditions. In addition, compared to their respective controls, the Gh4CL7-silencing cotton plants and the Gh4CL7-overexpressing Arabidopsis lines had a ~ 20% reduction and a ~ 10% increase in lignin content, respectively. The expression levels of genes related to lignin biosynthesis, including PAL, CCoAOMT, COMT, CCR and CAD, were lower in Gh4CL7-silencing plants than in controls. Taken together, these results demonstrated that Gh4CL7 could positively respond to drought stress and therefore might be a candidate gene for improvement of drought tolerance in cotton.
Conclusion
We characterized the 4CL gene family in upland cotton and revealed a role of Gh4CL7 in lignin biosynthesis and drought tolerance.
Keywords: Gossypium hirsutum, 4CL, Transgenic Arabidopsis, Drought stress, VIGS
Background
Cotton is an important cash crop in many developing countries and frequently grown in dry lands or on supplementary irrigation [1], because agricultural water consumption can no longer be expanded thanks to water competition among domestic, industrial and agricultural users [2]. The quantity and quality of fiber produced by cotton plants are directly related to water available to them during their developmental stages. When suffered from water deficits, especially during the period of flowering and fructification, cotton would show significant yield loss, sometimes up to 50% reduction compared to those that have been irrigated [3, 4].
In the long-term evolutionary history, plants have formed a complex gene-metabolic network to accommodate a variety of environmental changes. As an important metabolite, lignin plays vital roles in defense against biotic and abiotic stresses [5–7]. Lignin is synthesized through the phenylpropane pathway. 4-coumarate-CoA ligases (4CL, EC 6.2.1.12) is the main branch point enzyme of the phenylpropanoid pathway, which catalyzes cinnamic acid to generate corresponding CoA thioesters [8]. Products of 4CL subsequently serve as substrates of various oxygenases, reductases and transferases for biosynthesis of lignin, flavonoids, anthocyanins, aurones, stilbenes, coumarins, suberin, cutin, sporopollenin, and others [9]. The 4CL gene family has been characterized in many plants, such as Arabidopsis [10], rice [11] and aspen [12]. Genes of the 4CL family in dicots can be classified into two distinct groups, type I and type II [8]. Type I genes are mainly involved in lignin biosynthesis whereas type II genes are involved in biosynthesis of phenylpropanoids other than lignin. Some additional genes containing the same conserved motifs of 4CLs and showing high similarity with the 4CL proteins are classified as 4CL-like genes [13].
Studies have shown that 4CL genes play momentous roles in plants, such as regulation of growth and development, protection against biotic and abiotic stresses [11, 14, 15]. In Arabidopsis, At4CL1, At4CL2, and At4CL4 were found to be involved in lignin formation, the 4 cl1 4 cl2 double and 4 cl1 4 cl2 4 cl3 triple mutant plants exhibited a dwarf and bushy phenotype [10]. In rice, Os4CL2 was specifically expressed in anthers and induced by UV irradiation [16]. Plagiochasma appendiculatum thallus plants showed downregulation of Pa4CL1 when treated with abscisic acid (ABA), and showed upregulation of Pa4CL1 when treated with salicylic acid and MeJA [17]. In both poplars and Arabidopsis, the expression levels of 4CL genes were induced by salt stress and wounding [7]. The 4CL-like genes may also play a role in response to abiotic stresses [18, 19]. Overexpression of Fm4CL-like1 in tobacco increased drought tolerance due to increasing lignin accumulation and the activities of antioxidant enzymes, and upregulating the expression levels of stress-related genes [19]. Nevertheless, our knowledge of the 4CL gene family in cotton is very limited.
To gain insights into the cotton 4CL gene family and its role in abiotic stress tolerance, in this study, we did genome-wide identification of 4CL genes in G. hirsutum and analysed their expression changes in response to various abiotic stresses based on publically available RNA-seq datasets. We identified 34 putative Gh4CL genes in G. hirsutum and 26 of them were found to be induced by at least one stress, including Gh4CL7 that was up-regulated under polyethylene glycol (PEG) osmotic stress. We further investigated the function of Gh4CL7 in drought tolerance by silencing its expression in cotton using virus-induced gene silencing (VIGS) and generating transgenic Arabidopsis plants overexpressing Gh4CL7. Our results indicated that Gh4CL7 functions positively in response to drought stress and is a potential candidate gene for improving drought resistance of cotton by genetic engineering.
Results
Genome-wide identification and bioinformatics analysis of Gh4CL genes
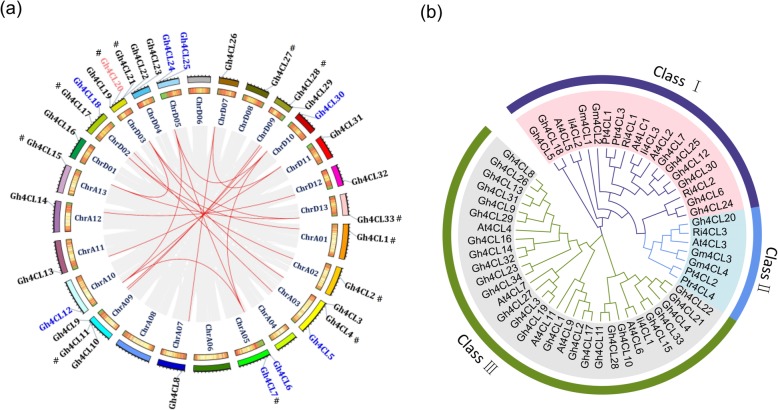
Using the approach described in Materials and Methods, we identified 34 Gh4CL genes in G. hirsutum. They are randomly distributed on 22 chromosomes and an unanchored scaffold that was not assigned to a particular chromosome (Fig. 1a, Table 1). We named them Gh4CL1 to Gh4CL34 based on their chromosomal location. Two pairs of Gh4CL genes, Gh4CL10/11 and Gh4CL21/22, are in tandem configuration on chromosomes A09 and D03, respectively. Segmental duplication could be involved in generation of 12 Gh4CLs based on MCScanX analysis. Phylogenetic analysis using the 34 Gh4CL genes and 4CL genes from A. thaliana, G. max, P. tremuloides, P. trichocarpa, R. idaeus, and I. tinctoria showed that they were clustered into three groups (Fig. 1b). The Ka/Ks ratio of each homologous/paralogous Gh4CL pair is < 1 (Additional file 2: Table S1), suggesting that these Gh4CL genes have experienced purifying selective pressure during their evolution history to eliminate deleterious mutations.
Fig. 1.
Phylogenetic, chromosomal distribution and interchromosomal relationships of the Gh4CL genes. a Chromosomal distribution of Gh4CL genes. The gray lines indicate all synteny blocks in the genome of G. hirsutum and the red lines indicate interchromosomal relationships of Gh4CLs. The first circle represents individual chromosomes length. The second circle represents the gene density of each chromosome by color-coded short bars. The chromosome number were shown inside the circle. b The neighbor-joining phylogenetic tree was generated using 4CL protein sequences from G. hirsutum and six other plants
Table 1.
Characteristics of the 34 Gh4CL genes in G. hirsutum
| Genes | Gene ID number | Genbank accession | Length/aa | MW/KDa | pI | subcellular localization |
|---|---|---|---|---|---|---|
| Gh4CL1 | Gh_A01G0870 | MN897786 | 552 | 60.36 | 8.66 | mitochondrial inner membrane |
| Gh4CL2 | Gh_A02G0862 | MN897787 | 549 | 59.56 | 8.70 | microbody (peroxisome) |
| Gh4CL3 | Gh_A03G1962 | MN897788 | 458 | 50.34 | 6.35 | chloroplast thylakoid membrane |
| Gh4CL4 | Gh_A03G0249 | MN897789 | 517 | 57.08 | 8.32 | plasma membrane |
| Gh4CL5 | Gh_A03G1091 | MN897790 | 555 | 61.09 | 5.73 | plasma membrane |
| Gh4CL6 | Gh_A05G3997 | MN897791 | 543 | 59.56 | 5.67 | plasma membrane |
| Gh4CL7 | Gh_A05G1188 | MN897792 | 543 | 59.51 | 5.84 | endoplasmic reticulum (membrane) |
| Gh4CL8 | Gh_A07G0468 | MN897793 | 555 | 61.02 | 8.75 | microbody (peroxisome) |
| Gh4CL9 | Gh_A09G2180 | MN897794 | 557 | 60.84 | 6.80 | microbody (peroxisome) |
| Gh4CL10 | Gh_A09G1370 | MN897795 | 129 | 14.11 | 9.77 | endoplasmic reticulum (membrane) |
| Gh4CL11 | Gh_A09G1371 | MN897796 | 459 | 50.53 | 8.54 | plasma membrane |
| Gh4CL12 | Gh_A10G0456 | MN897797 | 543 | 59.61 | 5.30 | endoplasmic reticulum (membrane) |
| Gh4CL13 | Gh_A11G0333 | MN897798 | 550 | 60.16 | 7.65 | microbody (peroxisome) |
| Gh4CL14 | Gh_A12G1362 | MN897799 | 521 | 56.42 | 8.30 | microbody (peroxisome) |
| Gh4CL15 | Gh_A13G2028 | MN897800 | 545 | 60.08 | 8.97 | plasma membrane |
| Gh4CL16 | Gh_D01G1584 | MN897801 | 524 | 54.36 | 7.16 | plasma membrane |
| Gh4CL17 | Gh_D02G0989 | MN897802 | 576 | 62.98 | 8.66 | microbody (peroxisome) |
| Gh4CL18 | Gh_D02G1514 | MN897803 | 555 | 61.10 | 5.80 | plasma membrane |
| Gh4CL19 | Gh_D03G1840 | MN897804 | 572 | 63.08 | 6.63 | chloroplast thylakoid membrane |
| Gh4CL20 | Gh_D03G0479 | MN897805 | 573 | 62.04 | 5.54 | plasma membrane |
| Gh4CL21 | Gh_D03G1317 | MN897806 | 452 | 49.64 | 8.40 | plasma membrane |
| Gh4CL22 | Gh_D03G1318 | MN897807 | 517 | 56.97 | 8.79 | plasma membrane |
| Gh4CL23 | Gh_D04G0054 | MN897808 | 568 | 61.94 | 6.17 | microbody (peroxisome) |
| Gh4CL24 | Gh_D05G3934 | MN897809 | 540 | 59.26 | 5.90 | endoplasmic reticulum (membrane) |
| Gh4CL25 | Gh_D05G1366 | MN897810 | 543 | 59.42 | 5.71 | endoplasmic reticulum (membrane) |
| Gh4CL26 | Gh_D07G0533 | MN897811 | 555 | 61.10 | 8.78 | microbody (peroxisome) |
| Gh4CL27 | Gh_D08G1670 | MN897812 | 559 | 61.33 | 6.60 | microbody (peroxisome) |
| Gh4CL28 | Gh_D09G1372 | MN897813 | 546 | 60.17 | 8.66 | plasma membrane |
| Gh4CL29 | Gh_D09G2385 | MN897814 | 557 | 60.94 | 6.50 | microbody (peroxisome) |
| Gh4CL30 | Gh_D10G0473 | MN897815 | 543 | 59.52 | 5.42 | endoplasmic reticulum (membrane) |
| Gh4CL31 | Gh_D11G0389 | MN897816 | 550 | 60.01 | 7.19 | microbody (peroxisome) |
| Gh4CL32 | Gh_D12G1488 | MN897817 | 547 | 59.54 | 8.74 | microbody (peroxisome) |
| Gh4CL33 | Gh_D13G2431 | MN897818 | 545 | 60.04 | 8.97 | plasma membrane |
| Gh4CL34 | Gh_Sca008083G01 | MN897819 | 568 | 61.77 | 6.09 | microbody (peroxisome) |
The protein length of Gh4CLs is between 129 and 576 amino acids (aa) with ORF from 390 to 1731 bp, molecular weight from 14.11 to 63.08 KD, and pI from 5.3 to 9.77. Most Gh4CLs seem to be associated with various biomembranes based on subcellular localization prediction (Table 1). Analyses of gene structures and motifs showed that each Gh4CL has multiple exons, introns and motifs (Additional file 1: Figure S1; Additional file 2: Table S2). All the Gh4CL proteins have two structural domains, a putative AMP-binding domain “SSGTTGLPKG” (Box I) and a conserved domain “GEICIRG” (Box II) (Additional file 1: Figure S2).
Cis-elements in combination with transcription factors regulate the transcription level of a gene. To identify potential cis-elements involved in regulation of transcription of Gh4CL genes, we scanned cis-elements in the promoter region (2 kb upstream of ATG) of each Gh4CL gene using the online tool PlantCARE [20]. Many Gh4CL genes harbored plant hormone-responsive and/or stress-responsive elements, including ABA responsive elements (ABREs), auxin responsive elements (AuxRR-core, TGA-elements and TGA-box), MeJA-responsive elements (CGTCA-motif, TGACG-motif), gibberellin-responsive elements (TATC-box, GARE-motif and P-box), salicylic acid responsive elements (TCA-elements), low-temperature responsive elements (LTR), defense and stress responsiveness elements (TC-rich repeats) and drought-responsive elements (MBS) (Additional file 1: Figure S3).
Tissue specific expression patterns of Gh4CL genes
The expression patterns in various tissues provide clue for the possible biological functions of genes of interest. We thus analysed the transcript abundance of the Gh4CL genes in different tissues (root, stem, leaves, flower, ovule and fibers at 5, 10, 15 and 20 days-post-anthesis (DPA)) under normal growth conditions using the publically available RNA-seq data (BioProject Accession: PRJNA248163) [21]. We found that 10 Gh4CL genes were expressed in all the tested tissues [base on fragments per kilobase of transcript per million mapped reads (FPKM) ≥ 1], and 4 genes (Gh4CL3, Gh4CL5, Gh4CL18 and Gh4CL27) showed weak or no expression in all tissues analysed (Fig. 2). In addition, 8 Gh4CL genes (Gh4CL2, Gh4CL4, Gh4CL8, Gh4CL12, Gh4CL17, Gh4CL24, Gh4CL29 and Gh4CL30) were highly expressed (FPKM ≥20) in stem, with the highest expression level observed for Gh4CL17 (FPKM ≥84) and 6 genes (Gh4CL7–8, Gh4CL12, Gh4CL20 and Gh4CL30–31) were strongly expressed in leaves, with the highest expression level observed for Gh4CL20 (FPKM ≥202).
Fig. 2.
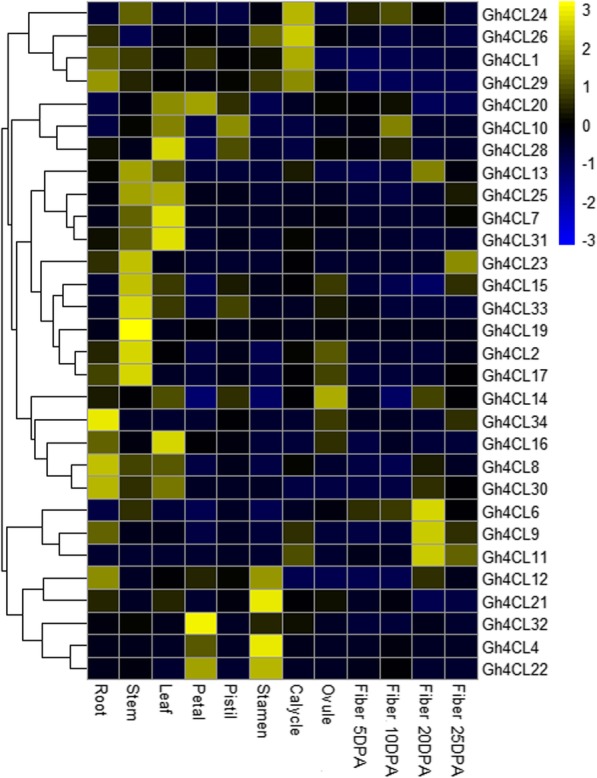
Expression pattern of the Gh4CL genes in different tissues of G. hirsutum based on the RNA-seq data. The heatmap was generated by using the pheatmap package
Expression analysis of Gh4CL genes under different abiotic stress conditions
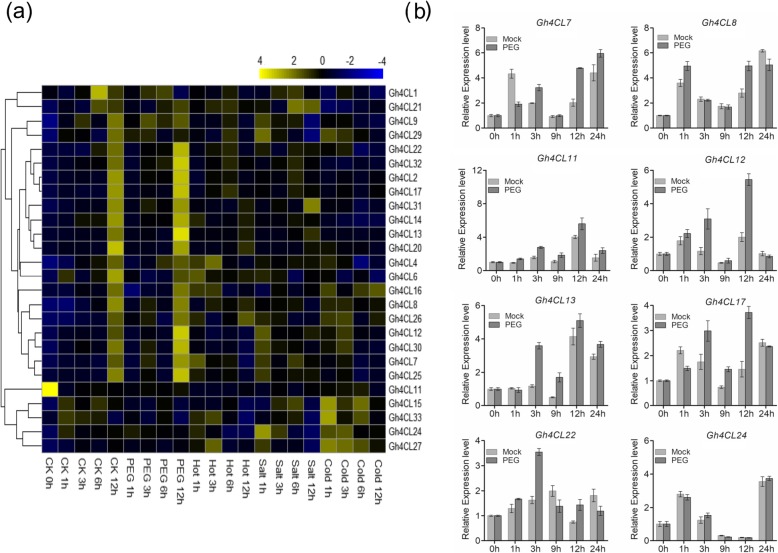
Since 4CL genes are capable of responding to biotic and abiotic stresses in various plant species, we further investigated the transcript abundance of the Gh4CL genes under cold, heat, PEG and salt stresses using the transcriptomic data of G. hirsutum (BioProject Accession: PRJNA248163) [21]. We found that 26 Gh4CL genes were induced significantly by one or more stresses, and the remaining 8 Gh4CL genes (Gh4CL3, Gh4CL5, Gh4CL10, Gh4CL18–19, Gh4CL23, Gh4CL28 and Gh4CL34) were not induced by either of the four stresses (Fig. 3a). Comparing the four stress conditions, more Gh4CL genes showed altered expression in response to salinity, cold and heat stresses than to PEG stress. Notably, ten Gh4CL genes (Gh4CL2, Gh4CL7–9, Gh4CL11–13, Gh4CL17, Gh4CL22, Gh4CL25 and Gh4CL31) showed increased expression (treatment FPKM/control FPKM ≥1.5) in response to PEG stress over the 3 h to 12 h time period. To verify these results, we investigated the expression patterns of the selected Gh4CL genes under the simulated drought treatment using quantitative real-time polymerase chain reaction (qRT-PCR). As shown in Fig. 3b, the expression levels of Gh4CL7–8, Gh4CL12–13, Gh4CL17, Gh4CL22 and Gh4CL24 were up-regulated in cotton leaves over the time period of 3 h to 24 h after PEG stress, consistent with the RNA-seq based results.
Fig. 3.
Expression profile of the Gh4CL genes in response to different abiotic stresses. a Heatmap showing the relative expression levels of each Gh4CL gene in each treatment showing at the bottom of the image. b The relative expression level of the selected Gh4CL genes under 20% PEG stress based on qRT-PCR. The GhUBQ7 gene was used as internal control. Error bars represents the standard deviation calculated from three biological replicates
Gh4CL7 plays an important role in lignin biosynthesis
Based on the above analysis results of promoter cis-elements and expression patterns under drought stress, three Gh4CL genes, including Gh4CL7, Gh4CL8 and Gh4CL13, were considered as candidate genes with a potential role in the regulation of drought stress response in cotton. In this study, we selected Gh4CL7 for further functional analysis by silencing its expression in cotton and overexpression in Arabidopsis thaliana.
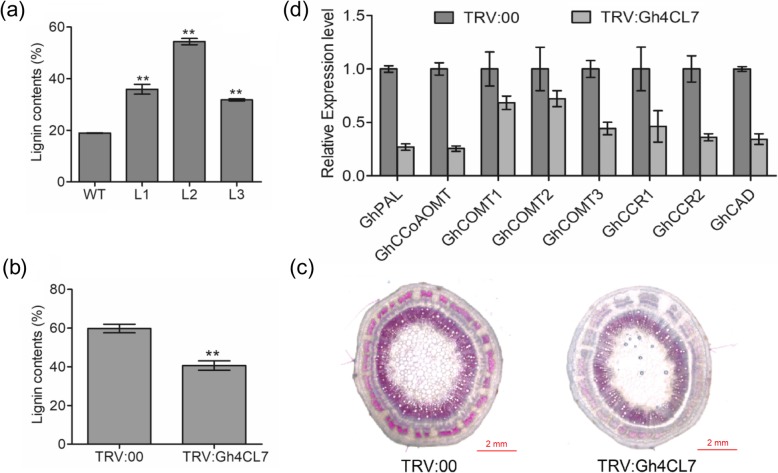
We used VIGS to silence the expression of Gh4CL7 using the TRV vector (TRV:Gh4CL7; Additional file 1: Figure S4). TRV:GhCHLI was used as a positive control of the VIGS experiment (Additional file 1: Figure S5). Arabidopsis thaliana plants overexpressing Gh4CL7 (Gh4CL7-OE) were obtained by the floral dip method. Gh4CL7 belongs to class I whose genes have been shown to regulate lignin biosynthesis [10, 22]. We thus first investigated whether or not Gh4CL7 is also involved in lignin biosynthesis by comparing the lignin contents of the Gh4CL7-OE Arabidopsis lines and TRV:Gh4CL7 cotton plants with that of their corresponding control plants. The lignin content increased by approximately 10% in the Gh4CL7-OE lines compared with wild-type (WT) plants (Fig. 4a), while decreased by approximately 20% in the TRV:Gh4CL7 plants compared with the TRV:00 plants (Fig. 4b). Additionally, the stem of the TRV:Gh4CL7 plants were sectioned and stained with phloroglucinol-HCl to detect the presence of lignin (Fig. 4c). We found that the stem section of the TRV:Gh4CL7 plants with reduced lignin content exhibited light red color, but the TRV:00 plants displayed typically purple-red color after phloroglucinol-HCl staining. These results suggested that Gh4CL7 is related to lignin synthesis. We also analysed the expression level of the phenylpropane pathway genes that are related to lignin biosynthesis, including GhPAL, GhCCoAOMT1, GhCOMT1, GhCOMT2, GhCOMT3, GhCCR1, GhCCR2, and GhCAD. The relative expression level of these genes were lower in the TRV:Gh4CL7 plants than in TRV:00 (Fig. 4d), indicating that Gh4CL7 could affect the accumulation of lignin by regulating the transcription level of these downstream genes of the lignin biosynthesis pathway.
Fig. 4.
Analyses of lignin contents and the expression level of genes related to lignin biosynthesis. a-b Comparison of lignin contents in Gh4CL7-OE Arabidopsis plants (a) and in Gh4CL7-silencing cotton plants (b). c Stem sections from the TRV:Gh4CL7 and TRV:00 cotton plants stained with phloroglucinol-HCl. d The relative expression levels of genes related to lignin biosynthesis. Data were represented as the mean ± SE of three biological replicates; asterisks indicate levels of significance based on t-test (* P < 0.05, ** P < 0.01)
Silencing of Gh4CL7 compromises tolerance of cotton to drought stress
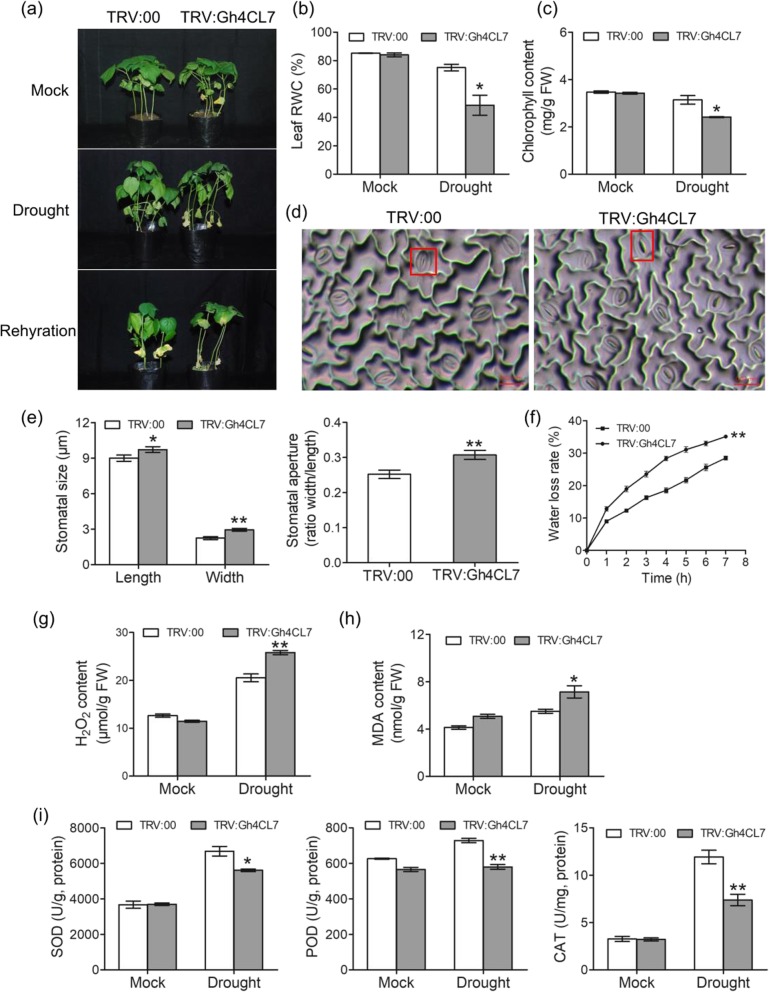
Phenotypic difference between the TRV:Gh4CL7 and TRV:00 plants was observed after 20 days of water deficiency treatment. Compared to the TRV:00 plants, the TRV:Gh4CL7 plants displayed severe wilting and yellowing leaves (Fig. 5a), consistent with a lower leaf relative water content (RWC) (Fig. 5b) and a decrease chlorophyll contents (Fig. 5c). Besides, it was also found that the size and the ratio of width to length of stomata significantly increased in the TRV:Gh4CL7 plants (Fig. 5d-f), which might accelerate the transpiration rate under drought conditions, consistent with the observed higher water loss relative (WLR) (Fig. 5g). The hydrogen peroxide (H2O2) content and malondialdehyde (MDA) level were measured to reflect the cell damage or injury in TRV:Gh4CL7 and TRV:00 plants. During drought stress, the TRV:Gh4CL7 plants accumulated more MDA (Fig. 5h) and H2O2 (Fig. 5i) compared to the TRV:00 plants. The activities of superoxide dismutase (SOD), peroxidase (POD) and catalase (CAT) in the TRV:Gh4CL7 and TRV:00 plants were also measured to explore the function of Gh4CL7 in the modulation of antioxidant enzymes (Fig. 5j). As expected, under drought stress conditions, the TRV:Gh4CL7 plants displayed a significantly reduced activity of SOD, POD and CAT as compared to the TRV:00 plants. Additionally, six stress-related genes (GhABI4, GhABF4, GhLEA14, GhRD22, GhRD29 and GhNCED1) were down-regulated in the TRV:Gh4CL7 plants after drought treatment (Additional file 1: Figure S6). These results suggested that silencing of Gh4CL7 decreases tolerance of cotton to drought stress.
Fig. 5.
Drought tolerance analysis of the Gh4CL7-silencing cotton plants. a Representative phenotypes of the TRV:00 (CK) and TRV:Gh4CL7 (VIGS) plants after 3 weeks of drought treatment. b-c Leaf RWC and chlorophyll contents of CK and VIGS plants under drought stress. d-e Comparison of stomata in the CK and VIGS plants. f Water loss rate of detached leaves from the CK and VIGS plants. g-h Comparison of H2O2 and MDA contents in the CK and VIGS plants. i Activities of antioxidative enzymes in the CK and VIGS plants. Mock, normal growth conditions; drought, 3 weeks of water deficit conditions. Statistical significance was determined by the student test. * and ** represent significant at P < 0.05 and P < 0.01, respectively
Overexpression of Gh4CL7 in Arabidopsis enhances drought tolerance
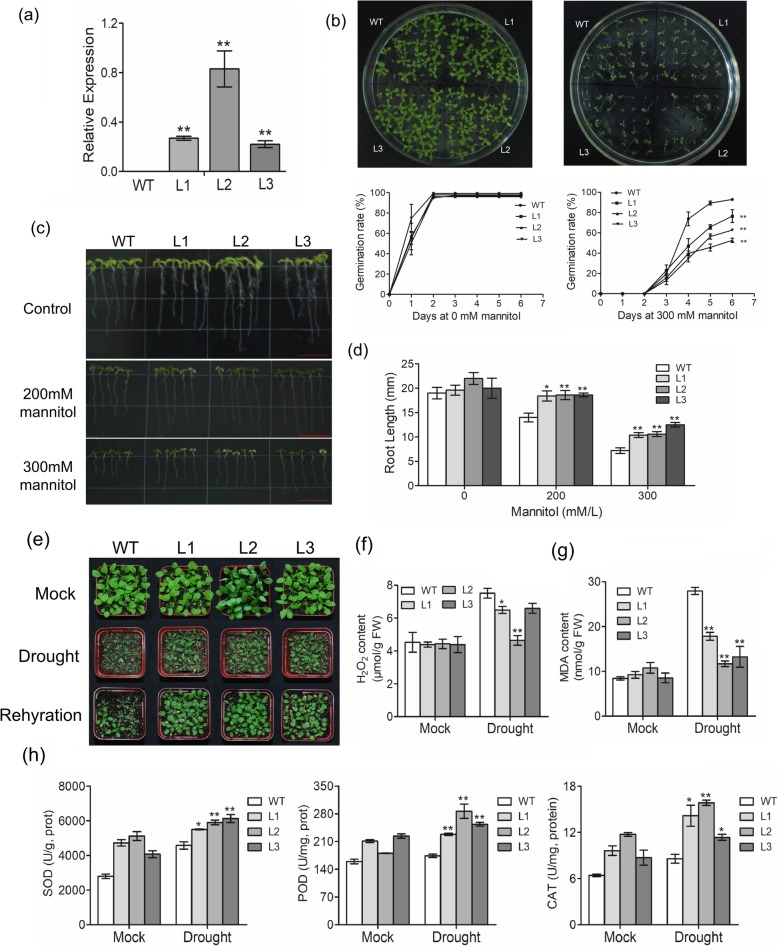
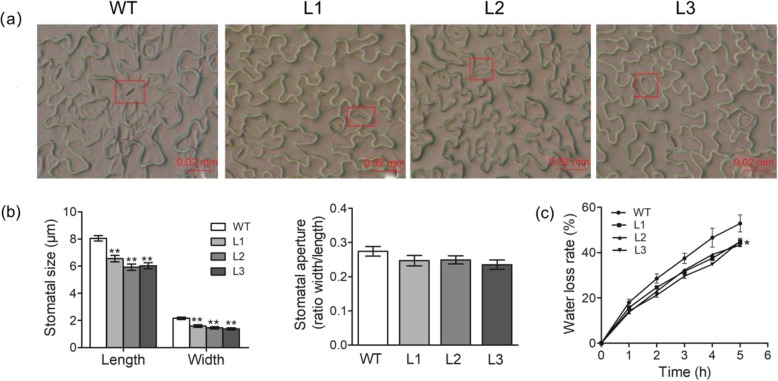
We further investigated the function of Gh4CL7 in response to drought stress using Arabidopsis plants overexpressing Gh4CL7. Three independent Gh4CL7-OE lines that showed an elevated level of Gh4CL7 (Fig. 6a) were selected for phenotyping under the drought stress conditions. Compared to the WT plants, the three Gh4CL7-OE lines had a decreased germination rate (Fig. 6b), but showed a significantly increased root length under the mannitol stress conditions (Fig. 6c, d). Three-weeks-old seedlings of Gh4CL7-OE and WT were used for water deficiency treatment. No obvious phenotypic difference was observed between Gh4CL7-OE and WT by the mock treatment. However, the Gh4CL7-OE plants showed much less damage than WT after 10 days of water deficiency (Fig. 6e). Under drought stress conditions, the H2O2 content and MDA level in the Gh4CL7-OE plants were relatively lower than that in WT (Fig. 6f-g), but the SOD, POD and CAT activities were significantly higher (Fig. 6h). Additionally, the size and the ratio of width to length of stomata significantly decreased in the Gh4CL7-OE Arabidopsis plants (Fig. 7a-b), consistent with a lower WLR observed in those plants (Fig. 7c). To further elucidate the possible mechanism of Gh4CL7 in response to drought stress, the transcript levels of four known ABA-responsive genes (AtRD29B, AtRD22, AtABI4, AtCOR15A) and two ABA-biosynthesis genes (AtNCED3 and AtNCED5) were analyzed in the Gh4CL7-OE lines and WT plants after drought stress treatment. The qRT-PCR data showed that the expression levels of these genes were induced in Gh4CL7-OE, but not or just slightly induced in WT by drought stress (Additional file 1: Figure S7). These results indicated that overexpression of Gh4CL7 could enhance the tolerance of transgenic Arabidopsis plants to drought stress.
Fig. 6.
Drought tolerance analysis of the Gh4CL7-overexpressing Arabidopsis plants. a The relative expression level of Gh4CL7 in three independent Gh4CL7-OE Arabidopsis plants. b Germination rate of the Gh4CL7-OE Arabidopsis seeds on 1/2 MS supplemented with 0 and 300 mM mannitol. c-d Root elongation of the 6-days-old Gh4CL7-OE Arabidopsis seedlings on 1/2 MS supplemented with 0, 200, and 300 mM mannitol. Bar = 1 cm. e Phenotypic assay of the Gh4CL7-OE and WT Arabidopsis plants during drought treatment. f-h The contents of H2O2 and MDA and antioxidative enzyme activity in Gh4CL7-OE and WT plants under normal and water deficit conditions. Ten-days-old seedlings were transplanted to soil and regularly watered for 2 weeks. For drought treatment, irrigation was terminated for 2 weeks. For the rehydration treatment, the plants were re-watered 3 days after the drought treatment
Fig. 7.
Stomatal opening in the WT and Gh4CL7-OE Arabidopsis plants under drought stress. a-b Comparison of stomata in the WT and Gh4CL7-OE plants after 14 days of water-withholding stress. c WLR of detached leaves from WT and Gh4CL7-OE Arabidopsis plants. Data were represented as the mean ± SE of three biological replicates. Statistical significance was determined by the student test. * and ** represent significant at P < 0.05 and P < 0.01, respectively
Discussion
The 4CL gene family has been characterized in several plants, including Arabidopsis thaliana, Populus trichocarpa, Oryza sativa and Glycine max [11, 23–25]. Genes of this family have been reported to function not only in plant growth and development [16, 22, 26], but also in response to biotic and abiotic stresses [7, 27]. However, no comprehensive analysis of the 4CL genes has been documented in G. hirsutum. In this study, we did genome-wide identification of 4CL genes in G. hirsutum and investigated their expression profiles in various tissues and under different stress conditions with an aim to identify 4CL gene(s) with a potential role in stress tolerance. In total, 34 Gh4CL members were identified in the G. hirsutum genome (Table 1). In other plant species, such as A. thaliana, 4CL genes were divided into three classes, i.e. class I, class II and 4CL-like [13]. The 34 Gh4CL genes could also be clustered into three classes. We named the 4CL-like class as class III, which contains the largest number of Gh4CL genes (in total 25) together with At4CL6–9, At4CL11, and Ii4CL1 (Fig. 1b). The class III Gh4CLs cannot catalyze any of the hydroxycinnamic acid substrates into the corresponding CoA esters, their function is different from that of class I (related to the lignin biosynthesis) and class II (related to the biosynthesis of flavonoids) 4CL genes [13, 18, 28]. Multiple sequence alignments revealed that all 4CL-like proteins contained similar structural components, e.g. conserved Box I and Box II domains without known specific biochemical function (Additional file 1: Figure S2) [29]. Gene structure analysis showed that Gh4CL genes of the same class share a similar intron-exon structure (Fig. 2b), similar to the observations made in other gene families [30–32].
Cis-elements located in the promoter region of genes play key roles in the developmental and environmental regulation of gene expression [33]. According to cis-element analysis, the promoter regions of Gh4CL genes possess elements related to stress responses, such as ABRE, TC-rich, LTR, MBS, TGA-element, TCA-element, CGTCA-motif and TGACG-motif [34–36], suggesting a potential role of the Gh4CL genes with these cis-element(s) in regulationof stresses, such as drought, salt, heat, ABA, and low temperature. 4CL genes have been shown to be involved in response to stresses in other plants [14, 27]. Based on transcriptome data, 26 Gh4CL genes were differentially expressed between the stress treatment and the mock, and many Gh4CL genes were induced by drought, including Gh4CL7 (Fig. 3a). The promoter of Gh4CL7 contains an MBS cis-acting element, which may be associated with up-regulation of Gh4CL7 upon drought treatment (Fig. 3b). In addition, the Gh4CL genes showed different expression profiles in different tissues (Fig. 2), suggesting that they perhaps play a broad range of roles in cotton growth and development.
Abiotic stresses often disrupt the balance between reactive oxygen species (ROS) production and clearance in cells, leading to increased ROS concentrations and oxidative damage to biofilms, proteins, DNA, and RNA, thereby inhibiting plant growth and development [37, 38]. Therefore, scavenging ROS is essential for plants to resist abiotic stresses. H2O2 is one of the ROS, and its over accumulation in the plant cell can cause oxidative damage, while a low level of H2O2 concentration correlates with drought tolerance [39]. In the Gh4CL7 gene-silenced plants, the H2O2 content was found to be increased significantly under drought stress, so was the MDA level, an indicator of ROS destructive effects [40]. SOD, POD, and CAT are antioxidant enzymes in plant cells which scavenge the toxic ROS and lead to enhanced tolerance under stress conditions [41]. We found that the activities of SOD, POD, and CAT were lower in the TRV:Gh4CL7 plants than in the TRV:00 plants indicating that the reduced ability of the Gh4CL7 silenced plants to scavenge ROS that might have led to membrane damages and chlorophyll content reduction. On the other hand, the Arabodopsis plants overexpressing Gh4CL7 had a lower level of H2O2 and MDA and a higher activity of antioxidant enzymes (SOD, POD, and CAT) compared to WT under the drought stress conditions. These results are consistent with previous finding that Fm4CL had a role in drought tolerance by modulating the level of ROS [19].
Drought stress affects crop yield and quality through its negative influence on seed germination, seedling growth, photosynthesis, and transpiration [1]. We found that A. thaliana plants overexpressing Gh4CL7 had longer roots but a lower germination rate than WT plants under the osmotic stress conditions, suggesting that Gh4CL7 played a negative role in seed germination, but a positive role in promoting root elongation under the osmotic stress conditions. Breeding crops with thriving and deeper root systems is the goal of geneticists and breeders, because it can increase productivity of crops under drought conditions [42]. Longer roots might be a result of changed ABA signaling pathway, which plays a crucial role in root development under drought stress [43, 44], as well as many other factors related to drought responses, including stomata closure and stress-gene regulation [45, 46]. Under drought stress conditions, the closed stomata can decrease transpiration rate that helps plants to resist adverse environmental conditions. Our results showed that the size of stomatas was bigger in the TRV:Gh4CL7 plants, suggesting a positive role of Gh4CL7 in reducing transpiration rate that allows cotton to maintain a more favourable water balance, and effectively improves drought tolerance. This was supported by the observation of a higher WLR in leaves of the TRV:Gh4CL7 plants than those of WT plants.
Lignin is the second largest polymer in plants after cellulose [47]. It provides mechanical support to plants by increasing cell wall hardness and enhancing compressive strength of cells [48, 49]. We found that repressing the expression level of Gh4CL7 in G. hirsutum reduced the lignin content and led to a reduction in drought resistance, consistent with the result of rice plants with a decreased lignin content being more prone to drought stress [50]. Studies in Fraxinus mandshurica also showed that decreased lignin content resulted in drought resistance reduction [19]. The hydrophobicity of lignin is thought to have an inhibitory effect on the transpiration of plant tissue under drought conditions [51], that could be the reason for Arabidopsis plants overexpressing Gh4CL7 with an increased level of lignin content being more resistant to drought.
Conclusions
The findings of this study demonstrate that the Gh4CL7-silencing cotton plants had an increased sensitivity of drought stress while overexpressing Gh4CL7 enhanced tolerance of drought stress in Arabidopsis. Gh4CL7 conferred tolerance to drought stress by increasing lignin content, improving the antioxidant system, closing stomata, and up-regulating the transcription levels of ABA-responsive genes. Although the exact mechanism of Gh4CL7-mediated drought tolerance is still yet to be uncovered, our results provide evidence for the role of Gh4CL7 in combating drought stress.
Methods
Identification of the 4CL family genes in Gossypium hirsutum
The annotated protein sequences of G. hirsutum [21] were downloaded from CottonGen (https://www.cottongen.org/). The hidden Markov model file corresponding to the AMP-binding domain (PF00501) was downloaded from the Pfam protein family database (http://pfam.xfam.org/) and used as query (P < 0.001) [52] to search for the 4CL genes in G. hirsutum with HMMER 3.0 [53]. The existence of the AMP-binding domain sequences was examined using the Pfam, SMART (http://smart.embl-heidelberg.de/), and National Center for Biotechnology Information (NCBI) Conserved Domains (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) databases [54, 55].
Gene structure, conserved motif and promoter analyses
The length, molecular weight (MW), and isoelectric point (pI) of the identified Gh4CL proteins were calculated using the ExPasy website tools (http://web.expasy.org/protparam/) [56]. PSORT software (https://psort.hgc.jp/) was used for predicting subcellular localization [57]. Gene Structure Display Server 2.0 (GSDS, http://gsds.cbi.pku.edu.cn/) was used for intron and exon analysis [58]. The conserved motifs in the Gh4CL protein sequences were identified using the Multiple Expectation Maximization for Motif Elicitation (MEME) program (version 5.0.5, http://meme-suite.org/tools/meme) [59]. The potential cis-elements in the promoter sequences (up to 2000-bp upstream ATG) of Gh4CL genes were identified using the PlantCARE program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Phylogenetic tree, chromosomal distribution and syntenic relationship analyses
The multiple sequence alignment of Gh4CLs was done by Clustal X [60] and DNAMAN (version 5.2.2). The 4CL homologous protein sequences of Arabidopsis thaliana (At4CL1: OAP14948; At4CL2: OAP07084; At4CL3: AEE34324; At4CL4: AY376731; At4CL5: AY250839; At4CL7: AY376733; At4CL9: AF360250 At4CL11: AY376735), Glycine max (Gm4CL1: AF279267; Gm4CL2: AF002259; Gm4CL3: AF002258; Gm4CL4: X69955), Rubus idaeus (Ri4CL1: AF239687; Ri4CL2: AF239686; Ri4CL3: AF239685), Populus tremuloides (Pt4CL1: U12012; Pt4CL2: U12013), and Isatis tinctoria (Ii4CL1: ADG46006; Ii4CL2: KC430622; Ii4CL3: KC430623) were downloaded from the NCBI (http://www.ncbi.nlm.nih.gov/, accessed on 7 May 2018) and used for the phylogenetic tree analysis by using the neighbor joining method (NJ) in MEGA 6.0 [61] with 1000 repetitions for the bootstrap test.
All the Gh4CL genes were mapped to G. hirsutum chromosomes, based on their physical location information, using TBtools [62]. Two or more homologous/paralogous 4CL genes located at a chromosomal region of < 200 kb were considered to be generated by tandem duplication events [63]. Multiple Collinearity Scan toolkit (MCScanX) was used to analyze the gene duplication events with the default parameters [64]. Non-synonymous (Ka) and synonymous (Ks) substitutions in each paralogous Gh4CL gene pair were calculated using KaKs_Calculator 2.0 [65].
Vector construction and genetic transformation
To generate Arabidopsis overexpressing lines, the coding sequence of Gh4CL7 was amplified using PrimeSTAR DNA polymerase (TaKaRa, Tokyo, Japan) with the gene-specific forward and reverse primers and ligated into the pCAMBIA2300 vector driven by the CaMV35S promoter. The expression vector pCAMBIA2300-Gh4CL7 was transformed into Agrobacterium tumefaciens strain GV3101. Arabidopsis ecotype Col-0 (wild-type, provided by Microbiology Institute of Chinese Academy of Sciences, Beijing, China) was used in genetic transformation by the floral dip method [66]. The harvested T0 generation seeds were selected on 1/2 Murashige and Skoog (MS) medium with 50 mg/L kanamycin, and the resistant plants were further validated by PCR. Single-copy lines with a segregation ratio of 3:1 were selected and planted until T3 generation. The transgenic Arabidopsis OE lines were grown in a growth chamber with 16 h light/8 h dark scheme, and the growth temperature was set at around 23 °C [67].
The pTRV1 and pTRV2 VIGS vectors were kindly provided by Prof. Longfu Zhu of Huazhong Agricultural University. A 287-bp fragment from the coding sequence of Gh4CL7 was amplified from cDNA of G. hirsutum cultivar stem and subsequently cloned into pTRV2 using BamH1 and Kpn1 double digestions to generate the TRV:Gh4CL7 vector. After PCR and double digestion confirmation, the TRV:Gh4CL7 construct was transformed into A. tumefaciens strain GV3101 by electroporation. G. hirsutum cv. Junmian-1 (provided by the Cotton Research Institute of Shihezi University, Shihezi, Xinjiang Province, China) was used in VIGS and was grown in pots filled with soil mix (3:1, humus:vermiculite) and placed in growth room at 24 ± 1 °C with 16 h light/8 h dark (200 μmol/m2/s photon flux density). VIGS was done as previously described by Xiong and colleagues [67].
Drought tolerance assays
PEG and natural drought treatments were conducted to investigate the function of Gh4CL7 in response to osmotic and drought stresses. For the PEG treatments, TRV:Gh4CL7 (VIGS) and TRV:00 (control) plants at three-leaf stage were subjected to stress by 20% PEG6000 (w/v). Leaf samples were collected at 0 (ck), 1, 3, 6, 9, 12, 24 h after treatment for RNA extraction. For the natural drought treatments, TRV:Gh4CL7 and TRV:00 plants were not irrigated for 3 weeks, followed by re-watering once. After 15 days of water deprivation, plant leaves were collected for RNA extraction and determination of physiological parameters.
For analyses of seed germination and root elongation, seeds or 3-day-old seedlings of Gh4CL7 -OE Arabidopsis lines and WT were grown on 1/2 MS plates supplemented with 0 (control), 200, 300 mM mannitol for 6 days. In the vegetative growth stage, 7 days-old transgenic Arabidopsis and WT plants were transplanted into soil and watered once every 3 days for 2 weeks, then kept without irrigation for 2 weeks, followed by re-watering once. After 10 days of drought treatment, the rosette leaves were collected for RNA extraction and determination of physiological parameters.
Determination of drought stress-related physiological parameters
The thiobarbituric acid (TBA) colorimetric method was used to measure the content of MDA according to the instruction of the malondialdehyde quantification kit (Suzhou Comin Biochemistry Co. Ltd., Su Zhou, China). H2O2 concentrations were determined by using the H2O2 determination kit (Suzhou Comin Biochemistry Co. Ltd., Su Zhou, China) by following the manufacturer’s instructions. Measurement of the activities of antioxidative enzymes was performed using 0.1 g leaf sample according to the instructions of the POD, SOD and CAT Assay Kit (Suzhou Comin Biochemistry Co. Ltd., Su Zhou, China). Total contents of chlorophyll were calculated according to the method described by Porra and colleagues [68]. Absorbances at different wavelengths were measured by the U-5100 UV/VIS spectrophotometer (HITACHI, Tokyo, Japan).
Measurements of WLR, RWC and stomatal aperture
For water loss assays, leaves from G. hirsutum and Arabidopsis were immediately weighed and placed in a growth room at room temperature with a humidity level of about 60%. The leaves were weighed once per hour. WLR was estimated as the percentage of fresh weight lost relative to the initial fresh weight [69]. For measurement of the RWC, fresh leaves were detached from plants and their fresh weights (FW) were immediately recorded. Then, the leaves were placed in distilled water for 8 h at 25 °C in the dark and measured the turgid weight (TW). Dry weights (DW) were recorded by drying samples at 65 °C until constant weight. RWC was calculated as (FW - DW)/(TW - DW) × 100 [70].
Stomatal pore area and size were determined using the rapid imprinting technique [71]. The abaxial leaf surfaces were covered with transparent nail polish and air dried at room temperature. The nail polish imprints were made into temporary slices and photographed by Zeiss microscope (SteREO Discovery.V20, Germany) with 300× magnifications. The length and width of stomatal pores were measured using the Image J software and the relative aperture area was calculated based on the ratio of width to length.
Lignin content measurement and histochemical staining
Lignin content was determined by the acetyl bromide method [72] and the phloroglucinol-HCl color-developing method. Transections of stem (above the cotyledons) of the Gh4CL7-silencing cotton plants at 28 days after sowing were made by hand cutting using razor blades. The transverse sections were immersed in 1 mL of 1 M hydrochloric acid for 3 min, followed by transferring into 1 ml 10% phloroglucinol ethanol-hydrochloric solution for 1 min, and visualized immediately under the microscope (Zeiss, SteREO Discovery.V20, Germany) for photographing [73].
RNA extraction and quantitative real-time PCR analysis
To investigate gene expression patterns, total RNA was extracted from leaves of G. hirsutum and Arabidopsis with the EASYspin Plus plant RNA kit (Aidlab, Beijing, China). RNA was reverse transcribed into cDNA using the M-mlv reverse transcript system (TAKARA, Da Lian, China). The qRT-PCR was performed using the Power SYBR Green PCR Master Mixture (Roche, Rotkreuz, Switzerland) on a Light Cycler® 480 II system (Roche, Rotkreuz, Switzerland) under the following conditions: initial pre-incubation at 95 °C for 5 min, followed by 40 cycles at 94 °C for 10 s, 59 °C for 10 s, and 72 °C for 10 s. The relative expression level of genes was analyzed by the 2-△△Ct method. The results were presented as the mean of three biological replications. The G. hirsutum histone3 gene and Arabidopsis EF-lα gene were used as the reference genes. All the primers used in this study were designed using the NCBI primer designing tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/, accessed on 27 August 2018) and listed in Additional file 2: Table S3.
Statistical analyses
Statistical analyses and data plotting were performed using SPSS and Graphpad Prism 5, respectively. ** and * represent significant differences at P < 0.01 and P < 0.05, respectively.
Supplementary information
Additional file 1: Figure S1. Gene structure and motif pattern of the Gh4CL genes. a Structural analysis. The left panel shows the neighbor-joining phylogenetic tree based on the amino acid sequences of the Gh4CLs. The classes I, II and III were marked correspondingly. The right panel shows the exon-intron structure of each Gh4CL gene with exons showing in orange boxes, introns in black lines between exons, and upstream/downstream UTRs in blue boxes. The number indicates the phase of the corresponding introns. The length of the Gh4CL genes is indicted by the scale line at the bottom. b Motif analysis. The motif analysis was performed by the MEME suite. Twenty motifs were detected and are displayed in color coded boxes. The length of proteins is indicted by the scale line at the bottom. Figure S2. Alignment of multiple Gh4CL and selected At4CL domain amino acid sequences. Multiple sequence alignment was performed using Clustal X. Box I and Box II represent the two conserved domains of the Gh4CL proteins. Figure S3. Analysis of cis-elements in the promoter of the Gh4CL genes. The numbers of different cis-elements are presented in the form of bar graphs. Figure S4. Relative expression levels of Gh4CL7 in plants infiltrated with TRV:00 and TRV:Gh4CL7 (n = 5). Total RNA was extracted from leaves at 2 weeks post-infiltration. Transcript levels were determined by qRT-PCR using GhUBQ7 as control. Figure S5. Silencing of the endogenous magnesium chelatase subunit I gene (GhCHLI) in cotton through VIGS. The leaf bleaching phenotype was observed 2 weeks after infiltration in TRV: GhCHLI plants. Figure S6. Transcription levels of stress responsive genes in CK and Gh4CL7-siliencing cotton plants. The normal condition plants were used as controls. GhUBQ7 gene was used as an internal control. All the gene expressions were normalized to the corresponding transcript levels in CK plants at normal condition. All the data represent mean ± SE for three biological replications. Figure S7. Transcription levels of stress responsive genes in transgenic and WT Arabidopsis plants. The normal condition plants were used as controls. AtEF-Lα gene was used as an internal control. All the gene expressions were normalized to the corresponding transcript levels in WT plants at normal condition. All the data represent mean ± SE for three biological replications.
Additional file 2: Table S1. Ka, Ks, Ka/Ks values for the Gh4CL paralogous gene pairs. Table S2. Information of the 20 motifs of Gh4CL proteins. Table S3. The primers used in qRT-PCR analysis.
Acknowledgements
Not applicable.
Abbreviations
- 4CL
4-coumarate-CoA ligases
- VIGS
Virus-induced gene silencing
- OE
Overexpression
- RWC
Relative water content
- MDA
Malondialdehyde
- H2O2
Hydrogen peroxide
- WLR
Water loss relative
- PEG
Polyethylene glycol
- DPA
Days post anthesis
- qRT-PCR
Quantitative reverse transcription polymerase chain reaction
- CAT
Catalase
- POD
Peroxidase
- SOD
Superoxide dismutase
- ROS
Reactive oxygen species
- WT
Wild type
- TRV:Gh4CL7
Gh4CL7 VIGS plants
- TRV:00
Empty vector VIGS plants
- FPKM
Fragments per kilobase of transcript per million mapped reads
Authors’ contributions
S-C S and X-P X designed the experiments. S-C S, X-P X and X-L Z performed the experiments, analysed data and prepared the manuscript. HF contributed with valuable discussions. Q-H Z, Y-J L and JS read and revised the manuscript. All authors provided helpful discussions and approved its final version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No: 31906438, 31360347), the National Key Research and Development Program of China (Grant No: 2016YFD0100200) and the Breeding Program of Shihezi University (Grant No:YZZX201601). The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The sequencing data are available in the NCBI Sequence Read Archive (SRA) database under the accession number PRJNA248163. All other data generated or analyzed during this study are included in this manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jie Sun, Email: sunjie@shzu.edu.cn.
Yan-Jun Li, Email: lyj20022002@sina.com.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-020-2329-2.
References
- 1.Hussain M, Farooq S, Hasan W, Ul-Allah S, Tanveer M, Farooq M, et al. Drought stress in sunflower: physiological effects and its management through breeding and agronomic alternatives. Agric Water Manag. 2018;201:152–166. doi: 10.1016/j.agwat.2018.01.028. [DOI] [Google Scholar]
- 2.Alexandratos N, Bruinsma J. World agriculture towards 2030/2050: the 2012 revision 12–03, 4. Rome: FAO; 2012. [Google Scholar]
- 3.Araújo AE, Silva CAD, Azevedo DMP, et al. Cultivo do algodão irrigado. Série Documentos, Sistemas de Produção. 3. 2003. pp. 1678–8710. [Google Scholar]
- 4.Pasapula V, Shen GX, Kuppu S, Paez-Valencia J, Mendoza M, Hou P, et al. Expression of an Arabidopsis vacuolar H+-pyrophosphatase gene (AVP1) in cotton improves drought- and salt tolerance and increases fibre yield in the field conditions. Plant Biotechnol J. 2011;9:88–99. doi: 10.1111/j.1467-7652.2010.00535.x. [DOI] [PubMed] [Google Scholar]
- 5.Hu Y, Li WC, Xu YQ, Li GJ, Liao Y, Fu FL. Differential expression of candidate genes for lignin biosynthesis under drought stress in maize leaves. J Appl Genet. 2009;50:213–223. doi: 10.1007/BF03195675. [DOI] [PubMed] [Google Scholar]
- 6.Hu Q, Min L, Yang XY, Jin SX, Zhang L, Li YY, et al. Laccase GhLac1 modulates broad-spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis. Plant Physiol. 2018;176:1808–1823. doi: 10.1104/pp.17.01628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang K, Cui H, Cao S, Yan L, Li M, Sun Y. Overexpression of CrCOMT from Carex rigescens increases salt stress and modulates melatonin synthesis in Arabidopsis thaliana. Plant Cell Rep. 2019. 10.1007/s00299-019-02461-7. [DOI] [PubMed]
- 8.Ehlting JR, Büttner DB, Wang Q, Douglas CJ, Somssich IE, Kombrink E. Three 4-coumarate:coenzyme a ligases in arabidopsis thaliana, represent two evolutionarily divergent classes in angiosperms. Plant J. 1999;19:9–20. doi: 10.1046/j.1365-313X.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- 9.Lavhale SG, Kalunke RM. Giri1Structural AP. Functional and evolutionary diversity of 4-coumarate-CoA ligase in plants. Planta. 2018;248:1063–1078. doi: 10.1007/s00425-018-2965-z. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Kim JI, Pysh L, Chapple C. Four isoforms of arabidopsis thaliana 4-coumarate: coa ligase (4cl) have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 2014;169:2409–2415. doi: 10.1104/pp.15.00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gui JS, Shen JH, Li LG. Functional characterization of evolutionarily divergent 4-Coumarate: coenzyme a ligases in Rice. Plant Physiol. 2011;157:574–586. doi: 10.1104/pp.111.178301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi R, Sun YH, Li Q, Heber S, Sederoff R, Chiang VL. Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 2010;51:144–163. doi: 10.1093/pcp/pcp175. [DOI] [PubMed] [Google Scholar]
- 13.Raes J, Rohde A, Christensen JH, VandePeer Y, Boerjan W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003;133:1051–1071. doi: 10.1104/pp.103.026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang CH, Ma T, Luo WC, Xu JM, Liu JQ, Wan DS. Identification of 4CL genes in desert poplars and their changes in expression in response to salt stress. Genes. 2015;6:901–917. doi: 10.3390/genes6030901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naik P, Wang JP, Sederof R, et al. Assessing the impact of the 4CL enzyme complex on the robustness of monolignol biosynthesis using metabolic pathway analysis. PLoS One. 2018;13:e0193896. doi: 10.1371/journal.pone.0193896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun HY, Li Y, Feng SQ, Zou WH, Guo K, Fan CF, et al. Analysis of five rice 4-coumarate: coenzyme a ligase enzyme activity and stress response for potential roles in lignin and favonoid biosynthesis in rice. Biochem Biophys Res Commun. 2013;430:1151–1156. doi: 10.1016/j.bbrc.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 17.Gao S, Yu HN, Xu RX, Cheng AX, Lou HX. Cloning and functional characterization of a 4-coumarate CoA ligase from liverwort Plagi ochasma appendiculatum. Phytochemistry. 2015;111:48–58. doi: 10.1016/j.phytochem.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 18.Di P, Hu YS, Xuan HJ, Xiao Y, Chen JF, Zhang L, et al. Characterisation and the expression profile of 4-coumarate:coa ligase (Ii4CL) from hairy roots of isatis indigotica. Afr J Pharm Pharmaco. 2012;6:2166–2175. [Google Scholar]
- 19.Chen XH, Wang HT, Li XY, Ma K, Zhan YG, Zeng FS. Molecular cloning and functional analysis of 4-Coumarate:CoA ligase 4(4CL-like 1) from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis. BMC Plant Biol. 2019;19:231–247. doi: 10.1186/s12870-019-1812-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Yves VDP, et al. Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang T, Hu Y, Jiang W, et al. Sequencing of allotetraploid cotton (gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 22.Rao GD, Pan X, Xu F, Zhang YZ, Cao S, Jiang XG, et al. Divergent and overlapping function of five 4-Coumarate: coenzyme a ligases from Populus tomentosa. Plant Mol Biol Rep. 2015;33:841–854. doi: 10.1007/s11105-014-0803-4. [DOI] [Google Scholar]
- 23.Lindermayr C, Möllers B, Fliegmann J, Uhlmann A, Lottspeich F, Meimberg H, et al. Divergent members of a soybean (glycine max l.) 4-coumarate:coenzyme a ligase gene family. Eur J Biochem. 2002;269:1304–1315. doi: 10.1046/j.1432-1033.2002.02775.x. [DOI] [PubMed] [Google Scholar]
- 24.Hamberger B, Hahlbrock K. The 4-coumarate:coa ligase gene family in arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. P Natl Acad Sci USA. 2004;101:2209–2214. doi: 10.1073/pnas.0307307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen HC, Song J, Williams CM, Shuford CM, Liu J, Wang JP, et al. Monolignol pathway 4-coumaric acid:coenzyme a ligases in populus trichocarpa: novel specificity, metabolic regulation, and simulation of coenzyme a ligation fluxes. Plant Physiol. 2013;161:1501–1516. doi: 10.1104/pp.112.210971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choudhary EK, Choi B, Cho BK, Kim JB, Park SU, Natarajan S, et al. Regulation of 4CL, encoding 4-coumarate: coenzyme a ligase, expression in kenaf under diverse stress conditions. Plant Omics J. 2013;6:254–262. [Google Scholar]
- 27.Shinde BA, Dholakia BB, Hussain K, Panda S, Meir S, Rogachev I, et al. Dynamic metabolic reprogramming of steroidal glycol-alkaloid and phenylpropanoid biosynthesis may impart early blight resistance in wild tomato (Solanum arcanum Peralta) Plant Mol Biol. 2017;95:411–423. doi: 10.1007/s11103-017-0660-2. [DOI] [PubMed] [Google Scholar]
- 28.Costa MA, Bedger DL, Moinuddin SGA, et al. Characterization in vitro and in vivo of the putativemultigene 4-coumarate: CoA ligase network in Arabidopsis: syringyl lignin and sinapate/sinapyl alcohol derivative formation. Phytochemistry. 2005;66:2072–2091. doi: 10.1016/j.phytochem.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 29.Schneider K, Hövel K, Witzel K, Hamberger B, Schomburg D, Kombrink E, et al. The substrate specificity-determining amino acid code of 4-coumarate: CoA ligase. Proc Natl Acad Sci U S A. 2003;100:8601–8606. doi: 10.1073/pnas.1430550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu G, Guo C, Shan H, Kong H. Divergence of duplicate genes in exon-intron structure. Proc Natl Acad Sci U S A. 2012;109:1187–1192. doi: 10.1073/pnas.1109047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao P, Wang DD, Wang RQ, Kong NN, Zhang C, Yang CH, et al. Genome-wide analysis of the potato Hsp20 gene family: identification, genomic organization and expression profiles in response to heat stress. BMC Genomics. 2018;19:61–73. doi: 10.1186/s12864-018-4443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J, Xu XY, Tian LL, Wang GL, Zhang XY, Guo WZ. Discovery and identification of candidate genes from the chitinase gene family for Verticillium dahliae resistance in cotton. Sci Rep. 2016;6:29022. doi: 10.1038/srep29022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z, Ge XY, Yang ZR, Zhang CJ, Zhao G, Chen EY, et al. Genome-wide identification and characterization of SnRK2gene family in cotton (Gossypium hirsutumL.) BMC Genet. 2017;18:54–68. doi: 10.1186/s12863-017-0517-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Browse J, Howe GA. New weapons and a rapid response against insect attack. Plant Physiol. 2008;146:832–838. doi: 10.1104/pp.107.115683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu WR, Yu YH, Ding JH, Hua ZY, Wang YJ. Characterization of a novel stilbene synthase promoter involved in pathogen and stress inducible expression from Chinese wild Vitis pseudoreticulata. Planta. 2010;231:475–487. doi: 10.1007/s00425-009-1062-8. [DOI] [PubMed] [Google Scholar]
- 36.Li W, Cui X, Meng ZL, Huang XH, Xie Q, Wu H, et al. Transcriptional regulation of Arabidopsis MIR168a and ARGONAUTE1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 2012;158:1279–1292. doi: 10.1104/pp.111.188789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hossain MA, Bhattacharjee S, Armin S-M, Qian PP, Xin W, Li HY, et al. Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: insights from ROS detoxification and scavenging. Front Plant Sci. 2015;6:420–439. doi: 10.3389/fpls.2015.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain G, Gould KS. Are betalain pigments the functional homologues of anthocyanins in plants? Environ Exp Bot. 2015;119:48–53. doi: 10.1016/j.envexpbot.2015.06.002. [DOI] [Google Scholar]
- 39.Parkhi V, Kumar V, Sunilkumar G, Campbell LM, Singh NK, Rathore KS. Expression of apoplastically secreted tobacco osmotin in cotton confers drought tolerance. Mol Breed. 2009;23:625–639. doi: 10.1007/s11032-009-9261-3. [DOI] [Google Scholar]
- 40.Sharma P, Jha AB, Dubey RS, Pessarakli M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J Exp Bot. 2012;12:1–26. [Google Scholar]
- 41.Ullah A, Sun H, Hakim YX, Zhang X. A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol Plant. 2018;162:439–454. doi: 10.1111/ppl.12651. [DOI] [PubMed] [Google Scholar]
- 42.Comas LH, Becker SR, Cruz VMV, Byrne PF, Dierig DA. Root traits contributing to plant productivity under drought. Front Plant Sci. 2013;4:442–458. doi: 10.3389/fpls.2013.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montillet JL, Leonhardt N, Mondy S, Tranchimand S, Rumeau D, Boudsocq M, et al. An abscisic acid-independent oxylipin pathway controls stomata closure and immune defense in Arabidopsis. PLoS Biol. 2013;11:e1001513. doi: 10.1371/journal.pbio.1001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowe JH, Topping JF, Liu J, Lindsey K. Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol. 2016;211:225–239. doi: 10.1111/nph.13882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu Y, Yu D. Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ Exp Bot. 2009;65:35–47. doi: 10.1016/j.envexpbot.2008.07.002. [DOI] [Google Scholar]
- 46.Tian YF, Gu HH, Fan ZX, Shi GY, Yuan JC, Wei F, et al. Role of a cotton endoreduplication-related gene, GaTOP6B, in response to drought stress. Planta. 2019;249:1119–1132. doi: 10.1007/s00425-018-3067-7. [DOI] [PubMed] [Google Scholar]
- 47.Penning BW, Hunter CT, Tayengwa R, Eveland AL, Dugard CK, Olek AT, et al. Genetic resources for maize cell wall biology. Plant Physiol. 2009;151:1703–1728. doi: 10.1104/pp.109.136804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vance C, And TK, Sherwood R. Lignification as a mechanism of disease resistance. Annu Rev Phytopathol. 2003;18:259–288. doi: 10.1146/annurev.py.18.090180.001355. [DOI] [Google Scholar]
- 49.Lin CY, Wang JP, Li QZ, Chen HC, Liu J, Loziuk P, et al. 4-Coumaroyl and caffeoyl shikimic acids inhibit 4-coumaric acid: coenzyme a ligases and modulate metabolic flux for 3-hydroxylation in monolignol biosynthesis of Populus trichocarpa. Mol Plant. 2015;8:176–187. doi: 10.1016/j.molp.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 50.Li WQ, Zhang MJ, Gan PF, Qiao L, Yang SQ, Miao H, et al. CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J. 2017;92:904–923. doi: 10.1111/tpj.13728. [DOI] [PubMed] [Google Scholar]
- 51.Jordan R. Structural changes and associated reduction of hydraulic conductance in roots of Sorghum bicolor L. following exposure to water deficit. Plant Physiol. 1992;99:203–212. doi: 10.1104/pp.99.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR. The pfam protein families database. Nucleic Acids Res. 2014;42:222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finn RD, Clements J, Eddy SR. Hmmer web server: interactive sequence similarit searching. Nucleic Acids Res. 2011;39:29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aron MB, Myra KD, Noreen RG, Shennan L, Farideh C, Lewis YG. Cdd: ncbi's conserved domain database. Nucleic Acids Res. 2015;43:D222. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Letunic I, Bork P. 20 years of the smart protein domain annotation resource. Nucleic Acids Res. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, et al., editors. The proteomics protocols handbook. Totowa: Humana Press; 2005. pp. 571–607. [Google Scholar]
- 57.Paul H, Keun-Joon P, Takeshi O, Naoya F, Hajime H, Adams-Collier CJ, et al. WOLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Hereditas. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- 59.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. Mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen CJ, Xia R, Chen H, He YH. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. 2018. [Google Scholar]
- 63.Holub EB. The arms race is ancient history in arabidopsis, the wildflower. Nat Rev Genet. 2001;2:516–527. doi: 10.1038/35080508. [DOI] [PubMed] [Google Scholar]
- 64.Wang YP, Tang HB, Debarry JD, Tan X, Li JP, Wang XY, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang D, Zhang Y, Zhang Y, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clough SJ, Bent AF. Floral dip: a simplifed method for agrobacterium-mediated transformat of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 67.Xiong XP, Sun SC, Li YJ, Zhang XY, Sun J, Xue F. The cotton WRKY transcription factor GhWRKY70 negatively regulates the defense response against Verticillium dahliae. Crop J. 2019;7:393–402. doi: 10.1016/j.cj.2018.10.005. [DOI] [Google Scholar]
- 68.Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents:verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta. 1989;975:384–394. doi: 10.1016/S0005-2728(89)80347-0. [DOI] [Google Scholar]
- 69.Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, et al. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006;45:309–319. doi: 10.1111/j.1365-313X.2005.02622.x. [DOI] [PubMed] [Google Scholar]
- 70.Sade N, Vinocur BJ, Diber A, Shatil A, Ronen G, Nissan H, et al. Improving plant stress tolerance and yield production: is the tonoplast aquaporin SlTIP2; 2 a key to isohydric to anisohydric conversion? New Phytol. 2009;181:651–661. doi: 10.1111/j.1469-8137.2008.02689.x. [DOI] [PubMed] [Google Scholar]
- 71.Geisler M, Nadeau J, Sack FD. Oriented asymmetric divisions that generate the stomatal spacing pattern in arabidopsis are disrupted by the too many mouths mutation. Plant Cell. 2000;12:2075–2086. doi: 10.1105/tpc.12.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morrison I. A semi-micro method for the determination of lignin and its use in predicting the digestibility of forage crops. J Sci Food Agric. 1972;23:455–463. doi: 10.1002/jsfa.2740230405. [DOI] [PubMed] [Google Scholar]
- 73.Pomar F, Merino F, Barceló AR. O-4-linked coniferyl and sinapyl aldehydes in lignifying cell walls are the main targets of the Wiesner (phloroglucinol-HCl) reaction. Protoplasma. 2002;220:17–28. doi: 10.1007/s00709-002-0030-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Gene structure and motif pattern of the Gh4CL genes. a Structural analysis. The left panel shows the neighbor-joining phylogenetic tree based on the amino acid sequences of the Gh4CLs. The classes I, II and III were marked correspondingly. The right panel shows the exon-intron structure of each Gh4CL gene with exons showing in orange boxes, introns in black lines between exons, and upstream/downstream UTRs in blue boxes. The number indicates the phase of the corresponding introns. The length of the Gh4CL genes is indicted by the scale line at the bottom. b Motif analysis. The motif analysis was performed by the MEME suite. Twenty motifs were detected and are displayed in color coded boxes. The length of proteins is indicted by the scale line at the bottom. Figure S2. Alignment of multiple Gh4CL and selected At4CL domain amino acid sequences. Multiple sequence alignment was performed using Clustal X. Box I and Box II represent the two conserved domains of the Gh4CL proteins. Figure S3. Analysis of cis-elements in the promoter of the Gh4CL genes. The numbers of different cis-elements are presented in the form of bar graphs. Figure S4. Relative expression levels of Gh4CL7 in plants infiltrated with TRV:00 and TRV:Gh4CL7 (n = 5). Total RNA was extracted from leaves at 2 weeks post-infiltration. Transcript levels were determined by qRT-PCR using GhUBQ7 as control. Figure S5. Silencing of the endogenous magnesium chelatase subunit I gene (GhCHLI) in cotton through VIGS. The leaf bleaching phenotype was observed 2 weeks after infiltration in TRV: GhCHLI plants. Figure S6. Transcription levels of stress responsive genes in CK and Gh4CL7-siliencing cotton plants. The normal condition plants were used as controls. GhUBQ7 gene was used as an internal control. All the gene expressions were normalized to the corresponding transcript levels in CK plants at normal condition. All the data represent mean ± SE for three biological replications. Figure S7. Transcription levels of stress responsive genes in transgenic and WT Arabidopsis plants. The normal condition plants were used as controls. AtEF-Lα gene was used as an internal control. All the gene expressions were normalized to the corresponding transcript levels in WT plants at normal condition. All the data represent mean ± SE for three biological replications.
Additional file 2: Table S1. Ka, Ks, Ka/Ks values for the Gh4CL paralogous gene pairs. Table S2. Information of the 20 motifs of Gh4CL proteins. Table S3. The primers used in qRT-PCR analysis.
Data Availability Statement
The sequencing data are available in the NCBI Sequence Read Archive (SRA) database under the accession number PRJNA248163. All other data generated or analyzed during this study are included in this manuscript.