Abstract
Duchenne muscular dystrophy is caused by mutations in the dystrophin-encoding DMD gene. While Duchenne is most commonly caused by large intragenic deletions that cause frameshift and complete loss of dystrophin expression, in-frame deletions in DMD can result in the expression of internally truncated dystrophin proteins and may be associated with a milder phenotype known as Becker muscular dystrophy. In this study, we describe two individuals with large in-frame 5’ deletions (exon 3–23 and exon 3–28) that remove the majority of the N-terminal region, including part of the actin binding and central rod domains. Both patients had progressive muscle weakness during childhood but are observed to have a relatively mild disease course compared to typical Duchenne. We show that in muscle biopsies from both patients, truncated dystrophin is expressed at the sarcolemma. We have additionally developed a patient-specific fibroblast-derived cell model, which can be inducibly reprogrammed to form myotubes that largely recapitulate biopsy findings for the patient with the exon 3–23 deletion, providing a culture model for future investigation of this unusual case. We discuss these mutations in the context of previously reported 5’ in-frame DMD deletions and relevant animal models, and review the spectrum of phenotypes associated with these deletions.
Keywords: dystrophin, Duchenne muscular dystrophy, Becker muscular dystrophy, dystrophin-glycoprotein complex, utrophin
1. Introduction
Duchenne muscular dystrophy (DMD) is a degenerative muscle disorder caused by mutations in DMD, the gene encoding the approximately 427 kDa dystrophin protein. Dystrophin is part of the dystrophin-glycoprotein complex (DGC), which provides structural stability at the sarcolemma during muscle contraction by linking the internal cell cytoskeleton and external extracellular matrix (1). Mutations in DMD result in the reduction or absence of the DGC at the sarcolemma, leading to sarcolemmal instability and progressive muscle damage.
With a prevalence of about 1 in 5,000 live male births, DMD is the most common pediatric muscular dystrophy (2). Mutations in DMD result in a large range of phenotypic variability. Most DMD mutations are large deletions that lead to frameshift and complete loss of dystrophin expression, resulting in the severe DMD phenotype: typically presenting by age 3–5 years, loss of ambulation around age 10–12 years, upper limb dysfunction in the late teen years and death due to cardiac or respiratory failure by their 20–30s. In some instances, large multi-exon in-frame mutations in DMD can result in the expression of an internally deleted dystrophin protein and lead to a milder form of dystrophy called Becker muscular dystrophy (BMD). BMD is clinically defined by loss of ambulation at 16 years or later, although BMD is a more heterogeneous group with some patients reported with loss of ambulation as late as the 6th or 7th decade of life (3, 4). Some individuals with large internal deletions are clinically described as having intermediate muscular dystrophy (IMD) to connote milder DMD or more severe BMD with loss of ambulation occurring between ages 13–16 years.
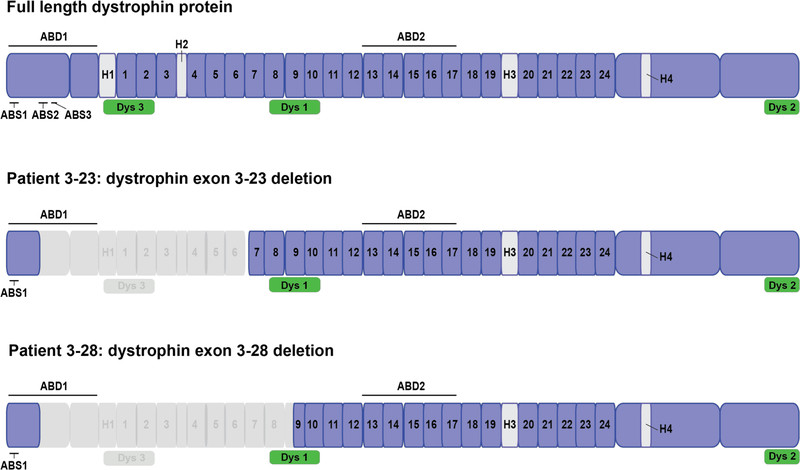
The main muscle isoform of dystrophin (dp427m) contains four major domains: a N-terminal actin binding domain (ABD1), a central rod domain containing a second actin binding domain (ABD2), a cysteine-rich domain (CR), and a C-terminal domain (CT) (Fig. 1) (reviewed in (5)). ABD1 consists of two calponin homology domains, which contain three actin-binding sites called ABS1, ABS2 and ABS3 (6). The central rod domain is the largest domain of dystrophin and contains 24 spectrin-like repeats (R1-R24), four hinge regions, and ABD2. In this study, we describe two individuals with large in-frame N-terminal deletions that remove a portion of ABD1, the hinge 1 and hinge 2 regions and a portion of the rod repeat domains (R1-R6, encoded by exons 10 to 23, for Patient 3–23 and R1-R9, encoded by exons 10 to 30, for Patient 3–28). Both patients were initially clinically diagnosed with DMD based on early childhood presentations, progressive proximal weakness and genetic findings predicting loss of ABD1 and protein stability at age of onset. However, over ten years of observation, both have followed a milder course of disease more consistent with BMD, but at the severe end of the BMD spectrum. Each of these unusual mutations have been previously reported, though limited information on clinical course and dystrophin expression is available. One patient with an exon 3–23 DMD deletion was described as DMD phenotype, and 3 patients with 3–28 deletion have been reported with DMD or IMD phenotype (7, 8). In order to better characterize the consequences of these mutations on dystrophin expression and disease severity, we performed muscle biopsies on both patients to assess amount of expressed mutant dystrophin protein that localizes to the sarcolemma. For Patient 3–23, we have additionally developed a patient fibroblast derived cell model that can be inducibly reprogrammed to myotubes in culture to form myotubes with similar dystrophin expression to the biopsy (9), validating this tool for future exploration of the functionality of this truncated dystrophin protein.
Figure 1. Schematic of dystrophin N-terminal deletions in Patients 3–23 and 3–28.
Dystrophin protein domains are shown in light/dark purple (Actin Binding Domain 1 and 2, Hinge 1–4, rod domains repeats 1–24). Predicted epitopes for antibodies against dystrophin are shown in green (NCL-Dys1, NCL-Dys2 and NCL-Dys3). Exon deletions 3–23 and 3–28 result in the loss of the majority of the N-terminal region, including part of ABD1, and a portion of the rod repeat domains (R1-R6 for Patient 3–23 and R1-R9 for Patient 3–28).
2. Materials and Methods
2.1. Muscle Biopsy Immunohistochemistry
Muscle biopsies were mounted in optimal cutting temperature compound (OCT - Tissue-Tek) and flash frozen in liquid nitrogen–cooled isopentane, and stored at - 80 °C until further processing. Transverse 10 μm cryosections were blocked with 3% bovine serum albumin (BSA) in phosphate buffered saline (PBS) for 30 min at room temperature. Avidin/biotin blocking kit (Vector Laboratories) was used according to manufacturer’s instructions. Sections were incubated in primary antibody in PBS at 4 °C overnight with the following antibodies: dystrophin rod domain (NCL-Dys1, 1:100, Leica Biosystems), dystrophin C-terminal (NCL-Dys2, 1:100, Leica Biosystems), dystrophin N-terminal (NCL-Dys3, 1:100, Leica Biosystems), utrophin (MANCHO3, 1:10, Developmental Hybridoma Studies Bank), α-SG (IVD3; 1:50; Developmental Hybridoma Studies Bank), and spectrin (PA1–46007, 1:200, Thermo Fisher Scientific). The location of the dystrophin antibody epitopes is depicted in Figure 1. Primary antibodies were detected by biotinylated anti–mouse (BA-9200; 1:500; Vector Laboratories). Fluorescein–conjugated avidin D (A-2001; 1:500; Vector Laboratories) was used to detect secondary antibodies. All sections were mounted in Vectashield (Vector Laboratories) and visualized using an Axioplan 2 fluorescence microscope with Axiovision 3.0 software (Carl Zeiss Inc).
2.2. Muscle Biopsy Immunoblot Analysis
Equal quantities of protein samples were resolved on 4–20% precise protein gels by SDS-PAGE (4–20% Precise Protein Gels, Thermo Fisher Scientific) and transferred to nitrocellulose membrane (Millipore). Membranes were blocked for 1 hour in 5% nonfat dry milk in tris buffered saline (TBS) with 0.2% Tween 20 and incubated in primary antibodies overnight at 4 °C. Incubations were performed with the following primary antibodies: dystrophin C-terminal (NCL-Dys2, 1:100, Leica Biosystems) and GAPDH (MAB374, 1:50,000, Chemicon). Horseradish peroxidase–conjugated anti-rabbit IgG and anti-mouse IgG (GE Healthcare) secondary antibodies were used at 1:5,000 dilutions in 5% nonfat dry milk and incubated at RT for 3 hours. Immunoblots were developed using enhanced chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate; Thermo Fisher Scientific).
2.3. Cell Culture and Transdifferentiation
Primary human dermal fibroblasts from patients were cultured from skin punch and were expanded in growth media (Dulbecco’s Modified Eagle Media with 15% fetal bovine serum, 1% nonessential amino acids, 1% penicillin/streptomycin) (9, 10). Fibroblasts were immortalized with lentiviral hTERT and subsequently transduced with previously described tamoxifen-inducible lentiviral MyoD-ERT (11) to create stable cell lines referred as inducible directly reprogrammed myotubes (iDRM). At approximately 70% confluence, the medium was changed for growth medium supplemented with 5uM of 4-OH tamoxifen. One day later, cells were differentiated into multinucleated myotubes by switching to a differentiation media composed of DMEM/Ham’s F-10 (v/v) supplemented with 1% penicillin/streptomycin, 2% horse serum, 2% of Insulin-transferrin-selenium solution, and 1uM of 4-OH tamoxifen for 7 days.
2.4. Cell Culture Immunofluorescence
Cells were grown as described above. After myotube differentiation, cells were fixed with acetone as previously described (12, 13). Dystrophin was detected using Dys1, Dys2 and Dys3 (Rod domain, C-term or N-term, 1:20 dilution Leica). Alpha-sarcoglycan was detected using IVD3 (1/20 dilution, Developmental Studies Hybridoma Bank (DSHB)). Utrophin was detected using Macho 3 (1:20 dilution, DSHB). migG1 and migG2b (BD Bioscience 554121 and Biologend 4002202) rabbit igG (Santa Cruz 2027) were used as controls. Secondary antibodies were used at a dilution of 1:500 (goat anti-mouse A32723 and A11004 and goat anti-rabbit A32731and A11036, Life Technologies). Pictures were obtained using a Zeiss microscope at a 20x or 40x magnification and processed with Axiovision software and/or ImageJ software.
3. Case Presentation and Results
3.1. Case presentation: Patient 3–23, exon 3–23 deletion
Patient 3–23 was diagnosed with DMD with an initial presentation of toe-walking, calf hypertrophy, hyperCKemia, and proximal weakness at age 5 years. Initial CK level was 12,030 U/L. Genetic Del/Dup testing of the DMD gene from Athena Diagnostics identified a de novo exon 3–23 deletion. The mutation was confirmed by exome sequencing and by a custom comparative genomic hybridization (CGH) microarray using 14,022 probes tiled across the DMD gene (14). CGH results indicated breakpoints located at approximately chrX:32395941 and chrX:32806573 (hg18 reference). The 5’ breakpoint is about 20kb away from exon 3 and the 3’ breakpoint is about 1500bp from exon 23.
Patient 3–23 was initiated on 0.75mg/kg prednisone daily, which was modified to deflazacort at 48mg Saturday/Sunday at age 14 years, because of his milder than typical DMD disease progression. At age 9 years, he was able to run, and presented no difficulties with rising from the floor. At age 15 years functional measures include a timed rise from floor of 10 seconds and timed 10M walk/run of 5 seconds. He lost the ability to rise from the floor at age 16, with a timed 10M walk of 9 seconds, and has maintained independent ambulation (now at 17 years of age) with a Northstar Ambulatory Assessment of 22/34 at age 16 years. He had no cardiac insufficiency at age 16, but forced vital capacity at age 16 was at 60% of normal with no breathing support. Muscle biopsy of the vastus lateralis was obtained at age 9 years and revealed diffuse perimysial and endomysial fibrosis, mild fiber size variation, and slightly increased central nuclei.
3.2. Case presentation: Patient 3–28, exon 3–28 deletion
Patient 3–28 was diagnosed with DMD at age 4 years with proximal weakness and an initial CK of 16,028 U/L. Genetic Del/Dup testing of the DMD gene from Athena Diagnostics identified an exon 3–28 deletion, and he was initiated on 0.9 mg/kg deflazacort daily at age 4 years. After an initial improvement in muscle function, he has had progressive proximal weakness with a timed rise from floor of 8.1 seconds and 10M walk/run of 5.1 seconds at age 11 years. At 12 years of age his timed 10M walk/run was 6.1 seconds and overall North Star Ambulatory Assessment was 21/34. He has normal cardiac and pulmonary function at age 11 years. Muscle biopsy of the vastus lateralis performed at age 7 revealed fiber size variation, increase in central nucleation, and rare necrotic fibers and slight endomysial fibrosis. Patient mutation was confirmed by exome sequencing, but precise deletion boundaries were not determined.
3.3. Truncated dystrophins are expressed at the sarcolemma in Patients 3–23 and 3–28
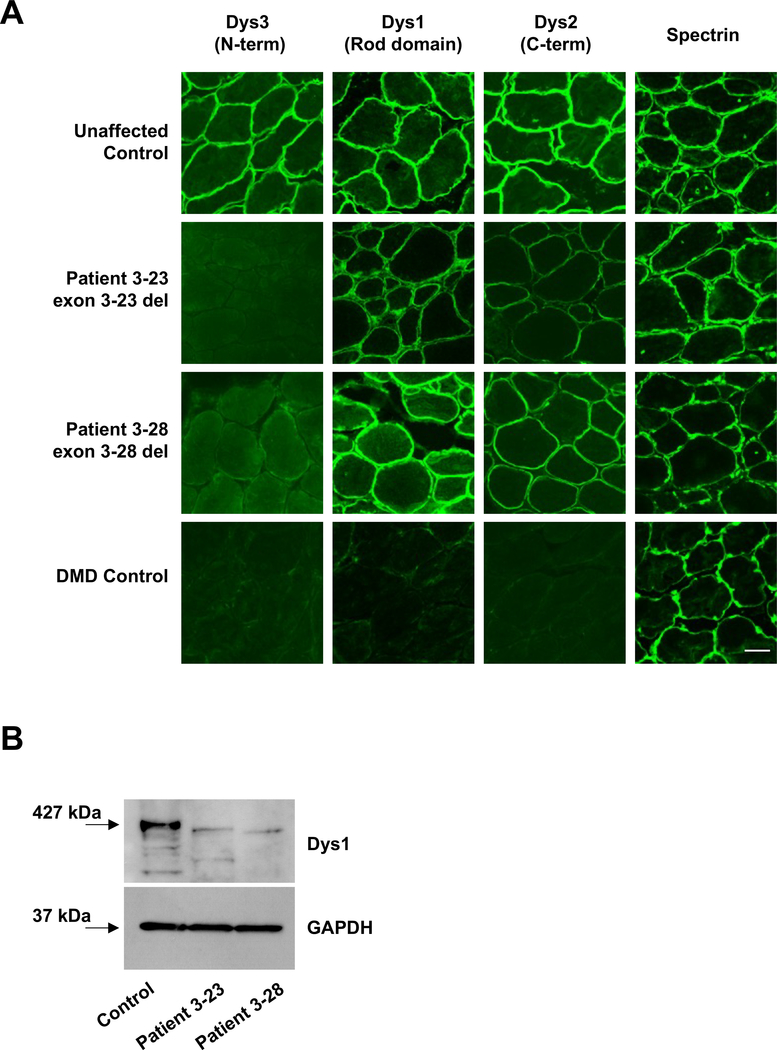
In order to determine whether the relatively mild phenotypes in Patient 3–23 and Patient 3–28 correlate with the expression of truncated dystrophin at the sarcolemma, we analyzed serial transverse skeletal muscle cryosections using a panel of antibodies that bind to the N-terminus (Dys3), central rod domain (Dys1), and C-terminus (Dys2) of dystrophin (Fig. 1). Dys1 staining revealed sarcolemmal localization of the truncated proteins in both individuals (Fig. 2A). While the epitope for the Dys1 antibody has been reported to be located within exons 26–30, data from patient samples indicates that exon 29 is essential for Dys1 antibody binding (15, 16). Exon 29 is present in both patients, which may account for the retained Dys1 signal at the sarcolemma. In both patients, Dys3 antibody (epitope exons 10–12) signal was not detected at the sarcolemma, which was expected since the Dys3 epitope is within the genetically deleted region of dystrophin.
Figure 2. Truncated dystrophins are expressed at the sarcolemma in biopsies from two patients with large 5’ in-frame DMD deletions.
(A) Transverse sections of biopsies were stained with antibodies against dystrophin and spectrin. Bar, 100 μm. (B) Immunoblot of control and patient biopsies with antibodies against dystrophin (Dys1) and GAPDH. Equal amounts of protein (30μg) were loaded in each lane. Predicted sizes at 427kDa (control, full length dystrophin), 314 kDa (Patient 3–23, 1023 amino acid deletion) and 287 kDa (Patient 3–28, 1276 amino acid deletion).
To further examine the relative abundance of dystrophin in the patient biopsies, we performed immunoblot analysis on total muscle extracts prepared from control and patient biopsies (Fig. 2B). The rod-domain antibody Dys1 revealed robust expression of full-length dystrophin polypeptide (427kDa) in control muscle. As expected, full-length dystrophin protein was not detected in the patient samples. Rather, truncated proteins consistent with the predicted protein sizes of approximately 314 kDa (Patient 3–23) and 287 kDa (Patient 3–28) were observed, consistent with the production of stable dystrophin proteins.
3.4. Utrophin expression is upregulated in Patients 3–23 and 3–28
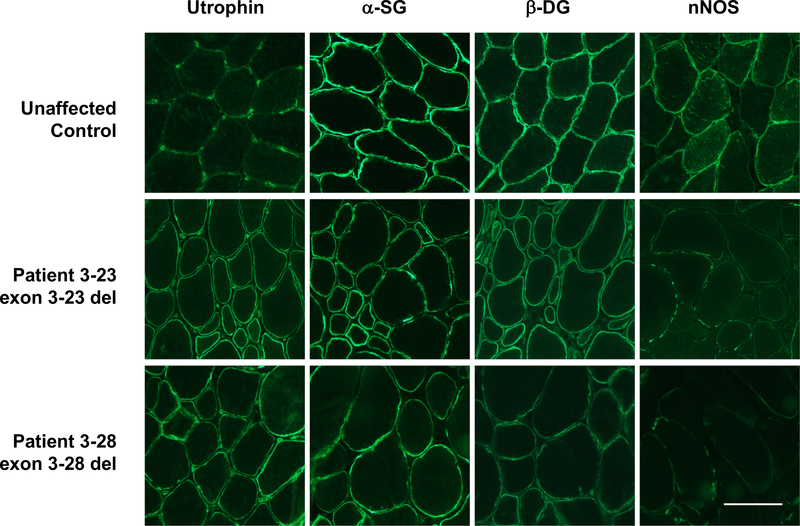
We used immunohistochemistry to evaluate expression and localization of other DGC proteins at the sarcolemma in both patients (Fig. 3). When dystrophin is not expressed, proteins associated with the DGC are typically reduced or absent at the sarcolemma, while sarcolemmal utrophin is increased. Consistent with this pattern, utrophin expression was upregulated in both Patients 3–23 and 3–28 relative to healthy unaffected muscle. Alpha-sarcoglycan and β-dystroglycan, proteins associated with both dystrophin and utrophin at the sarcolemma, are present in both Patients 3–23 and 3–28, although decreased relative to control muscle. By contrast, both biopsies had only very low levels of nNOS, a protein found in complex with dystrophin but not utrophin. These observations are consistent with the expected pathophysiology for these patients.
Figure 3. Sarcolemmal protein expression patterns are consistent with a BMD phenotype for both patients.
Transverse sections of biopsies were stained with antibodies against utrophin, α-sarcoglycan, β-dystroglycan, and neuronal nitric oxide synthase (nNOS). Bar, 100 μm.
3.5. Myotubes cultures derived from Patient 3–23 mirror observations made on biopsy
For Patient 3–23, a cell culture model was generated from dermal fibroblasts obtained from skin punch biopsy. Dermal fibroblasts were immortalized with hTERT and inducible MyoD-ERT vectors which can be propagated as fibroblasts and then terminally differentiation to myotubes in culture upon tamoxifen induction of ER-myoD nuclear translocation and fusion culture conditions (14). Due to the unexpectedly mild phenotype of this individual, we sought to develop and validate a cell model for future characterization of this in-frame mutation in the context of any potential modifier genes that may contribute to this phenotype. Inducible directly reprogrammed myotubes (iDRM) derived from Patient 3–23 and a healthy donor were differentiated to myotubes in culture (Fig. 4). Similar to the biopsy staining results, differentiated patient myotubes showed little or no signal Dys3 (N-term) signal and reduced levels of Dys2 (C-term) and Dys1 (central rod domain) relative to control myotubes. They additionally exhibited overall increased levels of utrophin signal, consistent with the increased expression seen in the biopsy from Patient 3–23.
Figure 4. Inducible directly reprogrammed myotubes from Patient 3–23 show reduced dystrophin expression.
For Patient 3–23 (exon 3–23 deletion, cell line CDMD1023) and an unaffected individual (cell line CDMD1002), inducible directly reprogrammed myotubes (iDRMs) were generated from dermal fibroblasts obtained from a skin punch biopsy. Dystrophin was detected using Dys1, Dys2 and Dys3 (Rod domain, C-term or N-term). Utrophin was detected using MANCHO3. Bar, 20 μm.
3.6. In-frame N-terminal deletions in the Universal Mutation Database-DMD database are associated with a spectrum of severity
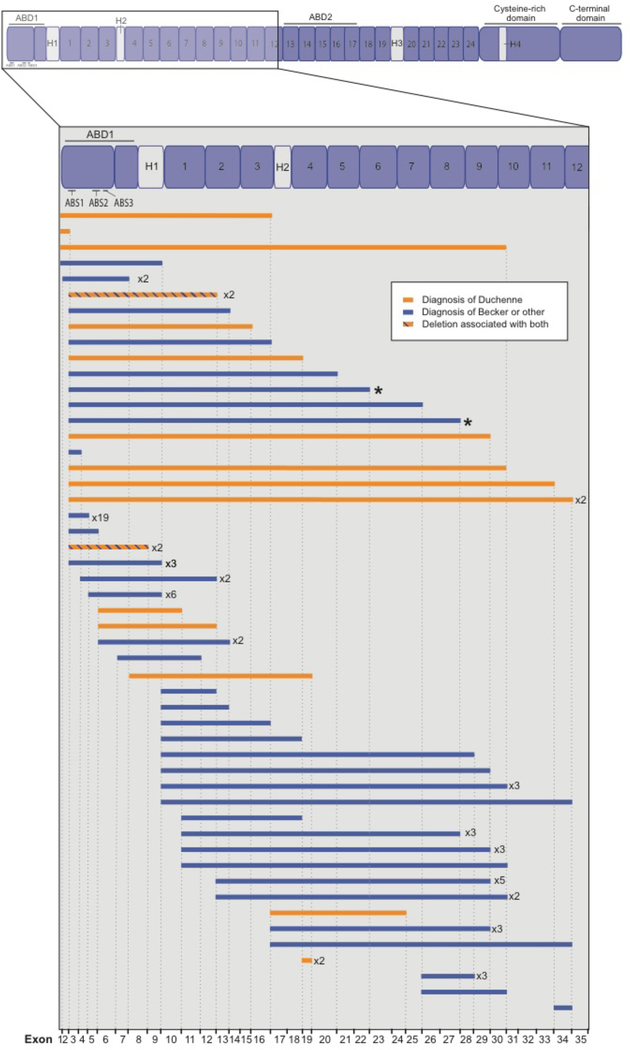
In a systematic search of the Universal Mutation Database for DMD (UMD-DMD, http://www.umd.be/DMD/W_DMD/index.html) database, we identified 97 individuals with in-frame deletions in the first 35 exons of the dystrophin gene (Fig. 5). Of these, 74 individuals (76%) were reported to have BMD (loss of ambulation greater than or equal to 16 years of age) or Intermediate Muscular Dystrophy (IMD, within the UMD-DMD database, defined as loss of ambulation between 13 and 16 years of age [10]). Five individuals were classified as asymptomatic at the time of the report.
Figure 5. Phenotypic subgroups associated with in-frame deletions in the first 35 exons of DMD.
The UMD-DMD database (http://www.umd.be/DMD/W_DMD/index.html) was searched for in-frame DMD deletions associated with dystrophinopathy occurring within exons 1–35 of the DMD gene. A total of 97 individuals with a reported phenotypic subgroup were identified. Of these, 74 individuals (76%) were reported to have BMD (loss of ambulation greater than or equal to 16 years of age) or Intermediate Muscular Dystrophy (within the UMD-DMD database, defined as loss of ambulation between 13 and 16 years of age [10]). Five individuals were classified as asymptomatic at the time of the report. Deletions shown in orange are associated with a diagnosis of Duchenne (within the UMD-DMD database, defined as loss of ambulation below 13 years of age). Deletions shown in blue are milder phenotypic groups, including BMD, IMD and asymptomatic at the time of report. Mutations associated with multiple diagnoses are reported in orange/blue hatch. Female carriers and database entries without a reported phenotypic subgroup were excluded. The two deletions reported in this study (exon 3–23 del and 3–28 del) are indicated by asterisks (*).
4. Discussion
Intragenic DMD deletions are the most common cause of DMD, with approximately 65–70% of patients having deletions of one or more exons (5, 17, 18). Two mutational hotspots have been described for deletions in DMD, the regions encompassing exons 2–20 and exons 45–55 (19). While patients with in-frame mutations in the 45–55 hotspot region often have a relatively mild disease phenotype, with an increased mean age at loss of ambulation (10, 20), those with in-frame mutations in the more N-terminal encoding region can present with highly variable phenotypes, with in-frame deletions sometimes leading to typical DMD phenotype. In this study, we describe two individuals with rare large in-frame N-terminal deletions with a disease course that is demonstrated to be milder than typical DMD but will nonetheless be on the more severe end of the BMD spectrum. Determining the exact contribution of mutation to disease severity is difficult in single cases, especially since treatment alters disease course. Both individuals are treated with glucocorticoid steroids from age 4–5 years, which have been shown to slow muscle degeneration and delay loss of ambulation in DMD patients. On average, untreated DMD patients lose the ability to walk between ages 9 and 11, while corticosteroid treatment prolongs ambulation between 2–3 years (10). Both patients were changed from daily dose steroids to weekend-only steroids, based on the unusually slow, but still progressive, childhood disease course. Both are likely to have some of their relatively slow progression attributed, at least in part, to the in-frame mutation and there is a more mild disease progression in the 3–23 patient, consistent with this in-frame mutation having greater function than the 3–28 mutant dystrophin. Few patients with typical DMD remain ambulant after age 17 years even with daily steroids. In these two patients, slower disease progression being caused by the in-frame mutations is consistent with our demonstration that both truncated dystrophin proteins are expressed in muscle samples from both patients, and are correctly localized, retaining sarcolemmal expression of proteins normally associated with dystrophin and utrophin complexes.
In-frame deletions that disrupt the N-terminal part of the dystrophin protein, encoding the first actin-binding domain, are commonly reported to lead to a more severe BMD presentation or a diagnosis of DMD when compared to in-frame deletions within the central rod domain (5, 17, 18, 21–25). However, there are a number of reports of N-terminal deletions associated with a moderately mild dystrophinopathy. One notable example is an individual with an in-frame exon 3–9 deletion reported as largely asymptomatic until diagnosed with BMD at age 67 (4). While this gene lesion maintains reading frame, it disrupts the ABD1 domain and removes actin binding sites ABS2 and ABS3. At least 10 other patients with exon 3–9 deletions have also been diagnosed with BMD (24). We identified 97 individuals from the UMD-DMD database with in-frame deletions in the first 35 exons of the dystrophin gene (Fig. 5). These deletions were associated with a range of reported phenotypes, including DMD, BMD, and individuals who were asymptomatic at the time of the report. In this cohort, we found several N-terminal deletions associated with a BMD phenotype. These mutations lack the actin binding site ABD1, supporting the idea that the loss of the ABD1 actin binding site within an in-frame protein can still be associated with some attenuation of the dystrophic phenotype.
In addition to in-frame N-terminal deletions, alternative translation initiation sites within exons 6 or 8 have been shown to allow the production of functional truncated dystrophin in patients with otherwise deleterious N-terminal mutations (10, 26, 27). For example, the relatively common out-of-frame deletion of exons 3–7 can lead to a range of dystrophic phenotypes including the milder BMD (10, 26, 28–31). Our recent analysis of the Duchenne Registry demonstrated that the large majority of boys amenable to exon 8 targeted therapy (N = 18) were still ambulatory at age 20; most of these boys reported an exon 3–7 deletion (10). In these cases, an alternative initiation site at exon 8, or exon 8 skipping is hypothesized to produce low levels of truncated dystrophin lacking all three actin-binding, sites within ABD1 (18, 26). Similarly, Wein and colleagues recently demonstrated that an internal ribosome entry site within exon 5 can drive synthesis of a smaller, N-terminal dystrophin protein starting at exon 6 (deleting ABS1 and ABS2) (27). Expression of this truncated dystrophin protein via adeno-associated virus (AAV) was associated with either a mild dystrophic phenotype or complete absence of symptoms in two patients with exon 2 mutation. This N-terminal truncated protein is also shown to ameliorate the dystrophic phenotype in a mouse model of DMD exon 2 duplication (27).
Along with several other lines of evidence, the mild dystrophic phenotype associated with some N-terminal deletions suggests that a largely functional truncated dystrophin protein without complete ABD1 domain can be stably expressed, albeit at levels lower than wild type, and retain significant functionality. In vitro studies have shown that some deletions in the N-terminal domain might lead to protein instability rather than a direct loss of protein function (25). In order to better understand the mild phenotype associated with exon 3–9 deletions, Kyrychenko and colleagues utilized multiple CRISPR-based correction strategies to restore the reading frame of human iPS cells carrying an out-of-frame exon 8–9 DMD deletion (25). When exons 3–7 were additionally deleted in these cells, the resulting exon 3–9 deleted iPSC-derived cardiomyocytes expressed truncated dystrophin and showed a partial correction of contractile defects. Intriguingly, they showed that a similar correction strategy deleting exons 7–11 led to a lower level of dystrophin expression and less functional improvement in cardiomyocytes, despite preserving the ABD1 domain. This deletion was prone to protein degradation, suggesting that the shorter exon 7–11 deletion is in fact less stable than the exon 3–9 deleted protein.
While point mutations in the DMD gene are less frequent in dystrophinopathy than deletions, they have provided insight into the role of the dystrophin N-terminal domain. Approximately 50% of known disease-causing missense mutations are located in the ABD1 (32, 33). Although it was initially suggested that ABD1 missense mutations might alter dystrophin’s affinity for F-actin and disrupt the connection between the cytoskeleton and sarcolemma, several groups observed early on that such mutations seemed to primarily affect protein stability (31, 34). In keeping with this suggestion, in vitro studies of point mutations in the ABD1 region do not substantially alter affinity for F-actin, and are rather associated with protein aggregation instability (32, 33, 35).
Animal studies looking at the functionality of truncated dystrophin constructs or “microdystrophins” provide some of the best evidence for understanding the role of the ABD1 and surrounding N-terminal region. In the dystrophin-null mdx model, Corrado and colleagues expressed a shortened dystrophin construct without the ABS2 and ABS3 portion of the ABD1, along with part of the Hinge 1 region (deletion from amino acids 45 to 273) (36). These mice exhibited a milder phenotype than mdx alone, and suggest that the retention of the central rod domain ABD2 region can partially compensate for the loss the N-terminal ABD1 domain. Consistent with this finding, a later study from the same group demonstrated that microdystrophin lacking the ABD2 domain function in mdx mice when the ABS1 or ABS1/ABS2 regions were deleted (23). Notably, the microdystrophins with full ABD2 and partial ABD1 deletions localized to the sarcolemma and significantly reduced degeneration in mdx muscle, suggesting that there may be additional actin binding domain(s).
The idea that the ABD1 is not absolutely required for dystrophin stability or function is further supported by a study in mdx mice demonstrating that expression of Dp260, the retinal isoform of dystrophin, can also reduce the dystrophin phenotype (37). The Dp260 isoform has an alternative exon 1 which splices into exon 30, and is expressed in retina, brain and heart (38). This isoform is highly relevant in considering the deletions described in our study. In particular, Patient 3–28 has a deletion from exon 3–28 that produces a protein very similar to the Dp260 isoform, lacking most of the ABD1 domain, hinge 1 and half of the spectrin repeat domain 9. The similarity of this deletion to a native isoform of dystrophin hints that it may be more stable and functional than other internally deleted dystrophin proteins. Notably, one in vitro study found that Dp260 have a similar protease resistance to full-length dystrophin, while some truncated dystrophin constructs exhibited reduced stability (35).
The two patients described in this study highlight the spectrum of phenotypes associated with N-terminal deletions in DMD, and demonstrate that the loss of ABD1 and other domains in the first third of the gene can result in a partially functional protein and a milder dystrophic phenotype. Taken together with in vitro and mouse model studies, these findings suggest that loss of protein stability may play a greater role than loss of function in determining the pathogenicity of N-terminal in-frame deletions. In particular, certain deletion-specific junctions may be unfavorable to transcript or protein stability, and lead to a more severe dystrophy. Studying the genotype-phenotype correlations in patients with these deletions, and generating relevant cell and animal models, can provide insight into the genetic complexity of dystrophinopathies and help guide the design of therapeutic strategies for dystrophin restoration.
Highlights.
5’ deletions in DMD present a wide phenotype spectrum from mild to severe
Two large 5’ DMD deletions show sarcolemmal expression of truncated dystrophin on biopsy
A new DMD delta 3–23 patient-derived cell model recapitulates biopsy findings
Acknowledgments
Funding
This work was supported by grants from the National Institutes of Health (NIH) R01 AR048179 and R01 HL126204 to RC; T32 AR059033 and F32 AR069469 to EMG, P30 AR057230 to MCM and SFN, and U54 AR052646 Wellstone Center of Excellence Training Fellowship to FB as well as support from the Muscular Dystrophy Association USA 274143 and 416364 to RC. U.S.; Patient-Centered Outcomes Research Institute PPRN-1306-04640; National Center for Advancing Translational Sciences, UCLA CTSI UL1TR000124 to SN and Department of Defense, DODAR060836, UCLA CTSI UL1TR000124 and Parent Project Muscular Dystrophy to MCM. Viral packaging was performed at the UCLA Vector Core. The UCLA Integrated Molecular Technologies Core is supported by CURE/P30 DK041301.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Declarations
Consent
Informed consent was obtained from the patient for the publication of this case report and any accompanying images.
References
- 1.Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993;122(4):809–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease C, Prevention. Prevalence of Duchenne/Becker muscular dystrophy among males aged 5–24 years - four states, 2007. MMWR Morb Mortal Wkly Rep. 2009;58(40):1119–22. [PubMed] [Google Scholar]
- 3.Ferreiro V, Giliberto F, Muniz GM, Francipane L, Marzese DM, Mampel A, et al. Asymptomatic Becker muscular dystrophy in a family with a multiexon deletion. Muscle Nerve. 2009;39(2):239–43. [DOI] [PubMed] [Google Scholar]
- 4.Heald A, Anderson LV, Bushby KM, Shaw PJ. Becker muscular dystrophy with onset after 60 years. Neurology. 1994;44(12):2388–90. [DOI] [PubMed] [Google Scholar]
- 5.Gao QQ, McNally EM. The Dystrophin Complex: Structure, Function, and Implications for Therapy. Compr Physiol 2015;5(3):1223–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarrett HW, Foster JL. Alternate binding of actin and calmodulin to multiple sites on dystrophin. J Biol Chem. 1995;270(10):5578–86. [DOI] [PubMed] [Google Scholar]
- 7.Luce LN, Dalamon V, Ferrer M, Parma D, Szijan I, Giliberto F. MLPA analysis of an Argentine cohort of patients with dystrophinopathy: Association of intron breakpoints hot spots with STR abundance in DMD gene. J Neurol Sci. 2016;365:22–30. [DOI] [PubMed] [Google Scholar]
- 8.Trimarco A, Torella A, Piluso G, Maria Ventriglia V, Politano L, Nigro V. Log-PCR: a new tool for immediate and cost-effective diagnosis of up to 85% of dystrophin gene mutations. Clin Chem. 2008;54(6):973–81. [DOI] [PubMed] [Google Scholar]
- 9.Barthelemy F, Wang D, Nelson SF, Miceli MC. Validation and Detection of Exon Skipping Boosters in DMD Patient Cell Models and mdx Mouse. Methods Mol Biol. 2018;1828:309–26. [DOI] [PubMed] [Google Scholar]
- 10.Wang RT, Barthelemy F, Martin AS, Douine ED, Eskin A, Lucas A, et al. DMD genotype correlations from the Duchenne Registry: Endogenous exon skipping is a factor in prolonged ambulation for individuals with a defined mutation subtype. Hum Mutat. 2018;39(9):1193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura E, Han JJ, Li S, Fall B, Ra J, Haraguchi M, et al. Cell-lineage regulated myogenesis for dystrophin replacement: a novel therapeutic approach for treatment of muscular dystrophy. Hum Mol Genet. 2008;17(16):2507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barthélémy F, Blouin C, Wein N, Mouly V, Courrier S, Dionnet E, et al. Exon 32 Skipping of Dysferlin Rescues Membrane Repair in Patients’ Cells. Journal of Neuromuscular Diseases. 2015;2(3):281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, et al. A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell Stem Cell. 2016;18(4):533–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kendall GC, Mokhonova EI, Moran M, Sejbuk NE, Wang DW, Silva O, et al. Dantrolene enhances antisense-mediated exon skipping in human and mouse models of Duchenne muscular dystrophy. Sci Transl Med 2012;4(164):164ra0. [DOI] [PubMed] [Google Scholar]
- 15.Ginjaar IB, Kneppers AL, v d Meulen JD, Anderson LV, Bremmer-Bout M, van Deutekom JC, et al. Dystrophin nonsense mutation induces different levels of exon 29 skipping and leads to variable phenotypes within one BMD family. Eur J Hum Genet. 2000;8(10):793–6. [DOI] [PubMed] [Google Scholar]
- 16.van den Bergen JC, Wokke BH, Janson AA, van Duinen SG, Hulsker MA, Ginjaar HB, et al. Dystrophin levels and clinical severity in Becker muscular dystrophy patients. J Neurol Neurosurg Psychiatry. 2014;85(7):747–53. [DOI] [PubMed] [Google Scholar]
- 17.Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34(2):135–44. [DOI] [PubMed] [Google Scholar]
- 18.Aartsma-Rus A, Ginjaar IB, Bushby K. The importance of genetic diagnosis for Duchenne muscular dystrophy. J Med Genet. 2016;53(3):145–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeshima Y, Yagi M, Okizuka Y, Awano H, Zhang Z, Yamauchi Y, et al. Mutation spectrum of the dystrophin gene in 442 Duchenne/Becker muscular dystrophy cases from one Japanese referral center. J Hum Genet. 2010;55(6):379–88. [DOI] [PubMed] [Google Scholar]
- 20.Bello L, Morgenroth LP, Gordish-Dressman H, Hoffman EP, McDonald CM, Cirak S, et al. DMD genotypes and loss of ambulation in the CINRG Duchenne Natural History Study. Neurology. 2016;87(4):401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beggs AH, Hoffman EP, Snyder JR, Arahata K, Specht L, Shapiro F, et al. Exploring the molecular basis for variability among patients with Becker muscular dystrophy: dystrophin gene and protein studies. Am J Hum Genet. 1991;49(1):54–67. [PMC free article] [PubMed] [Google Scholar]
- 22.Novakovic I, Bojic D, Todorovic S, Apostolski S, Lukovic L, Stefanovic D, et al. Proximal dystrophin gene deletions and protein alterations in becker muscular dystrophy. Ann N Y Acad Sci. 2005;1048:406–10. [DOI] [PubMed] [Google Scholar]
- 23.Banks GB, Gregorevic P, Allen JM, Finn EE, Chamberlain JS. Functional capacity of dystrophins carrying deletions in the N-terminal actin-binding domain. Hum Mol Genet. 2007;16(17):2105–13. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura A, Fueki N, Shiba N, Motoki H, Miyazaki D, Nishizawa H, et al. Deletion of exons 3–9 encompassing a mutational hot spot in the DMD gene presents an asymptomatic phenotype, indicating a target region for multiexon skipping therapy. J Hum Genet. 2016;61(7):663–7. [DOI] [PubMed] [Google Scholar]
- 25.Kyrychenko V, Kyrychenko S, Tiburcy M, Shelton JM, Long C, Schneider JW, et al. Functional correction of dystrophin actin binding domain mutations by genome editing. JCI Insight. 2017;2(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winnard AV, Mendell JR, Prior TW, Florence J, Burghes AH. Frameshift deletions of exons 3–7 and revertant fibers in Duchenne muscular dystrophy: mechanisms of dystrophin production. Am J Hum Genet. 1995;56(1):158–66. [PMC free article] [PubMed] [Google Scholar]
- 27.Wein N, Vulin A, Falzarano MS, Szigyarto CA, Maiti B, Findlay A, et al. Translation from a DMD exon 5 IRES results in a functional dystrophin isoform that attenuates dystrophinopathy in humans and mice. Nat Med. 2014;20(9):992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malhotra SB, Hart KA, Klamut HJ, Thomas NS, Bodrug SE, Burghes AH, et al. Frame-shift deletions in patients with Duchenne and Becker muscular dystrophy. Science. 1988;242(4879):755–9. [DOI] [PubMed] [Google Scholar]
- 29.Baumbach LL, Chamberlain JS, Ward PA, Farwell NJ, Caskey CT. Molecular and clinical correlations of deletions leading to Duchenne and Becker muscular dystrophies. Neurology. 1989;39(4):465–74. [DOI] [PubMed] [Google Scholar]
- 30.Muntoni F, Gobbi P, Sewry C, Sherratt T, Taylor J, Sandhu SK, et al. Deletions in the 5’ region of dystrophin and resulting phenotypes. J Med Genet. 1994;31(11):843–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winnard AV, Klein CJ, Coovert DD, Prior T, Papp A, Snyder P, et al. Characterization of translational frame exception patients in Duchenne/Becker muscular dystrophy. Hum Mol Genet. 1993;2(6):737–44. [DOI] [PubMed] [Google Scholar]
- 32.Henderson DM, Lee A, Ervasti JM. Disease-causing missense mutations in actin binding domain 1 of dystrophin induce thermodynamic instability and protein aggregation. Proc Natl Acad Sci U S A. 2010;107(21):9632–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh SM, Kongari N, Cabello-Villegas J, Mallela KM. Missense mutations in dystrophin that trigger muscular dystrophy decrease protein stability and lead to cross-beta aggregates. Proc Natl Acad Sci U S A. 2010;107(34):15069–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts RG, Gardner RJ, Bobrow M. Searching for the 1 in 2,400,000: a review of dystrophin gene point mutations. Hum Mutat. 1994;4(1):1–11. [DOI] [PubMed] [Google Scholar]
- 35.Henderson DM, Belanto JJ, Li B, Heun-Johnson H, Ervasti JM. Internal deletion compromises the stability of dystrophin. Hum Mol Genet. 2011;20(15):2955–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corrado K, Rafael JA, Mills PL, Cole NM, Faulkner JA, Wang K, et al. Transgenic mdx mice expressing dystrophin with a deletion in the actin-binding domain display a “mild Becker” phenotype. J Cell Biol. 1996;134(4):873–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warner LE, DelloRusso C, Crawford RW, Rybakova IN, Patel JR, Ervasti JM, et al. Expression of Dp260 in muscle tethers the actin cytoskeleton to the dystrophin-glycoprotein complex and partially prevents dystrophy. Hum Mol Genet. 2002;11(9):1095–105. [DOI] [PubMed] [Google Scholar]
- 38.D’Souza VN, Nguyen TM, Morris GE, Karges W, Pillers DA, Ray PN. A novel dystrophin isoform is required for normal retinal electrophysiology. Hum Mol Genet. 1995;4(5):837–42. [DOI] [PubMed] [Google Scholar]