Abstract
Despite the controversy regarding the existence and physiological relevance of class A G protein-coupled receptor dimerization, there is substantial evidence for functional interactions between the dopamine D2 receptor (D2R) and the adenosine A2A receptor (A2AR). A2AR-D2R complexes have been detected in rodent brains by proximity ligation assay; however, their existence in the human brain has not been demonstrated. In this study, we used Brightfield proximity ligation assay, combined with a systematic sampling and a parameter-free naive Bayesian classifier, and demonstrated proximity between the D2R and the A2AR in the adult human ventral striatum, consistent with their colocalization within complexes and the possible existence of D2R-A2AR heteromers. These methods are applicable to the relative quantification of proximity of two proteins, as well as the expression levels of individual proteins.
Keywords: : adenosine, adenosine A2A receptor, dopamine, dopamine receptor type 2, G protein-coupled receptor, proximity ligation assay, machine learning, naive Bayesian classifier
METHOD SUMMARY
Brightfield proximity ligation assay was used to assess the expression of G protein-coupled receptors and their proximity in postmortem adult human brains. A novel automated machine learning method (Bayesian optimized PLA signal sorting) was developed to automatically quantify Brightfield proximity ligation assay data.
The dopamine receptor type 2 (D2R) is an extensively studied class A G protein-coupled receptor (GPCR) that has been shown to play a critical role in various brain functions and has been implicated in a variety of neuropsychiatric disorders, including schizophrenia [1,2], Parkinson's [3,4], Alzheimer's [5] and Huntington's disease [7] as well as addiction [8]. In addition, the D2R is the common target of all current antipsychotic medications [9], which has led to extensive publication of literature regarding its pharmacological and signaling properties [10]. Previous studies in cell lines suggest that D2Rs may function as a part of heteromeric complexes, in which D2R activity and signaling can be modulated by another receptor subunit [11–14]. The putative heteromer formed by the D2R with the adenosine A2A receptor (A2AR) is one of the most studied among the class A GPCRs [15–19], and it has been hypothesized that pharmacological targeting of A2AR could be an efficient strategy to modulate D2R activity [20,21]. However, the structural properties of class A receptor heteromers, their existence in vivo and their relevance to receptor physiology or pathophysiology remain unclear and a topic of active study and debate [22–24]. The signaling properties of putative D2R-A2AR heteromers have been mostly studied in heterologous systems, and it is important to study these complexes in native mammalian brain.
Both time-resolved fluorescence resonance energy transfer-based assays [25,26] and antibody-based in situ proximity ligation assays (PLAs) [27,28] have been used to study receptor complexes in native tissue. Using fluorescent PLA, we and others have successfully detected endogenous D2R-A2AR complexes in the rodent striatum, which provided ex vivo evidence for the existence of D2R-A2AR heteromeric complexes composed of native receptors [28,29]. However, the existence of D2R-A2AR complexes in human brains has yet to be established.
PLA has been widely used to assess protein–protein interaction, protein expression and post-translational modification both in vitro and ex vivo [30–33]. PLA puncta are generated when a pair of oligonucleotide-conjugated antibodies bind to neighboring antigens, followed by ligation of the oligonucleotides and subsequent rolling cycle amplification, leading to DNA structure that can be detected by fluorophore- or horseradish peroxidase-labeled oligonucleotide probes. In contrast to standard immunohistochemistry and immunofluorescence, both of which rely on a field of precipitate or fluorescence that can only be quantified by total intensity, PLA results in individual puncta, allowing relative quantification of proteins (single PLA) or complexes (dual PLA) with higher spatial resolution. To date, PLA has been applied to postmortem human brain in a limited number of studies. PLA was used to assess α-synuclein oligomers in Parkinson's disease [34] and to detect the interaction between SORL1 and APP in Alzheimer's disease [35]. A limiting factor in applying these approaches to the human brain relates to the size of the samples, which make it challenging to obtain representative data while minimizing the number of samples, as classical stereology techniques are extremely time consuming. Further, PLA is highly sensitive to tissue processing [28], and its use in postmortem human brain tissue requires additional consideration and optimization. Notably, human brains are classically fixed by immersion in fixative and paraffin-embedded, a method that is not typically used with rodent brain tissue.
Here, we optimized in situ PLA to detect D2R, A2AR and D2R-A2AR complexes with Brightfield microscopy in the human ventral striatum and developed a new approach, combining whole-slide scanning, systematic random sampling and parameter-free automated image analysis that employs a naive Bayesian classifier for faithful and robust signal separation. This study constitutes a proof-of-concept for relative quantification of individual proteins and antigen complexes in situ using postmortem human brain samples.
Materials & methods
PLAs & antibodies
Information about human brain specimens is described in Supplementary Table 1, and tissue processing is described in the Supplementary materials and methods. Anti-D2R (rabbit polyclonal, ABN462, MilliporeSigma, MO, USA) was used at a final concentration of 1.67 μg/ml for single PLA (recognition of one antigen), and 5 μg/ml for dual PLA (proximity between two antigens). Anti-A2AR (mouse monoclonal, 05-717, MilliporeSigma) was used at 1 μg/ml for single PLA and at 1.67 μg/ml for dual PLA. The specificity of anti-D2R and anti-A2AR antibodies was previously validated using brain tissue from D2R and A2AR knockout mice [28,36]. PLA was performed with anti-rabbit and anti-mouse secondary antibody-conjugated PLA probes and the Brightfield detection kit (MilliporeSigma Duolink®, MO, USA) according to the user's manual (Supplementary materials and methods). A detailed protocol is available online.
Microscopy & sampling
For fluorescent staining, images were taken with a Zeiss LSC510 confocal laser-scanning microscope (Zeiss, Germany) using a 63× oil objective and z-stack scanning with a step interval of 0.5 μm. Brightfield whole-slide images were taken with a Leica SCN400 (Leica Biosystems, IL, USA) at the Herbert Irving Comprehensive Cancer Center (HICCC), using tube lens magnification (2×) in addition to a 20× objective lens (Numerical Aperture 0.65) to achieve 40× magnification and z-stack scanning with a step interval of 1 μm. Whole-slide scanned virtual images were viewed with a Leica SCN400 image viewer (version 2.2), and a single layer with the most PLA puncta in focus was used to export images (zoom at 40×) for PLA signal quantification.
A systematic random sampling method adapted from Gundersen et al. [37] was performed to choose areas in the brain region of interest (ROI) to quantify the PLA signal (Supplementary Figure 1).
Quantification with the Bayesian optimized PLA signal sorting package
We designed a custom MATLAB package Bayesian optimized PLA signal sorting (BOPSS) to analyze PLA signal images [38]. To use the program, we input a folder containing Brightfield images for single and dual PLA samples and negative controls. In the first step of BOPSS, clusters of noncellular foreground pixels (putative puncta [PP]) are located in all of the images using an approach typically used in the processing of histological samples [39]. The PP are then randomly split into two groups (training set and test set). The test set of PP is used to calculate two distributions (negative control distribution and PLA signal distribution) for each of the 11 intensity-based and morphological features (11 pairs of distributions in total are generated). Next, these 11 pairs of distributions are used to categorize PP in the test set. At this step, for each feature, each punctum is determined to be more similar to the trained PLA signal distribution or the trained noise distribution. Some puncta may be more similar to the PLA signal in a few features, but more similar to the noise in the rest of the features. To account for all 11 features, a product (the likelihood) of the punctum being PLA signal is calculated and compared with the likelihood of it being noise. If the likelihood of the punctum being a PLA signal is higher than the likelihood of the punctum being noise, it is preserved as a PLA signal.
Results & discussion
Optimized PLA assay for rodent & human brain sections
Whereas PLA on rodent brain fixed by transcardial perfusion has been widely validated [28,33], human brain tissue is classically fixed by direct immersion in formalin and paraffin-embedding, which could alter the detection and quality of the signal. Tissue preparation, especially fixation, is known to alter morphology and immunostaining results [40–42]. To assess how alternative fixation approaches may affect PLA imaging results in human brain sections, we compared the results of immunofluorescent PLA (PLA-FL) on mouse brain sections following different fixation approaches. We performed single-recognition PLA (single PLA), which allows detection of an antigen with only one primary antibody, for D2R on sections of mouse brains that underwent two different fixation protocols: 1 fixation in 4% PFA overnight after perfusion (perfusion fixation), or 2 fixation by direct immersion in 4% PFA for 4 days without perfusion (direct immersion fixation). With both fixation protocols, we obtained a clear PLA signal (puncta) of D2R distributed throughout the striatum, with one major difference in the nonspecific background. As described previously [28,33,36] we observed a fluorescent signal in the nuclear compartments in samples fixed through perfusion (Figure 1A & C), whereas this background was much lower in sections from brains fixed without perfusion (Figure 1B & D). These results suggested that the PLA signal for D2Rs from tissue fixed using the direct immersion method is preserved, whereas the nuclear background is greatly reduced. Although the exact cause of this nuclear background is unclear, it was previously described with PLA-FL from perfused adult animals [28,43] and on brain slices from mouse pups (P0-P1) fixed by direct immersion in fixative [33]. In addition, we performed PLA-FL on sections from snap-frozen mouse brains, which were fixed with 4% PFA after cryosectioning, and observed a similar strong nuclear background (data not shown). It is therefore tempting to propose that it is the rapid fixation process, which occurs through intracardial perfusion of fixative, or rapid penetration of the fixative in the much smaller pup brain or a thin cryosection, which is responsible for the nuclear background. This signal does not result from specific or nonspecific binding of primary or secondary antibodies, since it is not detectable in regular immunostaining with anti-D2R or anti-A2AR antibodies. Moreover, the nuclear background is still present when antibodies are omitted in the PLA process [28]. We speculate that the crosslinking of chromatin by rapid fixation could result in nonspecific binding of the labeled oligonucleotides used during the detection step of the PLA. Regardless of the underlying causes, our results help to explain the dramatic differences in background described in previously published studies [28,43,44].
Figure 1. . The effect of fixation on the nonspecific nuclear signal in single-recognition PLA-FL in mouse brain tissue.
Representative images of D2R single-recognition PLA in coronal striatal section from mouse brains fixed by transcardial perfusion of fixative (A & C) or by direct immersion in fixative (B & D). DAPI counterstaining (blue) (C & D) labels nuclei. Note that in the nuclear area, the hazy background signal is almost absent and the puncta overlapping the nuclei are much reduced in the non-perfused tissue compared with the perfused tissue. Scale bar, 50 μm.
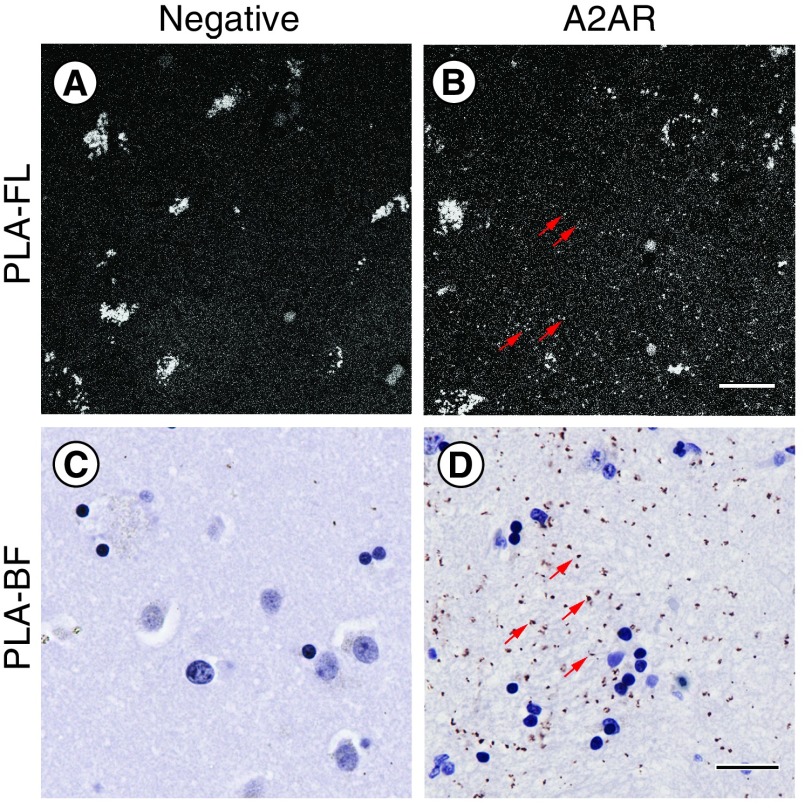
Next, we performed PLA-FL on paraffin-embedded human brain sections. In marked contrast to mouse brain sections, a prominent nonspecific signal was observed, detectable with either 488 or 564-nm excitation (data not shown) in the form of dense aggregates that often accumulated in the soma (Figure 2A-B), suggesting a nonspecific autofluorescence signal, in addition to detection of specific PLA puncta. To address this issue, we replaced the original PLA-FL with a Brightfield-compatible detection method, Brightfield PLA (PLA-BF), which allows visualization of the PLA signal with a chromogenic substrate of horseradish peroxidase. The PLA-BF for single-recognition of the A2AR showed a similar PLA punctate pattern, however, without the nonspecific signal observed with the PLA-FL method (Figure 2C & D).
Figure 2. . Differences between PLA-FL and PLA-BF detection methods.
Representative images of negative controls (A & C) and A2AR single-recognition PLA-FL (B), PLA-BF (D) performed on paraffin-embedded human brain sections of the ventral striatum from the same human subject. The arrows in B and D indicate representative PLA puncta. Hematoxylin counterstaining (blue) labels nuclei (C & D). Scale bar, 20 μm. Note the high autofluorescence background in PLA-FL and the clean background in the PLA-BF.
Autofluorescence is a common issue for imaging the adult human brain but not with transmitted light. Therefore, PLA-BF avoids the autofluorescence that confounds PLA-FL (Figure 2) and even standard immunofluorescence in brain samples from human adults and aged animals [45]. Notably, PLA-BF and PLA-FL on mouse brain sections yielded comparable results (data not shown). Altogether, these observations suggest that PLA-BF is a superior alternative to PLA-FL for paraffin sections from human autopsy brains. Moreover, the stability of the Brightfield-based signal compared with fluorescence-based methods allows a better conservation of the samples without the photobleaching inherent to the imaging of fluorescent samples.
Assessing the expression of D2R, A2AR & D2R-A2AR in the human brain
To assess the expression of D2R, A2AR and D2R-A2AR complexes, we performed single PLA-BF for each individual receptor, and dual PLA-BF for D2R-A2AR complexes on coronal sections of adult human ventral striatum that included the nucleus accumbens (NAcc) and limited parts of the putamen (Ptm) and caudate (Cdt). The anatomical structure was confirmed by Luxol fast blue and cresyl violet (LFB/CV) staining, which stains myelin and Nissl substance, respectively (Supplementary Figure 1). Considering the low number of PLA puncta for D2R-A2AR complexes relative to individual D2R and A2AR in mouse brains [28], we used different concentrations of primary antibodies for dual and single PLAs to obtain optimal staining results, avoiding saturation in single PLA that can lead to overcrowded signal instead of countable puncta. In all tested samples, we observed strong signal for single PLA of D2R and A2AR and a clear but weaker signal for dual PLA of D2R-A2AR complexes in the NAcc (Figure 3 A–I), Ptm and Cdt (data not shown). The dual PLA for negative controls that omitted anti-A2AR antibody showed no or very few detectable puncta in the same areas (Figure 3 J–L and data not shown). These results support the existence of D2R-A2AR complexes in native human brains. We note that interpretation of the puncta from single PLA is complex with our current reagents, since the polyclonal secondary antibodies can bind to either two epitopes on a single primary antibody bound to a single antigen, or to two different primaries bound to proximal antigens. We expect that the former will be more efficient. In addition, we had to use different primary antibody concentrations for single and dual PLA so as to not saturate the single PLA signal. For these reasons, the absolute number of single PLA and dual PLA puncta cannot be compared directly but must be used for relative quantitation.
Figure 3. . Detection of single PLA for A2AR and D2R, and dual PLA for D2R-A2AR by PLA-BF in the NAcc.
The expression of A2AR (A–C), D2R (D–F), D2R-A2AR (G–I) and negative controls (J–L) in the NAcc was detected in three human subjects, PI12260, PI12270 and PI12277. Scale bar: 25 μm.
Systematic random sampling & automated quantification of the PLA-BF signal in human brain sections
Because the PLA signal appears as puncta, quantitative analysis of proteins is more plausible than in traditional immunostaining. However, the large size of the human brain sections makes it challenging to quantify the PLA signal from an entire brain region. For example, the brain ROI in this study, the NAcc, was approximately 40 mm2 (Supplementary Table 2). Therefore, as described in the Supplementary materials and methods, we obtained virtual images by whole-slide scanning, divided the ROI into a series of sampling areas and only quantified the PLA signal in the areas (named counting loci in this study) that were selected by a systematic random sampling method adapted from Gundersen et al. [37] (Supplementary Figure 1F). Therefore, the total analyzed area represented only ∼3% of the ROI (Supplementary Table 2) but was composed of a set of representative counting loci evenly distributed within the entire ROI.
To efficiently and reproducibly quantify PLA puncta across our large dataset in an unbiased way, we applied and compared three different quantification approaches. Two of these approaches, Analyze Particles (ImageJ) [46] and Spot detector (ICY) [47], rely on the user to set intensity and size parameters to distinguish clusters of pixels against the background of an image. In contrast, our custom MATLAB package BOPSS, which we designed to automatically quantify PLA-BF signal, is an unsupervised machine-learning approach and requires no parameters to be set by the user.
Analyze particles (Image J) depends on threshold and particle size to detect PLA puncta. The automated threshold (Figure 4: ImageJ_auto) worked well for the dual PLA signal, but underestimated the signal for single PLA, and showed high counts in the negative controls (Figure 4D–F, P-Q), which resulted in a low signal (the dual PLA signal)/background noise (the negative control signal) (S/N = 2.14). Most of the nonspecific puncta in the negative controls detected by Analyze Particles corresponded to background in the nuclear area, typical of the signal obtained with classical DNA intercalating reagents like DAPI or Hoechst. Manually optimizing the threshold reduced the nonspecific detection in the negative controls but caused severe undercounting for both dual and single PLA and failed to improve S/N (data not shown). Adjusting the size of particles also failed to improve S/N or undercounting of specific PLA signal (data not shown). These results suggested that Analyze Particles (Image J) cannot properly discriminate specific and nonspecific signals and underestimated specific PLA signal.
Figure 4. . Comparison of automated quantification methods.
Three counting images of the single (A), dual (B) and negative PLA (C) from subject PI12277 were used to test puncta detection and quantification approaches with selected parameters: Particle Analysis (Image J) with auto threshold (Image J_Auto, D-F), Spot Detector (ICY) with parameters favoring detection of either dual and negative (ICY_D, G–I) or single (ICY_S, J–L) PLA signals, and BOPSS (BOPSS, M–O). Image J and ICY quantified the puncta in the transformed and contrast enhanced images (D–L) as described in Supplementary data. The counted puncta were marked in red dots (D-F & M-O) or labeled in red numbers (G-L) in the analyzed images. Single PLA puncta density results were analyzed with repeated one-way ANOVA; there was a significant effect of quantification method (p = 0.014) (P). Dual PLA and its negative control were analyzed by repeated two-way ANOVA, as they shared the same PLA conditions except for the omission of one of the two primary antibodies; the interaction between quantification method and PLA signal was not significant (accounts for 3.73% for the total variance, p = 0.21); both the quantification method (accounts for 56.9% of the total variance, p = 0.005) and the PLA condition (accounts for 26.22% of the total variance, p = 0.009) had significant effects on the variance (Q). Bonferroni's multiple comparisons were performed for both sets of analyses: to compare BOPSS and the other quantification methods: *Multiplicity-adjusted p < 0.05 (P & Q); to compare the results of dual PLA and negative PLA results: #Multiplicity-adjusted p < 0.05 (Q). Data were plotted as mean ± SEM.
PLA: Proximity ligation assay.
Spot detector (ICY) detects PLA puncta through a series of functions: 1, Scales that determine the sensitivity and the size of spots to detect, and 2, a Filtering that determines the maximum and minimum size of accepted objects. When parameters were optimized for dual PLA, this method showed very little – if any – nonspecific detection in the negative controls and good detection for dual PLA, but noticeable underestimation for single PLA (Figure 4G–I & P-Q: ICY_D). With optimized parameters for single PLA, Spot Detector showed increased detection in every PLA assay, including nonspecific detection in the negative controls (Figure 4J–L & P–Q: ICY_S). Therefore, two different sets of parameters were necessary for specific and efficient detection for single PLA (puncta of high density) and dual/negative PLA signal (puncta of low density).
In contrast to ImageJ and ICY, BOPSS uses a machine-learning algorithm to learn the properties of the puncta based on the data. It measures a variety of features for each cluster of pixels and uses the distributions of these features to contrast between negative controls and samples. It processes color images containing cells and PLA signal without relying on any user-defined parameters, but rather on the specification of negative control and experimental images. On a standard laptop PC (2.6 GHz CPU), we analyzed the output results for 10 megabytes of images in 5 s. In a pretest with three full counting images of each PLA condition, single PLA, dual PLA and negative control, this all-in-one script completed machine learning-based optimization and quantification automatically. Compared with the pre-optimization counting (BOPSS_0) that only selected puncta based on size and color, the optimized quantification BOPSS improved the signal (the dual PLA signal)/background noise (the negative control signal) ratio for dual PLA from 1.63 to 8.41, reduced the averaged counts in the negative controls from 1165 ± 300 to 96 ± 29 (puncta/mm2), a 92% reduction in false positives, while maintaining efficient detection in both dual and single PLA (Figure 4 & data not shown).
To assess the accuracy of BOPSS, we randomly selected three areas (Supplementary Figure 2A–C) covering 43% of a full counting image and quantified the PLA puncta using manual counting (Supplementary Figure 2J–L) or with BOPSS (BOPSS_0 and BOPSS, Supplementary Figure 2D–I). The results showed that BOPSS achieved remarkably similar results for dual PLA and negative control compared with the manual counting (Supplementary Figure 2N). For single PLA, BOPSS counted slightly more PLA puncta than the mean of four independent manual counting attempts (Supplementary Figure 2M). However, manual counting, although highly reproducible for the low-density signals, was extremely difficult for the high density puncta and the results were somewhat variable. Post hoc manual analysis verified that BOPSS was overall remarkably accurate but not perfect, as it occasionally undercounted PLA puncta for high density signals (Supplementary Figure 2G), but also occasionally overcounted signal in nuclear areas (Supplementary Figure 2G–I).
Compared with the other two automated quantification methods described previously, BOPSS avoided time-consuming and potentially biased parameter optimization, but nonetheless resulted in comparable quantification results for the dual PLA and negative controls as ICY_D (Figure 4Q). At the same time, BOPSS significantly improved detection of dense single PLA signal, leading to comparable results for single PLA as ICY_S (with optimized parameters for single PLA) (Figure 4P: BOPSS). Therefore, BOPSS achieved a favorable S/N for dual PLA similar to that of ICY_D (Figure 4Q) by reducing nonspecific detection without underestimating real PLA puncta, and simultaneously overcame the underestimation issue for single PLA. In addition, BOPSS led to comparable results as manual counting of PLA puncta (Supplementary Figure 2), and was much more consistent than manual quantification, which is an advantage for processing a large dataset in an unbiased manner. Thus, overall, BOPSS seems to be a straightforward approach for achieving completely automated, efficient and specific detection of PLA signals and worked equally well for both single PLA (puncta of high density) and dual (or negative) PLA signal (puncta of low density).
PLA has been previously applied to postmortem human brains in a very limited number of studies [34,35] and optimization of unbiased quantification methods has not been achieved. Combining whole slide scanning microscopy, the systematic random sampling method, and automated quantification with BOPSS, we developed here a systematic automated approach for quantitative study of PLA-BF, which enhanced our data analysis capacity to the level required for processing image data from the human brain.
Quantifying the expression of D2R, A2AR & D2R-A2AR in the human brain
Because the ventral striatum tissue blocks contained only partial Ptm and Cdt of varied sizes, we focused their quantification on the NAcc. The quantification results from the three human subjects (Supplementary Table 1) showed varied counts of D2R, A2AR and D2R-A2AR complex in the NAcc (Supplementary Figure 3A–C). The averaged relative fractions of D2R-A2AR complex were 12.3 ± 1.8% relative to total A2AR and 13.4 ± 5.3% relative to total D2R. In this small preliminary proof-of-concept analysis, we cannot address whether the differences in the absolute levels of complex and their levels relative to the two single receptors represent true individual variation or differences in the exact regions quantified (Supplementary Figure 3E and F). Nonetheless, the results show that our method can allow relative quantification for both numbers of antigens as well as the proximity between the two antigens.
The existence of heteromers composed of A2AR and D2R has been supported by multiple studies using different approaches, including co-immunoprecipitation in vitro, fluorescence resonance energy transfer and bioluminescence resonance energy transfer in living cells, and PLA-FL in rodent striatum, making it the most characterized class A GPCR heteromer [15–18]. To our knowledge, our study is the first to detect D2R-A2AR complexes in situ in postmortem human brains, suggesting that the physical proximity between D2R and A2AR is conserved from rodent to human. Whether this signal represents true heteromerization of two functional receptors that communicate directly or close proximity of receptors in microdomains or a larger complex remains to be determined. We have explored systematic sampling and automated quantification methods to assess the expression of D2R, A2AR and D2R-A2AR among individuals. Although the number of samples here was very small, the results demonstrate that single and dual PLA assays can be carried out in paraffin sections of human autopsy brains, that the signal can be measured by computerized image analysis and that the results can be used reliably to explore differences in protein expression in a way that is vastly more quantitative than traditional immunostaining. Our study constitutes a proof-of-concept for the feasibility of combining PLA and stereology for the relative quantitation of GPCRs and their complexes in postmortem human brains.
Future perspective
We optimized PLA for use with human brain tissue and demonstrated in striatum the existence of GPCR complexes composed of dopamine receptor D2R and adenosine receptor A2AR (D2R-A2AR) that are potential therapeutic targets for various neurological and psychiatric diseases. Our work highlights the utility of PLA in human brain sections for semi-quantitative analysis of GPCRs and their complexes in pathophysiological conditions.
Supplementary Material
Acknowledgments
The authors would like to thank G Rosoklija and the staff at the Core Facility, Department of Pathology, HICCC, Columbia University Medical Center for technical support.
Footnotes
Author contributions
Y Zhu, P Trifilieff and JA Javitch designed the experiments and wrote the manuscript, with input from all the authors. AJ Dwork prepared all human samples and helped to design the sampling method. Y Zhu performed the PLA assays, designed the sampling method, and performed data analysis. J Mészáros developed BOPSS. Z Sun performed image sampling. R Walle performed data analysis with ICY. R Fan and Y Zhu performed manual quantification.
Financial & competing interests disclosure
This study was supported by National Institutes of Health grants MH54137, MH060877 and MH090964. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The study was approved by the Institutional Review Board (IRB) of New York State Psychiatric Institute (NYSPI, Protocol 6477R). All mice were handled in accordance with the National Institute of Health (NIH) Guide for the Care and Use of Laboratory Animals. Experimental protocol (NYSPI1388) was approved by the Institutional Animal Care and Use Committee (IACUC) at NYSPI.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Howes OD, Kapur S. The dopamine hypothesis of schizophrenia: version III – the final common pathway. Schizophr. Bull. 35(3), 549–562 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel NH, Vyas NS, Puri BK, Nijran KS, Al-Nahhas A. Positron emission tomography in schizophrenia: a new perspective. J. Nucl. Med. 51(4), 511–520 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Niccolini F, Su P, Politis M. Dopamine receptor mapping with PET imaging in Parkinson's disease. J. Neurol. 261(12), 2251–2263 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Politis M, Wilson H, Wu K, Brooks DJ, Piccini P. Chronic exposure to dopamine agonists affects the integrity of striatal D2 receptors in Parkinson's patients. Neuroimage Clin. 16, 455–460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell RA, Herrmann N, Lanctot KL. The role of dopamine in symptoms and treatment of apathy in Alzheimer's disease. CNS Neurosci. Ther. 17(5), 411–427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dauer W, Przedborski S. Parkinson's disease: mechanisms and models. Neuron 39(6), 889–909 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Galvan L, Andre VM, Wang EA, Cepeda C, Levine MS. Functional differences between direct and indirect striatal output pathways in Huntington's disease. J. Huntingtons Dis. 1(1), 17–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trifilieff P, Martinez D. Imaging addiction: D2 receptors and dopamine signaling in the striatum as biomarkers for impulsivity. Neuropharmacology 76, Pt B 498–509 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman JA, Stroup TS, Mcevoy JP. et al. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N. Engl. J. Med. 353(12), 1209–1223 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Urs NM, Peterson SM, Caron MG. New concepts in dopamine D2 receptor biased signaling and implications for schizophrenia therapy. Biol. Psychiatry 81(1), 78–85 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouvier M. Oligomerization of G-protein-coupled transmitter receptors. Nat. Rev. Neurosci. 2(4), 274–286 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Rocheville M, Lange DC, Kumar U, Patel SC, Patel RC, Patel YC. Receptors for dopamine and somatostatin: formation of hetero-oligomers with enhanced functional activity. Science 288(5463), 154–157 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Ng GY, O'dowd BF, Lee SP. et al. Dopamine D2 receptor dimers and receptor-blocking peptides. Biochem. Biophys. Res. Commun. 227(1), 200–204 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Asher WB, Mathiasen S, Holsey MD, Grinnell SG, Lambert NA, Javitch JA. Extreme vetting of dopamine receptor oligomerization. G-Protein-Coupled Receptor Dimers. Springer; (2017).99–127 [Google Scholar]
- 15.Chen JF, Moratalla R, Impagnatiello F. et al. The role of the D(2) dopamine receptor (D(2)R) in A(2A) adenosine receptor (A(2A)R)-mediated behavioral and cellular responses as revealed by A(2A) and D(2) receptor knockout mice. Proc. Natl Acad. Sci. USA 98(4), 1970–1975 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferre S, Bonaventura J, Tomasi D. et al. Allosteric mechanisms within the adenosine A2A-dopamine D2 receptor heterotetramer. Neuropharmacology 104, 154–160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Proposes a model for the functional properties of adenosine A2A receptor -dopamine D2 receptor oligomers and provides evidence regarding their physiological relevance
- 17.Casado-Anguera V, Bonaventura J, Moreno E. et al. Evidence for the heterotetrameric structure of the adenosine A2A-dopamine D2 receptor complex. Biochem. Soc. Trans. 44(2), 595–600 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Canals M, Marcellino D, Fanelli F. et al. Adenosine A2A-dopamine D2 receptor-receptor heteromerization: qualitative and quantitative assessment by fluorescence and bioluminescence energy transfer. J. Biol. Chem. 278(47), 46741–46749 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Diaz-Cabiale Z, Hurd Y, Guidolin D. et al. Adenosine A2A agonist CGS 21680 decreases the affinity of dopamine D2 receptors for dopamine in human striatum. Neuroreport 12(9), 1831–1834 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Cunha RA, Ferre S, Vaugeois JM, Chen JF. Potential therapeutic interest of adenosine A2A receptors in psychiatric disorders. Curr. Pharm. Des. 14(15), 1512–1524 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuxe K, Borroto-Escuela DO, Romero-Fernandez W. et al. Moonlighting proteins and protein-protein interactions as neurotherapeutic targets in the G protein-coupled receptor field. Neuropsychopharmacology 39(1), 131–155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert NA, Javitch JA. CrossTalk opposing view: weighing the evidence for class A GPCR dimers, the jury is still out. J. Physiol. 592(12), 2443–2445 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Discusses the advantages and limitations of available methods for the study of the structure and functions of class A GPCR dimers.
- 23.Vischer HF, Castro M, Pin JP. G protein-coupled receptor multimers: A question still open despite the use of novel approaches. Mol. Pharmacol. 88(3), 561–571 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Gurevich VV, Gurevich EV. GPCR monomers and oligomers: it takes all kinds. Trends Neurosci. 31(2), 74–81 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albizu L, Cottet M, Kralikova M. et al. Time-resolved FRET between GPCR ligands reveals oligomers in native tissues. Nat. Chem. Biol. 6(8), 587–594 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nobis M, Herrmann D, Warren SC. et al. A RhoA-FRET biosensor mouse for intravital imaging in normal tissue homeostasis and disease contexts. Cell Rep. 21(1), 274–288 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Borroto-Escuela DO, Hagman B, Woolfenden M. et al. In situ proximity ligation assay to study and understand the distribution and balance of GPCR homo- and heteroreceptor complexes in the brain. Receptor and Ion Channel Detection in the Brain: Methods and Protocols. Springer; 109–124 (2016). [Google Scholar]
- 28.Trifilieff P, Rives ML, Urizar E. et al. Detection of antigen interactions ex vivo by proximity ligation assay: endogenous dopamine D2-adenosine A2A receptor complexes in the striatum. BioTechniques 51(2), 111–118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• First study to apply PLA on fixed brain tissue and to provide in situ evidence for the existence of D2R-A2AR complexes in the mouse brain.
- 29.Borroto-Escuela DO, Narvaez M, Wydra K. et al. Cocaine self-administration specifically increases A2AR-D2R and D2R-sigma1R heteroreceptor complexes in the rat nucleus accumbens shell. Relevance for cocaine use disorder. Pharmacol. Biochem. Behav. 155, 24–31 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Fredriksson S, Gullberg M, Jarvius J. et al. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20(5), 473–477 (2002). [DOI] [PubMed] [Google Scholar]; • Seminal study that describes the use and validity of proximity ligation assay.
- 31.Soderberg O, Gullberg M, Jarvius M. et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3(12), 995–1000 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Gullberg M, Gustafsdottir SM, Schallmeiner E. et al. Cytokine detection by antibody-based proximity ligation. Proc. Natl Acad. Sci. USA 101(22), 8420–8424 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]; • First paper demonstrating the feasibility of antibody-based proximity ligation assay.
- 33.Biezonski DK, Trifilieff P, Meszaros J, Javitch JA, Kellendonk C. Evidence for limited D1 and D2 receptor coexpression and colocalization within the dorsal striatum of the neonatal mouse. J. Comp. Neurol. 523(8), 1175–1189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts RF, Wade-Martins R, Alegre-Abarrategui J. Direct visualization of alpha-synuclein oligomers reveals previously undetected pathology in Parkinson's disease brain. Brain 138(6), 1642–1657 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thonberg H, Chiang HH, Lilius L. et al. Identification and description of three families with familial Alzheimer disease that segregate variants in the SORL1 gene. Acta. Neuropathol. Commun. 5(1), 43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frederick AL, Yano H, Trifilieff P. et al. Evidence against dopamine D1/D2 receptor heteromers. Mol. Psychiatry 20(11), 1373–1385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gundersen HJ, Jensen EB, Kieu K, Nielsen J. The efficiency of systematic sampling in stereology–reconsidered. J. Microsc. 193(Pt 3), 199–211 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Mészáros J. Bayesian Optimized PLA Signal Sorting (BOPSS) (2019). https://github.com/neurojojo/BOPSS [Google Scholar]
- 39.Wu M-N, Lin C-C, Chang CC. Brain Tumor Detection Using Color-Based K-Means Clustering Segmentation. Presented at: Third International Conference on Intelligent Information Hiding and Multimedia Signal Processing. Kaohsiung, Taiwan, 26–28 November, 2007, 2, 245–250 (2007). [Google Scholar]
- 40.Alshammari MA, Alshammari TK, Laezza F. Improved methods for fluorescence microscopy detection of macromolecules at the axon initial segment. Front. Cell. Neurosci. 10, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lavenex P, Lavenex PB, Bennett JL, Amaral DG. Postmortem changes in the neuroanatomical characteristics of the primate brain: hippocampal formation. J. Comp. Neurol. 512(1), 27–51 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan F, Wu X, Crawford M. et al. The search for an optimal DNA, RNA, and protein detection by in situ hybridization, immunohistochemistry, and solution-based methods. Methods 52(4), 281–286 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The authors systematically compared nine fixation methods for their ability to preserve cell morphology and DNA/RNA/protein quality for histological applications.
- 43.Taura J, Fernandez-Duenas V, Ciruela F. Visualizing G protein-coupled receptor-receptor interactions in brain using proximity ligation in situ assay. Curr. Protoc. Cell Biol. 67(1), 17.17.11–16 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Borroto-Escuela DO, Romero-Fernandez W, Narvaez M, Oflijan J, Agnati LF, Fuxe K. Hallucinogenic 5-HT2AR agonists LSD and DOI enhance dopamine D2R protomer recognition and signaling of D2-5-HT2A heteroreceptor complexes. Biochem. Biophys. Res. Commun. 443(1), 278–284 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Davis AS, Richter A, Becker S. et al. Characterizing and diminishing autofluorescence in formalin-fixed paraffin-embedded human respiratory tissue. J. Histochem. Cytochem. 62(6), 405–423 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schindelin J, Arganda-Carreras I, Frise E. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9(7), 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Chaumont F, Dallongeville S, Chenouard N. et al. Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods 9(7), 690–696 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.