Abstract
Long noncoding RNA (lncRNA) biology is a new and exciting field of research, with the number of publications from this field growing exponentially since 2007. These studies have confirmed that lncRNAs are altered in almost all diseases. However, studying the functional roles for lncRNAs in the context of disease remains difficult due to the lack of protein products, tissue-specific expression, low expression levels, complexities in splice forms, and lack of conservation among species. Given the species-specific expression, lncRNA studies are often restricted to human research contexts when studying disease processes. Since lncRNAs function at the molecular level, one way to dissect lncRNA biology is to either remove the lncRNA or overexpress the lncRNA and measure cellular effects. In this article, a written and visualized protocol to overexpress lncRNAs in vitro is presented. As a representative experiment, an lncRNA associated with inflammatory bowel disease, Interferon Gamma Antisense 1 (IFNG-AS1), is shown to be overexpressed in a Jurkat T-cell model. To accomplish this, the activating clustered regularly interspaced short palindromic repeats (CRISPR) technique is used to enable overexpression at the endogenous genomic loci. The activating CRISPR technique targets a set of transcription factors to the transcriptional start site of a gene, enabling a robust overexpression of multiple lncRNA splice forms. This procedure will be broken down into three steps, namely (i) guide RNA (gRNA) design and vector construction, (ii) virus generation and transduction, and (iii) colony screening for overexpression. For this representative experiment, a greater than 20-fold enhancement in IFNG-AS1 in Jurkat T cells was observed.
Keywords: Genetics, Issue 145, lncRNA, CRISPR, IFNG-AS1, overexpression, lentiviral, real-time PCR
Introduction
While most biomedical research has focused on protein-coding transcripts, the majority of transcribed genes actually consists of noncoding RNAs (Ensembl release 93). Current research is beginning to explore this field, with the number of publications on lncRNAs in disease processes rising exponentially between 2007 and 20171. These publications demonstrate that many lncRNAs are associated with disease. However, the molecular mechanisms of these lncRNAs are difficult to study due to their diverse functions as compared to mRNAs. Compounding the problem of understanding the role of lncRNAs in disease, lncRNAs are often expressed at lower levels than coding RNAs2. Additionally, lncRNAs are poorly conserved, which limits the functional studies in human-cell-line-based techniques3. One method to study the mechanism of these novel genes is to endogenously overexpress them in cultured cells. Overexpression studies can provide key information as to the function of specific genes and enable researchers to dissect key molecular pathways.
A new method to activate the transcription of lncRNA genes is based on CRISPR technologies developed initially by the Gersbach laboratory4. This protocol has been adapted for use in lncRNA biology and for the expression of these genes in other model systems. In the CRISPR overexpressing technique, a protein called CRISPR-associated protein 9 (Cas9) can be directed toward a DNA sequence via an antisense gRNA that is recognized by Cas9. Normally, Cas9 will induce DNA cleavage; however, mutations in Cas9, previously developed for the overexpression technique, deactivate this step5. When a transcriptional activator is fused to a “dead” Cas9 (dCas9) and transduced into cell lines, the endogenous overexpression of lncRNAs can be achieved4,6. The addition of additional modifications to the gRNA, enabling additional transcriptional factors to bind to the gRNA, increased the efficacy of the dCas9 gene activation system 10-fold7. Importantly, it was demonstrated that transcriptional activation requires close proximity (<200 bp) to the transcriptional start site genes, enabling a specific upregulation in gene-rich areas7. Unlike CRISPR knockout technologies, dCas9 and gRNA cassettes need to be integrated into the genome to allow for cells to retain an overexpression over multiple generations. One method to achieve this is to use lentiviruses to integrate the dCas9- and gRNA-containing cassettes. After integration, the lncRNA gene expression can be determined.
In this article, a protocol for lncRNA overexpression in a Jurkat T-cell model will be demonstrated. A step-by-step procedure is shown and can also be adapted to adherent cells.
Protocol
NOTE: It is important to note that this protocol uses replication-deficient lentiviruses. Perform viral handling only after appropriate lab safety training. Bleach all items and surfaces that come in contact with live viruses for a minimum of 10 min after handling. Use a disposable lab coat and face/eye protection, as well as double gloves, at all times. Perform virus work in biosafety level 2+ labs with viral certification. Dedicate tissue culture hoods and incubators to viral work.
1. Vector Design and Generation
NOTE: The best way to identify gRNA sequences is to use online design tools (e.g., http://crispr-era.stanford.edu) (Supplemental Figure 1: gRNA design). For the accompanying representative experiment, a company designed and created the gRNA vectors used in the representative experiment. The dCas9 plasmid was also purchased online.
The gRNA sequence is located upstream of the nucleotide sequence “NGG”, where “N” is any nucleotide, which also is within 100 basepairs of the transcriptional start site. Search genetic databases such as BLAT (https://genome.ucsc.edu/cgi-bin/hgBlat?command=start) for the gRNA sequence, to make sure there are no other sites in the genome with similar sequences (Supplemental Figure 2: BLAT). This will ensure that the gRNA sequence is unique to the gene of interest (GOI).
Store the gRNA and dCas9 plasmids, which come as bacterial stubs, at 4 °C. Streak out Escherichia coli on a Luria broth (LB) agar plate with ampicillin (Supplemental Table 1) and grow the bacteria overnight at 37 °C.
Pick a colony and grow the bacteria in 5 mL of LB with ampicillin (Supplemental Table 1) overnight, vigorously shaking the plate in a 37 °C incubator. The next day, add 2 mL of bacteria to 200 mL of LB with ampicillin grow in a 2 L flask, vigorously shaking the flask overnight in a 37 °C incubator.
Spin down the bacteria at 3,724 × g for 20 min. Remove the media and place the tube containing the bacteria on ice.
-
Purify bacterial DNA using a plasmid purification kit per the manufacturer’s protocol.
NOTE: The plasmid purification kits contain proprietary resuspension buffer, lysis buffer, wash buffer, equilibration buffer, and elution buffer.
Add 10 mL of resuspension buffer from the kit to the bacteria and pipet the bacteria into the solution. Then, add 10 mL of lysis buffer from the kit to the resuspended bacteria and mix them by inverting the tube. Wait 5 min to lyse the bacteria; then, add 10 mL of ice-cold neutralization buffer from the kit to the lysed bacteria and mix them by inverting the tube.
Equilibrate the DNA column from the kit with 10 mL of equilibration buffer for 5 min. Pour the samples into the DNA column filter and let the solution pass through the filter.
Wash the samples 2x with 30 mL of wash buffer and, then, elute the DNA with 30 mL of elution buffer in a 15 mL conical tube. Slowly layer 10.5 mL of isopropanol at room temperature onto the eluted DNA, invert the tube, and immediately spin it at 16,200 × g for 30 min at 4 °C.
Remove the supernatant and add 1 mL of 70% ethanol to the DNA pellet. Resuspend the pellet by flicking the tube. Spin it at 16,200 × g for 10 min at 4 °C and use a 200 μL pipet tip to remove most of the ethanol. Air-dry the pellet for 5 min and resuspend it in 200 μL of water, and store the DNA at −20 °C. The expected concentration will be >1 μg/μL.
2. Virus Generation and Particle Count
When ready to create the dCAS9-containing lentivirus, coat 100 mm tissue culture dishes with 5 mL of 0.01% poly-L-lysine (Supplemental Table 1). Remove the excess liquid thoroughly from the plates and let them air-dry for a few minutes in a biosafety cabinet.
Plate 5 × 106 HEK 293T cells per dish in 10 mL of complete Dulbecco’s modified Eagle’s medium (DMEM) (Supplemental Table 1) and incubate them overnight at 37 °C with 5% CO2. The next day, remove the medium from the HEK 293T dishes and add 10 mL of complete DMEM.
Mix 6.5 μg of plasmid pMDLg/pRRE containing the gag/pol (components of the virus), 3.5 μg of the plasmid pMDG2.G containing the VSVG (a component of the virus), 2.5 μg of the plasmid pRSV-Rev containing Rev (a component of the virus), 10 μg of an long terminal repeat (LTR)-containing gRNA vector or an LTR-containing dCas9 vector and add water to a total volume of 450 μL. Filter the mixture through a 0.2 μm filter tip attached to a syringe.
Add 50 μL of 2.5 M CaCl2 (Supplemental Table 1) to each transfection sample of DNA and mix gently. Filter the calcium/DNA mixture through a 0.2 μm filter tip attached to a syringe. Pipet 500 μL of 2x HBS (Supplemental Table 1) into a 5 mL polystyrene tube. Add the 500 μL DNA/CaCl2 mix dropwise and gently vortex. Incubate at room temperature for 3 min.
Slowly add 1 mL of the DNA/CaCl2/HBS suspension to each dish. Immediately swirl the dishes to distribute the contents evenly. Incubate each dish overnight in a 5% CO2 incubator at 37 °C.
On day 3, slowly remove and discard the media from the dishes. Carefully wash the cells 1x with phosphate-buffered saline (PBS). Add 6 mL of complete DMEM supplemented with 20 mM HEPES and 10 mM sodium butyrate. Incubate the cells for 5–6 h in a 5% CO2 incubator at 37° C.
Wash the cells 1x with PBS and add 5 mL of complete DMEM with 20 mM HEPES to the HEK 293T cells. Incubate for 12 h overnight in a 5% CO2 incubator at 37 °C.
On day 4, collect the HEK 293T cell supernatants and filter the supernatant. Freeze 1 mL aliquots at −80 °C.
When ready to use the virus, thaw an aliquot of the viral particle containing conditioned media (stored as described in step 2.8) on ice. Bring all reagents to room temperature.
-
Use a p24 Enzyme-Linked Immunosorbent Assay (ELISA) kit to quantify the number of viral particles per milliliter.
Dilute the wash concentrate from the kit by adding 19 parts of distilled deionized water. Dilute the positive control from the kit to 200 ng/mL, using RMPI 1640 as the diluent, and make the dilutions for standard curve in 1.5 mL tubes according to the p24 ELISA dilution table (Supplemental Table 2).
Add 20 μL of 5% Triton X-100 to all wells except the substrate blank. Add 200 μL of RPMI 1640 to the negative control wells. Add 200 μL of each standard (in duplicate) to the designated wells.
Using a spectrophotometer, measure the concentration of the virus within the samples, using a standard curve. Start the sample dilution at 1:1,000 and modify the volume as necessary in order to be within the range of the standard curve. Dilute the samples with Triton X-100 to a final concentration of 0.5% and add 200 μL of each sample in RPMI 1640 to designated wells. Seal the plate and incubate it for 2 h at 37 °C.
Wash the plate 6x with 300 μL of 1x wash buffer per well. Remove any excess fluid by inverting the plate and tapping it on a paper towel. Add 100 μL of detector antibody from the kit to all wells except the substrate blank. Seal the plate and incubate it for 1 h at 37 °C. Wash the plate and, then, remove any excess fluid by inverting the plate and tapping it on a paper towel.
In order to measure the detector antibody, prepare streptavidin-horseradish peroxidase (SA-HRP) within 15 min of use. Dilute the SA-HRP at 1:100 with SA-HRP diluent. Mix the diluted SA-HRP thoroughly and add 100 μL to all wells except the blank. Seal and incubate the plate for 30 min at room temperature.
Wash the plate with 1x wash buffer and tap away any excess liquid as in step 2.10.4. Use ortho-phenylenediamine (OPD) substrate solution, which provides the peroxidase the necessary substrates to produce chemiluminescence, within 15 min of preparation. Use one OPD tablet to 11 mL of substrate diluent for each plate.
Vortex the OPD solution vigorously to dissolve it completely and protect it from light. Add 100 μL of OPD substrate solution to all wells, including the blank. Using a spectrophotometer, read absorbance at 450 nm immediately, repeat this 10x at 1 s intervals, and take the average measurement.
3. dCas9-VP64 Transduction
Culture Jurkat T cells in complete DMEM in a T75 flask with a density of 2 × 106 cells in 15 mL of media. Culture the cells in a 37 °C incubator with 5% CO2.
Spin down the cells for 5 min at 233 × g and, then, resuspend them in 10 mL of reduced serum media. Count the cells with a hemocytometer or an automated cell counter.
Resuspend and plate 1 × 106 Jurkat cells in 5 mL of reduced serum media with polybrene (Supplemental Table 1) in a T75 flask. Add HEK 293T-conditioned media containing 1 × 106 dCas9-containing viral particles. The number of viral particles can be calculated roughly from the p24 ELISA because 1 μg/mL p24 equals 1 × 107 viral particles. Put the flasks in 37 °C incubator with 5% CO2.
4. Selection and Clone Creation
Three days post-infection, spin down the cells at 233 × g for 5 min and resuspend them in 10 mL of complete DMEM with puromycin (Supplemental Table 1). Plate the cells in T75 flasks and culture them at 37 °C with 5% CO2. Every third day, for a period of 9 days, spin down the cells at 233 × g for 5 min and replace the media with complete DMEM with puromycin.
Count the cells using a hemocytometer or an automated cell counter. On a 96-well plate treated with tissue culture, plate 10,000 cells in 100 μL of complete DMEM with puromycin in the first well and serially dilute 1:1 the contents of the following wells with complete DMEM with puromycin. Perform 24 dilutions and repeat this 4x per plate in order to ensure that there is an adequate number of cells per well. Incubate the cells at 37 °C with 5% CO2.
Expand clonal cells into two 24-well plates, then into a 6-well plate, and eventually into a T75 flask. In order to focus on three of the clones, plate 2 × 106 cells per well of a 6-well plate for three of the clones. Plate the other three wells with nontransduced Jurkat cells as controls. Put the cells in a cell culture incubator overnight.
For RNA extraction, use a commercially available RNA isolation kit. Spin down the cells at 233 × g for 5 min at room temperature and remove the media. Use a 1 mL pipet tip to resuspend the cells in 350 μL of lysis buffer. Add 350 μL of 70% ethanol to the lysed cells, mix thoroughly with a 1 mL pipet, and transfer the sample to the RNA column.
Spin the lysates at 10,000 × g for 1 min, remove the flow-through, and add 700 μL of low-stringency wash buffer from the kit to the column. Spin the lysates at 10,000 × g for 1 min, remove the flow-through, and add 80 μL of DNase I solution from the kit to the column; let the samples incubate for 15 min at room temperature.
Spin the lysates at 10,000 × g for 1 min, remove the flow-through, and add 700 μL of high-stringency wash buffer from the kit to the column. Spin the lysates at 10,000 × g for 1 min, remove the flow-through, and add 700 μL of low-stringency wash buffer to the column.
Remove the column from the collection tube and transfer it to a new collection tube. Spin the lysates at 10,000 × g for 1 min and, then, transfer the RNA column to a 1.5 mL tube. Add 50 μL of water to the column and spin at 10,000 x g for 1 min.
Use a spectrophotometer to quantify the concentration of RNA. Expect the concentration to be between 100–500 ng/μL, with a 260/280 absorbance between 1.9–2.1. Store the RNA at −80 °C.
In 250 μL tubes, add 1 μg of RNA, 4 μL of complementary DNA (cDNA) synthesis buffer, 1 μL of reverse transcriptase, and water to a final volume of 20 μL. Include no reverse transcriptase controls. In a thermal cycler, synthesize cDNA at 42 °C for 30 min and at 95 °C for 5 min to inactivate the reverse transcriptase. Dilute the cDNA with 60 μL of water after the synthesis.
-
Prepare polymerase chain reactions (PCRs) containing 5 μL of SYBR green, 4 μL of cDNA, 0.5 μL of forward primer (10 μM), and 0.5 μL of reverse primer (10 μM). Run the PCR as follows: step 1 = 95 °C for 3 min, step 2 = 95 °C for 10 s, step 3 = 60 °C for 10 s, and then, repeat steps 2 and 3 for 30x.
NOTE: The sequence for the dCas9 forward primer is 5’-TCGCCACAGCATAAAGAAGA and the sequence for the dCas9 reverse primer is 5’-CTTTTCATGGTACGCCACCT. The forward primer for Hypoxanthine Phosphoribosyltransferase 1 (HPRT1) is 5’-GACCAGTCAACAGGGGACAT and the reverse primer is 5’-GCTTGCGACCTTGACCATCT.
5. gRNA Transduction, Selection, Clone Creation, and Screening
Retransduce the validated Jurkat-dCas9 cells with gRNA-containing viruses as outlined in section 3. Select cells in DMEM with hygromycin (Supplemental Table 1) for 10 days. Spin down the cells and change the media every 3 days.
Perform a serial dilution of cells (as described in step 4.2) and expand individual colonies (as described in step 4.3). Purify RNA from the colonies exactly as described in step 4.5 and perform cDNA synthesis as described in step 4.9. Perform real-time PCR against the GOI and HPRT1 or an appropriate housekeeping gene, similarly to the PCR described in step 4.10, except in this case, image the wells each cycle after step 3.
Use the comparative cycle threshold (Ct) method to determine fold change in the GOI8. Briefly, use the following equation to calculate relative transcript levels (RTLs): 2−(average Ct of GOI – average Ct of housekeeping gene). Then, calculate the average RTL of the controls and divide all RTL values by the average control RTL to generate a fold change compared to the control samples.
Representative Results
A dual vector system to overexpress the lncRNA IFNG-AS1
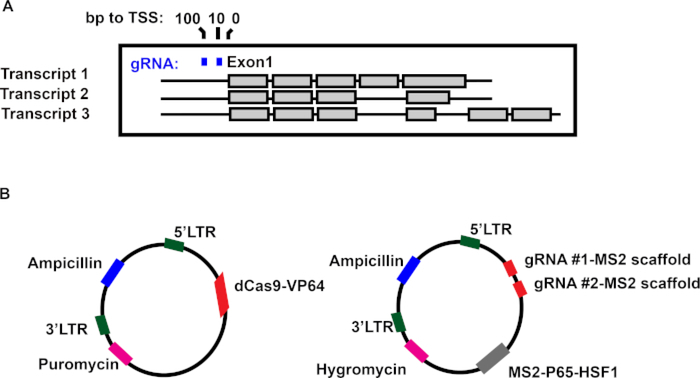
The example experiment in this manuscript is the overexpression of a Jurkat T-cell model system expressing the lncRNA IFNG-AS19. IFNG-AS1 is an lncRNA associated with inflammatory bowel disease, that has been seen to regulate Interferon Gamma10. The IFNG-AS1 gene contains three splice variants that all use the same transcriptional start site (Figure 1A). Therefore, gRNA sequences that were 10 and 100 bp away from the transcriptional start site and had an “NGG” sequence upstream were used for directing the Cas9-activating complex to the transcriptional locus of IFNG-AS1 (Figure 1A). A two-plasmid system was used to transduce either dCas9 or gRNAs/enhancers into cells as a single plasmid alone makes viral particle generation difficult due to plasmid size (Figure 1B). To enable the selection of double-transduced cells, the dCas9 vector contained a puromycin (aminonucleoside) resistance gene, and the gRNA-containing plasmid contained a hygromycin (aminoglycoside) resistance gene. gRNAs were fused to an MS2 scaffold sequence that enabled the MS2 scaffold protein to bind to the gRNAs. Fused to the MS2 protein are additional transcriptional activators to enhance the overexpression of IFNG-AS17. Using these vectors, viral particles can be created to transduce this overexpression system into most human cell lines.
Figure 1: A dual vector system to overexpress the lncRNA IFNG-AS1.
(A) A schematic of the IFNG-AS1 gene structure and the relationship between guide RNA (gRNA) binding sites and the transcriptional start site (TSS). (B) The features of the gRNA and dCAS9 vectors.
Viral titering and colony screening
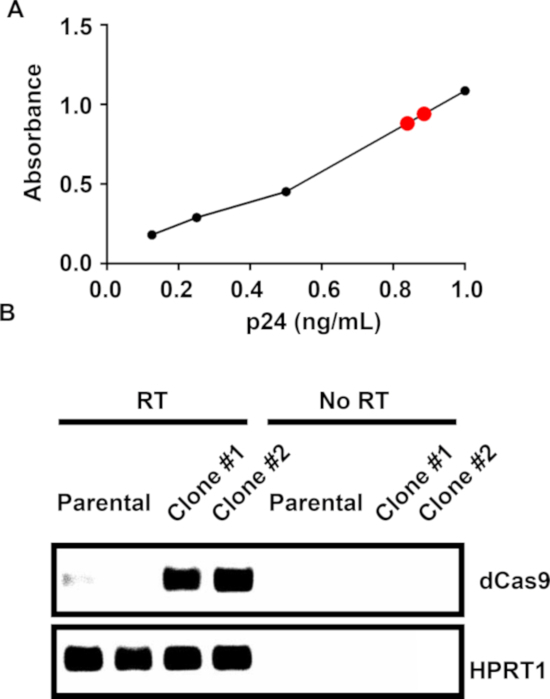
After generating the dCas9- and gRNA-containing plasmids, plasmid purifications were performed and lentiviruses were created. As lentiviruses randomly integrate their cassettes into the genome, a quantification of the number of viral particles enables the least number of integrations possible. To accomplish this, conditioned-media-containing viruses were measured with a p24 ELISA kit, allowing for the calculation of the number of virions per milliliter. After measuring, both viral supernatants had nearly 1 μg/mL of p24, which allowed the use of 100 μL of virus to transduce the Jurkat cells. After transduction, antibiotic-selected cells were serially diluted and clones were expanded. Cas9 expression was then analyzed by real-time PCR and agarose gel electrophoresis (Figure 2B). Both clones selected were positive for Cas9 expression. To confirm mRNA expression, reverse transcriptase was omitted from the cDNA reaction (Figure 2B). Primers against HPRT1 were used to confirm the presence of RNA in the nontransduced cells.
Figure 2: Viral titering and colony screening.
(A) An example p24 ELISA standard curve for lentivirus-containing, conditioned media. The black dots represent standard samples and the red dots unknown samples. (B) After transducing, selecting, and generating colonies, real-time PCR and gel electrophoresis against dCas9 were performed on the RNA from either nontransduced Jurkat cells (parental) or dCas9-transduced clones. No reverse transcriptase (No RT) controls were performed.
Measuring IFNG-AS1 gene expression
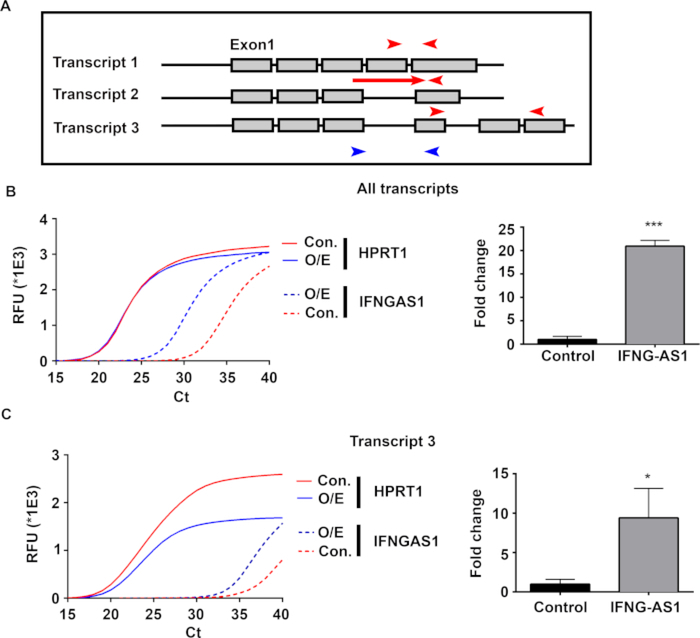
Using the dCas9 clones as a parental cell line, cells were transduced with either a nontargeting gRNA-containing virus or a virus containing gRNAs against IFNG-AS1. After gRNA transduction, selection, and clone creation analogous to dCas9 cell line creation, the IFNG-AS1 expression was measured to verify overexpression. IFNG-AS1 produces three splice variants, each of which can be detected individually with transcript-specific primers (red) or against all known IFNG-AS1 transcripts (blue) (Figure 3A). All fluorescence curves were exponential with HPRT1 reaching the exponential phase (or, Ct) within a half cycle between control and IFNG-AS1-gRNA-expressing cells (Figure 3B,C). Primers against all known IFNG-AS1 transcripts were the most detectable between experiments with measurements of 20-fold increases in IFNG-AS1 (Figure 3B). While primers against transcripts against 1 and 2 successfully amplified in peripheral blood mononuclear cells (PBMCs), these transcripts were not detectable in Jurkat cells (data not shown). However, when the third transcript of IFNG-AS1 was detectable in concentrated RNA, a five- to tenfold significant increase in IFNG-AS1 levels was seen. These data suggest either alternative splicing of IFNG-AS1 in Jurkat cells exist compared to primary cells. Primers against IFNG-AS1 detected large increases in this gene, thereby validating the activating CRISPR overexpression system.
Figure 3: Measuring IFNG-AS1 gene expression.
(A) A diagram of the relationship between primer sets and transcript variants. The red arrows represent transcript-specific primers, while the blue arrows indicate primers against all known transcripts. (B and C) Representative average PCR curves and fold-change quantifications for control and IFNG-AS1-overexpressing cells. RFU = relative fluorescence units. N = 3 samples per group. Mean ± SD. *p < 0.05, ***p < 0.001. A Student’s t-test was used to calculate the p-values.
Discussion
This manuscript presents a protocol for using activating CRISPR to overexpress lncRNAs in vitro. This is an especially important technique when studying long noncoding RNAs as the transcriptomic product is the functional unit. After overexpression, the researcher can, then, use these cells to increase the signal-to-noise ratio when studying binding partners and even measure the cellular consequences of increased levels of lncRNAs. Additionally, as lncRNAs frequently act on cis-genes, this endogenous overexpression technique enables these events to be studied11,12.
This manuscript highlights a generalized protocol for gRNA creation and lentivirus generation, transduction, and selection, and a representative example of lncRNA overexpression in a peripheral blood T cell line was outlined. Additionally, this technique has already been shown to be successful in bone-marrow-derived immune cells13, neurons7, mouse embryonic stem cells14, and kidney epithelial cells4. While this protocol is generally applicable for most cell lines, cells that are hard to transduce might require individual titers as to enable higher infection rates. Additionally, it is important to perform antibiotic kill curves when using new cell lines, as this protocol utilizes a positive selection strategy.
Of note, this protocol has several technical aspects that require special attention. Reproducible and consistent pipetting for real-time PCR is critical to establish reproducible results. Even small differences in volumes will cause highly variable values, thereby making interpretation difficult. In addition to a proper quantitative PCR primer design, controls, including the no-reverse-transcriptase control for the real-time PCR primers, are critical as they confirm that the RNA being quantified is not genomic DNA. The selection of housekeeping genes for real-time PCR is also important in order to adequately perform these experiments. While HPRT1 was chosen in this protocol, other genes of interests targeted by this technique might alter HPRT115. A careful selection of housekeeping genes based on these factors should be taken into consideration.
There are a few drawbacks to activating CRISPR and to particular aspects of this protocol. One potential limitation is the expression of transcripts in highly gene-rich regions as other neighboring genes could be turned on. It is possible that neighboring genes in close proximity may be activated and controls should be performed to assess this. In addition, other limitations include the lack of binding for particular gRNAs to a given DNA sequence. The selection of multiple gRNAs may be required to identify the appropriate gRNA to drive expression. Another caveat to the system is that the cell of interest has to have the capacity to drive the promoters in the cassettes in order to drive the expression of the gRNAs and dCas9. For the studies presented here, the Jurkat T-cell model was able to drive the expression of these components for a successful overexpression system.
Ideally, the protocol described here should be paired with other corroborating information, such as single transcript overexpression data and functional effects from knockdown or knockout studies, to bolster the case for any of the subsequent findings. This technique offers a robust way of overexpressing any gene in a particular cell type. Given the limitations of lncRNA biology, such as poor species conservation, techniques such as the one described here are critical to exploring their function in relevant cell types. This study focused on one lncRNA that has been implicated in inflammatory bowel disease pathophysiology but serves as a model of studying the function of other lncRNAs in human disease biology.
Supplementary Material
Supplemental Figure 1: gRNA design
Supplemental Figure 2: BLAT
Supplemental Table 1: Solutions
Supplemental Table 2: ELISA dilutions
Acknowledgments
C.P. is supported by RO1 DK60729 and P30 DK 41301-26. D.P. is supported by a Crohn’s & Colitis Foundation (CCFA) career development award, CURE: Digestive Diseases Research Center (DDRC) DK41301, and UCLA Clinical and Translational Science Institute (CTSI) UL1TR0001881. The UCLA virology core was funded by the Center of AIDS Research (CFAR) grant 5P30 AI028697. The UCLA Integrated Molecular Technologies core was supported by CURE/P30 grant DK041301. This work was also supported by the UCLA AIDS Institute.
Footnotes
Disclosures
The authors have nothing to disclose.
Video Link
The video component of this article can be found at https://www.jove.com/video/59233/
References
- 1.Miao Y et al. Trends of long noncoding RNA research from 2007 to 2016: a bibliometric analysis. Oncotarget. 8 (47), 83114–83127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derrien T et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Research. 22 (9), 1775–1789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnsson P, Lipovich L, Grander D, Morris KV Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochimica et Biophysica Acta. 1840 (3), 1063–1071 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez-Pinera P et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nature Methods. 10 (10), 973–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337 (6096), 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeder ML et al. CRISPR RNA-guided activation of endogenous human genes. Nature Methods. 10 (10), 977–979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konermann S et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 517 (7536), 583–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmittgen TD, Livak KJ Analyzing real-time PCR data by the comparative C(T) method. Nature Protocols. 3 (6), 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Gomez JA et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 152 (4), 743–754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Padua D et al. A long noncoding RNA signature for ulcerative colitis identifies IFNG-AS1 as an enhancer of inflammation. American Journal of Physiology-Gastrointestinal and Liver Physiology. 311 (3), G446–457 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collier SP, Henderson MA, Tossberg JT, Aune TM Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. Journal of Immunology. 193 (8), 3959–3965 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotake Y et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 30 (16), 1956–1962 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert LA et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 159 (3), 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng AW et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Research. 23 (10), 1163–1171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matouskova P et al. Reference genes for real-time PCR quantification of messenger RNAs and microRNAs in mouse model of obesity. PLoS One. 9 (1), e86033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: gRNA design
Supplemental Figure 2: BLAT
Supplemental Table 1: Solutions
Supplemental Table 2: ELISA dilutions