Abstract
The SARS-CoV nucleocapsid (N) protein serves multiple functions in viral replication, transcription, and assembly of the viral genome complex. Coronaviruses specifically package genomic RNA into assembled virions, and in SARS-CoV, it is reported that this process is driven by an interaction between the N-protein and a packaging signal encoded within the viral RNA. While recent studies have uncovered the sequence of this packaging signal, little is known about the specific interaction between the N-protein and the packaging signal sequence, and the mechanisms by which this interaction drives viral genome packaging. In this study, we developed a novel in vivo cell-based assay for examining this interaction between the N-protein and packaging signal RNA for SARS-CoV, as well as other viruses within the coronaviridae family. Our results demonstrate that the N-protein specifically recognizes the SARS-CoV packaging signal with greater affinity compared to signals from other coronaviruses or non-coronavirus species. We also use deletion mapping to identify a 151-nt region within the packaging signal sequence that is critical for N-protein-RNA binding, and conversely, we show that both the N-terminal and C-terminal domains of the N protein are necessary for recognizing the packaging RNA. These results describe, for the first time, in vivo evidence for an interaction between the SARS-CoV N-protein and its packaging signal RNA, and demonstrate the feasibility of using this cell-based assay to further probe viral RNA-protein interactions in future studies.
Keywords: SARS, Coronavirus, Packaging signal sequence, Nucleocapsid, Cell-based assay, Protein-RNA interaction
Highlights
-
•
Development of a novel assay to examine interactions between the SARS-CoV N-protein and viral RNA in cells in vivo.
-
•
N-protein alone is able specifically recognize the CoV packaging signal RNA in vivo.
-
•
Both the N-terminal and C-terminal are required for the N-protein to interact with the viral RNA.
1. Introduction
Severe acute respiratory syndrome (SARS) is a severe respiratory disease caused by infection with the SARS-associated coronavirus (SARS-CoV), a member of the coronaviridae family [[1], [2], [3]]. SARS-CoV is an enveloped, plus-sense, single-stranded RNA virus that can infect and replicate in a number of human cell types, including pneumocytes and enterocytes in the respiratory system, intestinal mucosal cells, immune cells, and neurons. A defining property of the virus is that it produces a nested set of subgenomic RNAs via discontinuous transcription, which are in turn translated into viral structural proteins as well as additional accessory proteins [3,4]. The four structural proteins include the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins, which are encoded in the 3’ end of the viral genome; of these, the N-protein is the most abundant protein in virus-infected cells, and is the only protein found in the nucleocapsid.
Following translation, these SARS proteins enter the secretory pathway in the endoplasmic reticulum-Golgi intermediate compartment (ERGIC), where they are assembled into virions. While the M protein coordinates the majority of protein-protein interactions required for coronavirus assembly, the coronavirus packaging signal, a cis-regulatory element encoded within the viral RNA, plays an essential role in packaging the viral genome into the capsid. Previous studies have identified the packaging signals of the human coronavirus and the mouse hepatitis virus as a conserved region embedded within the coding sequence for nonstructural protein 15 [[5], [6], [7], [8], [9], [10], [11], [12], [13], [14]]. Meanwhile, the 580-nucleotide SARS-CoV packaging sequence lies within the coding sequence for nsp12 [14]. The mechanism by which this sequence regulates viral packaging, however, remains elusive.
Recently, studies have demonstrated that packaging of the SARS-CoV viral genome may require the N-protein, which has been shown to interact with a 318-nucleotide sequence within the packaging signal in a ribonucleoprotein complex to form virus-like particles in Vero E6 cells. The N-protein harbors two distinct RNA-binding domains, a potentially unique characteristic among capsid proteins of other RNA viruses. While the N-terminal domain (NTD) functions as a monomer, the C-terminal domain (CTD) of the N-protein forms oligomers that may drive helical nucleocapsid formation [[15], [16], [17], [18], [19]]. Outside of viral RNA packaging, the N-protein has been implicated in maintaining viral structure and viability, as well as viral RNA transcription and translation.
Although crystallography and nuclear magnetic resonance studies have determined the structures for the RNA-binding domains of the various coronoaviruses, no studies to date have characterized the structure of the N-protein-viral RNA complex, or have otherwise elucidated the mechanism by which the N-protein recognizes and interacts with the SARS-CoV viral RNA [15,[20], [21], [22]]. Thus far, studies that have characterized this interaction have relied on systems using either unnatural synthetic RNA or non-viral RNA substrates in vitro. Nonetheless, investigating the ability of the N-protein to interact with the packaging signal in living cells is of particular importance, as this would address whether and if so, how the N-protein alone might be able to recognize and interact specifically with the packaging signal amongst a pool of diverse cellular RNAs in vivo. Here, we address this gap in the literature by developing an in vivo cell-based platform to probe the specific interaction between the SARS-CoV N-protein and packaging RNA sequence. We demonstrate that the SARS-CoV N-protein is specific for the SARS-CoV packaging RNA sequence, with lower affinity for packaging sequences of other coronaviruses and even lower for non-coronavirus species. We also show that a 151-nucleotide sequence (positions 168–318) of the SARS-CoV packaging sequence is critical for N-protein binding. Conversely, deletion mapping of the N-protein revealed that both the NTD and the CTD are necessary for binding CoV packaging sequences, and that neither alone were sufficient. These results represent the first time in which the N-protein-viral RNA interaction was examined in vivo, and provides a framework for further elucidating the exact mechanism by which this interaction occurs in order to coordinate viral packaging.
2. Materials and method
2.1. Plasmids
-
i)
Construction of reporter plasmids
The LacZ reporter plasmids used in this study and the primers used to generate them are listed in Supplementary Table 1. The packaging signal sequence for SARS-CoV, human coronavirus (HCoV), mouse hepatitis virus (MHV) and influenza A/WSN/33 virus (H1N1) were chemically synthesized (Bio Basic Inc., Canada). Each packaging sequence segment was digested with Hind III and Avr II, and subsequently inserted into pLacZ (pLacZ-Basic; Clontech, Shiga, Japan) to generate pLacZ-SARS-PS318, pLacZ-HCoV-PS100, pLacZ-MHV-PS97 and pLacZ-Flu-N(F)-PS57. pLacZ-SARS-PS230 and pLacZ-SARS-PS151 reporter plasmids were generated by inserting PCR amplified DNA products, using pLacZ-SARS-PS315 as the template. Packaging signal sequences for all reporter plasmids are listed in Supplementary Table 2.
-
ii)
Construction of protein expression plasmids
The N-protein sequence was amplified via PCR from pCMV-SARS-N (Sino Biological Inc., Beijing, China), a vector encoding the full-length human SARS coronavirus, following codon-optimization to increase the level of protein expression in mammalian cells. The amplicon was then digested with Xba I and Hind III prior to insertion in a pTAC plasmid (Sigma Aldrich, MO, USA) to generate pTAC-SARS-N. The N-protein deletion constructs, pTAC-SARS-N1(45–181), pTAC-N2(1–181), pTAC-N3(1–365), pTAC-N4(45–365), pTAC-N5(182–365), pTAC-N6(182–422) and pTAC-N7(248–365) were generated by inserting PCR amplified DNA products using the primers shown in Supplementary Table 1 pCMV-SARS-N was used as the template. All plasmids used in this study are available upon request.
2.2. Transformation
Individual reporter plasmids containing various packaging signal sequences along with either the pTAC-SARS-N or N-protein deletion plasmids described above were doubly transformed with individual pLacZ reporter plasmids into Escherichia coli strain JM109 (Promega, WI, USA). Co-transformants were selected from LB agar plates containing 100 μg/ml of ampicillin (Sigma Aldrich, MO, USA) and 50 μg/ml of kanamycin (Sigma Aldrich, MO, USA).
2.3. In vivo cell-based assay platform (ONPG and CPRG assay)
The ONPG assays in this study were performed based on previously described protocols [[23], [24], [25]]. Briefly, transformed bacterial colonies were grown at 37 °C in liquid LB broth containing 100 μg/ml ampicillin and 50 μg/ml of kanamycin until the culture reached an OD600 of 0.4. Isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma Aldrich, MO, USA) was added at varying concentrations (0.1 mM–0.5 mM) to induce reporter gene expression, after which absorbance at OD600 was measured at specific time points. 50 μL of cells were then harvested and lysed with 20 μL of 0.1% of SDS, 20 μL of chloroform, and 450 μL of Z-buffer (0.06 M Na2HPO4, 0.04 M NaHPO4, 0.01 M KCL, 1 mM MgSO4, and 50 mM β-mercaptiethanol). Subsequently, β-galactosidase activity was determined by adding 100 μL of O-nitrophenyl-β-d-thiogalactopyranoside (ONPG; Sigma Aldrich, MO, USA) or 1X chlorophenol red-β-d-galactopyranoside solution (CPRG; Sigma Aldrich Co., MO, USA). After incubation at room temperature for 5 min, the reaction was terminated with 200 μL of 1 M Na2CO3. A portion of the supernatant was taken to measure the absorbance at OD420, OD550, and OD575 with a spectrophotometer (VersaMax Microplate Reader, Molecular devices, CA, USA), and results were analyzed via SoftMax® Pro Software (Molecular devices, CA, USA). Miller units of β-galactosidase activity were calculated using the following formula: 1000 x (OD420–1.75 x OD550)/(ΔT x OD600), where ΔT is the time interval of the ONPG hydrolysis reaction in minutes. All optical images of results were taken using a digital camera (EOS 100D, Canon Inc., Tokyo, Japan).
2.4. Supplementary Methods
Details of other assays used are provided in the Supplementary Materials and Methods.
3. Results
3.1. Development of an in vivo cell-based assay for measuring protein-RNA binding
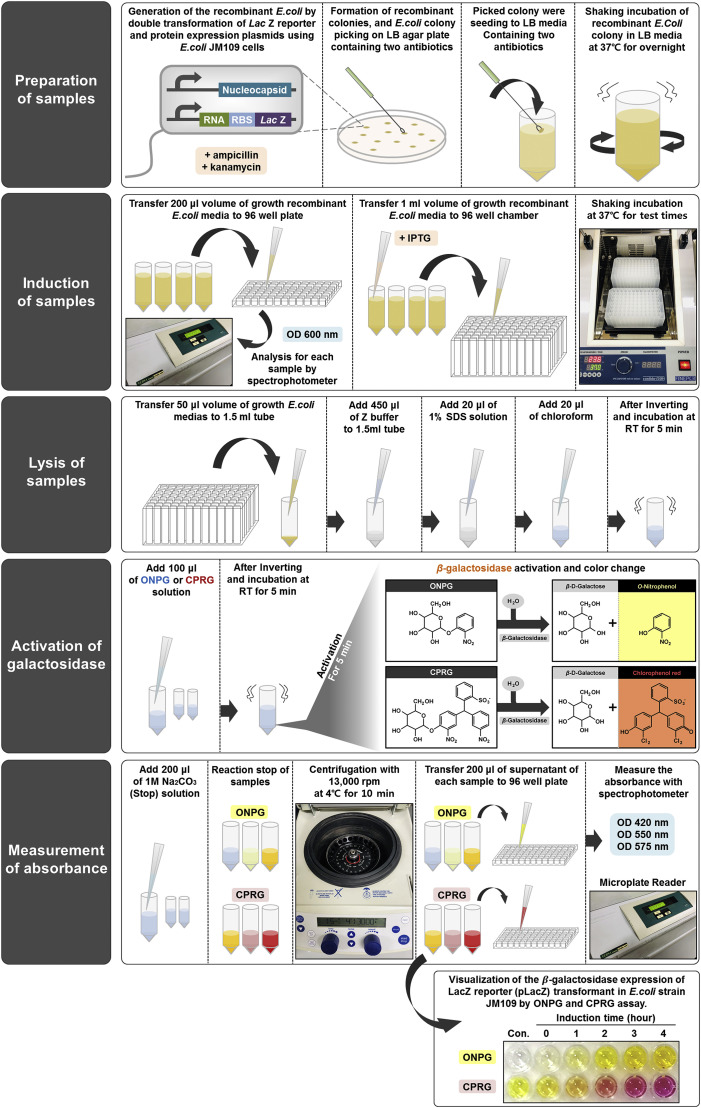
To examine whether the N-protein interacts with coronavirus (CoV) packaging signals, we modified a previously described in vivo cell-based LacZ translation inhibition assay that probes for specific interactions between RNA-binding proteins and their target sequences [23,24]. In this assay, putative target sequences are placed upstream of a LacZ reporter gene; if an RNA binding protein expressed in the cell recognizes and binds to a target sequence, LacZ translation is inhibited. Moreover, the strength of the interaction between the RNA binding protein and its target sequence is directly proportional to the degree of inhibition of LacZ translation. Fig. 1 depicts the workflow for this assay.
Fig. 1.
Schematic representation of the in vivo cell-based assay platform. The in vivo cell-based assay, including the ONPG and CPRG assays, were performed in the following manner: 1) Preparation of samples, 2) IPTG induction, 3) Lysis process, 4) ONPG and CRPG assays to measure β-galactosidase activity, 5) Absorbance measurement.
E. coli JM109 cells were first doubly transformed with one vector consisting of the target RNA sequence upstream of the LacZ reporter gene, and another encoding the SARS-CoV N-protein (pTAC-SARS-N, GenBank accession number AY307165), each containing resistance genes for a different antibiotic. Following bacterial transformation, initial OD 600 nm values were measured prior to IPTG induction. Cells were then lysed, and obtained lysates were incubated with either ONPG or CPRG to activate β-galactosidase until reactions were stopped with sodium carbonate. Final absorbance values of sample supernatants were measured with a spectrophotometer. As shown in Fig. 2 A, upon addition of ONPG or CPRG, lysates from cells doubly transformed with both the N-protein and LacZ reporter vectors showed gradual changes in color from either clear to yellow-orange (ONPG) or yellow to red-magenta (CPRG) over time. Meanwhile, transformation with the N-protein vector and a control LacZ reporter vector with a nontarget sequence upstream of the LacZ open reading from failed to demonstrate this change in color over time, demonstrating the ability of this technique to probe for specific RNA-protein interactions.
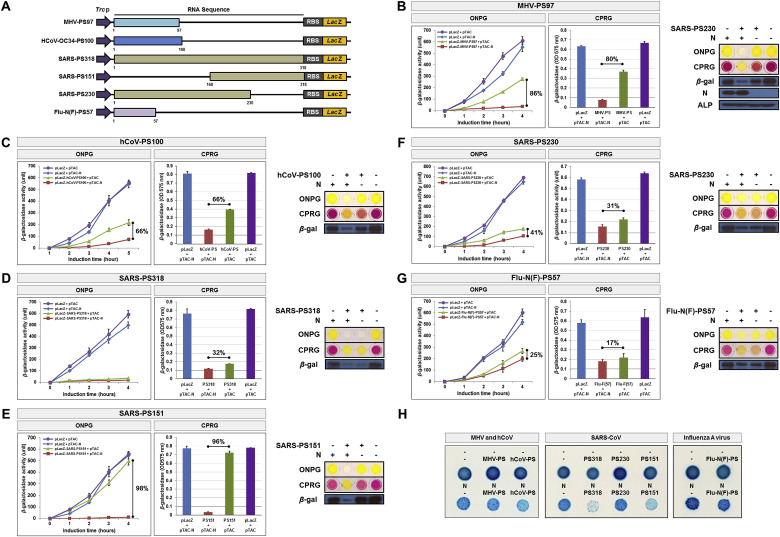
Fig. 2.
The N-protein specifically recognizes viral RNA packaging signals from the coronaviridae family. (A) Schematic representation of the packaging signal (PS) sequence regions of coronaviruses (mouse hepatitis virus, human coronavirus, SARS-CoV) and the influenza A virus, as well as the deletion constructs used to probe regions necessary for N-protein binding. Numbers in parenthesis indicate nucleotide lengths of the sequences designated. (B) Effect of N-protein on mouse hepatitis virus PS-lacZ reporter gene expression, and various regions of the PS-RNA sequence (C–G). β-galactosidase activities achieved upon transformation with individual reporter constructs or control vectors, examined in the presence (red; square) and absence (green; triangle) of N-protein, respectively. pLacZ and pTAC are control plasmids that lack expression of either the packaging signal sequence or N-protein, respectively. Results of the ONPG and CPRG assays measuring the interaction of the SARS-CoV N-protein and each packaging signal RNA are shown on the right, as well as a Western blot demonstrating the expression of LacZ (β-galactosidase) at 4 h after IPTG induction. Alkaline phosphatase (ALP) was probed as a loading control. All experiments were performed in triplicate, and data are presented as the mean ± SD (standard deviation) of three separate experiments. (H) Assay for LacZ translation on X-gal/IPTG agar plates upon co-transformation of JM109 cells with SARS-CoV N-protein and CoV packaging sequences. Minus (-) and N show JM109 transformants containing CoV packaging signal RNAs in the absence and presence of N-protein, respectively. All experiments were performed three times. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. In vivo interaction between the SARS-CoV N-protein and coronavirus RNA packaging signals
We first investigated the interaction between the N-protein and the packaging RNA sequences for the mouse hepatitis virus (MHV-PS97) and the human coronavirus OC43 (hCoV-PS100). As shown in Fig. 2B, cells co-expressing the N-protein and the MHV-PS97 packaging sequence showed a 86% reduction of β-galactosidase activity in response to ONPG and CRPG, respectively, compared to cells expressing the packaging sequence alongside an empty vector that did not encode the N-protein. Meanwhile, co-expression of the N-protein and hCoV-PS100 led to 66% inhibition of LacZ expression. Cells that expressed the N-protein alone or expressed neither the N-protein nor a CoV packaging sequence showed markedly higher β-galactosidase activity. These results demonstrate that the N-protein specifically interacts with various CoV packaging RNA sequences, such that its mere expression without a target RNA sequence is insufficient to inhibit LacZ translation.
We then examined the potential interaction between the N-protein and the SARS-CoV packaging sequence by doubly transforming JM109 cells with the full-length SARS-CoV packaging RNA sequence (pLacZ-SARS-PS318; nt 19175–20032) and the N-protein vector. This approach failed to identify any positive interactions between the N-protein and the packaging sequence because the expression of LacZ with the reporter vector alone was too low (Fig. 2D). Thus, we used a deletion-mapping approach to probe for specific regions within the SARS-CoV packaging sequence that might be responsible for mediating its interaction with the N-protein. As shown in Fig. 2E, cells expressing a short 151-nt sequence (168–310 position; SARS-PS151) of the full-length packaging sequence alongside the N-protein showed the greatest inhibition (98%) of β-galactosidase activity. We also investigated the interaction between the N-protein and a 230-nt region (1–230 position; SARS-PS230) of the full-length packaging sequence, which yielded a 41% inhibition of β-galactosidase activity (Fig. 2F). These results indicate that this 151-nucleotide sequence at the 168–318 position is the major determinant sequence for the interaction between the N-protein and the SARS-CoV packaging signal.
Following these results, we sought to investigate the potential for the N-protein to interact with packaging RNA sequences from other non-coronavirus species, such as the influenza A/WSN/33 virus. Co-expression of the N-protein with a reporter vector encoding the packaging signal of the influenza A/WSN/33 virus (pLacZ-Flu-N(F)-PS57) resulted in 25% inhibition of β-galactosidase activity compared to cells expressing the packaging sequence alone (Fig. 2G). Similar results were obtained when we visualized blue/white colonies to screen for interactions between the N-protein and packaging sequences (Fig. 2H). This demonstrates the specificity of the N-protein for packaging signals of the coronaviridae family, as significant inhibition of β-galactosidase activity is observed only in the presence of both the N-protein with a CoV packaging RNA sequence, and not with the RNA sequence of a non-CoV virus.
3.3. Characterization of the specific RNA-binding determinant region of the SARS-CoV N-protein
Conversely, we sought to probe the regions of the N-protein that are responsible for mediating its interactions with the MHV-PS97 and hCoV-PS100 packaging signals. Thus, we performed our cell-based assay using deletion constructs of the N-protein, which harbored various combinations of deletions in the N-terminal domain (NTD), C-terminal domain (CTD), linker region (LKR), or the intrinsically disordered regions (IDR) flanking the N-protein RNA sequence (Fig. 3 A). We confirmed the expression of truncated N-protein via Western blot, as shown in Supplementary Fig. 1.
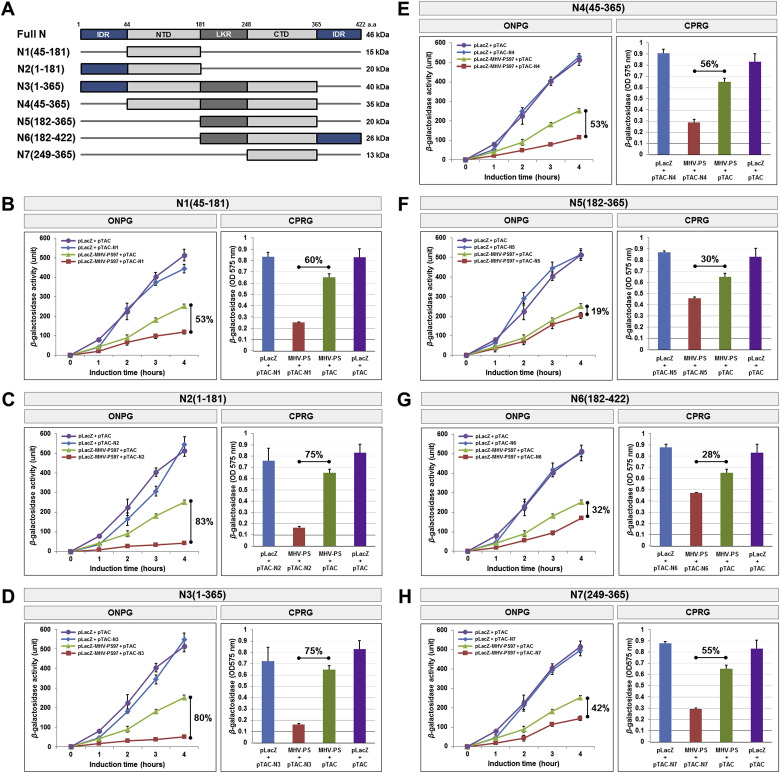
Fig. 3.
The effect of various N-protein deletion mutants on N-protein binding to the mouse hepatitis virus packaging sequence.
(A) Schematic diagram of various deletion constructs of the SARS-CoV N-protein. The molecular weights of each deletion construct are indicated on the right. (B–H) The truncated forms of N-protein inhibited the translation of the LacZ reporter to varying degrees, indicating an interaction between the protein and the mouse hepatitis virus packaging sequence. Results of the ONPG and CPRG assays are shown on the right. All experiments were performed in triplicate, and data are presented as the mean ± SD (standard deviation) of three separate experiments.
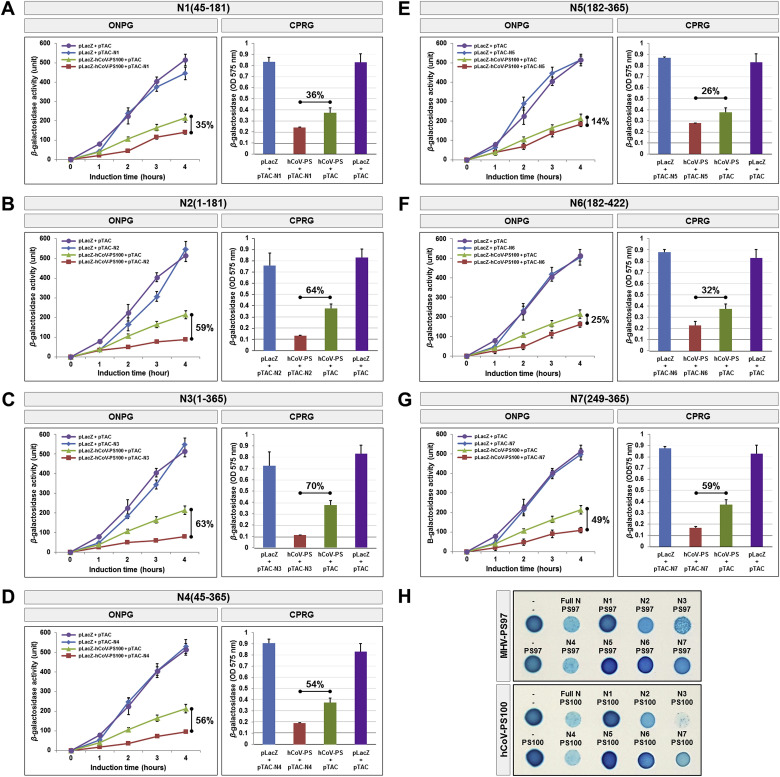
Co-expression of the MHV and hCoV PS reporter plasmids with either N5(182–365) (Fig. 3, Fig. 4 E) and N6(182–422) (Fig. 3, Fig. 4F) despite including the CTD region showed minimal inhibition of β-galactosidase activity (14–19% and 25–32%, respectively). Notably, all three of these constructs lack the NTD, suggesting that this region is critical for the interaction between the N-protein and the MHV or hCoV packaging signals. Nonetheless, co-expression of the packaging signal with N1(45–181) or N7(248–365), both of which encode the NTD (Fig. 3, Fig. 4A), or CTD (Fig. 3, Fig. 4G) alone, was unable to fully recapitulate the inhibitory effects observed with full length N-protein. Meanwhile, co-expression of the packaging signal with N2(1–181) (Fig. 3, Fig. 4B) or N3(1–365) (Fig. 3, Fig. 4E), which include the N-term IDR and NTD, was able to fully recapitulate the inhibitory effects observed with full length N-protein (Table 1 ). A similar result was also obtained via screening for blue/white colonies (Fig. 4H). Overall, these results indicate that while the NTD and CTD, including the N-term IDR, are both necessary for the N-protein to interact with CoV packaging sequences, protein-RNA binding is also in part mediated by other domains of the N-protein.
Fig. 4.
Deletion mapping of the N-protein reveals regions critical for human coronavirus packaging sequence binding.
(A–F) Our in vivo cell-based assay was performed with various truncated forms of N-protein with the human coronavirus packaging signal sequence. Results of the ONPG and CRPG assays are shown on the right, and all experiments were formed in triplicates. Data are presented as the mean ± SD (standard deviation) of three separate experiments. (G) Assay for LacZ translation on X-gal/IPTG agar plates upon co-transformation of JM109 cells with SARS-CoV N-deletion mutants and CoV packaging sequences (MHV-PS97 and hCoV-PS100). Minus (-) show JM109 transformants in the absence and presence of N-protein and packaging sequence, respectively. All experiments were performed three times.
Table 1.
Binding activity of packaging signal RNAs to the N-protein or deletion mutants by ONPG assay.
| Protein | Region (amino acid) | LacZ Translation inhibition efficiency (%) |
Binding Efficiency | |
|---|---|---|---|---|
| MHV-PS97 | hCoV-PS100 | |||
| Full N | 1–365 | 86 | 66 | +++++ |
| N1(45-181) | 45–181 | 53 | 35 | +++ |
| N2(1-181) | 1–181 | 83 | 59 | +++++ |
| N3(1-365) | 1–365 | 80 | 63 | +++++ |
| N4(45-365) | 45–365 | 53 | 56 | ++++ |
| N5(182-365) | 182–365 | 19 | 14 | + |
| N6(182-422) | 182–422 | 32 | 25 | ++ |
| N7(249-365) | 249–365 | 42 | 49 | ++++ |
4. Discussion
In the present study, we developed an in vivo cell-based assay to measure protein-RNA interactions in living cells for understanding the mechanism of SARS-CoV viral RNA packaging. Notably, this study represents the first in vivo demonstration of the interaction between the N-protein and CoV packaging RNA sequences. We found that the N-protein is specific for the SARS-CoV packaging sequence and interacts with this sequence at a greater affinity than packaging signals derived from other coronaviruses or viruses of other species, such as the mouse hepatitis virus, the human coronavirus, and the influenza A virus. Furthermore, deletion mapping analysis revealed that a 151-base region in the 318-nt sequence of the SARS-CoV packaging signal is a key mediator of its interaction with the N-protein; however, further experiments are required to explore the exact mechanism by which the N-protein recognizes and binds to this sequence for viral genome packaging.
We also performed deletion mapping of the N-protein, which demonstrated that amino acids 1–182 of the N-terminal domain, which include the N-term IDR and the NTD region, are critical for the N-protein to bind to the CoV packaging sequence. Our results also demonstrated that the CTD and linker regions are necessary for this interaction. These findings support previously published results in which a two-domain fragment containing both the NTD and CTD demonstrated the greatest nucleotide-binding affinity to the oligonucleotides in vitro [15]. Furthermore, studies of Middle East respiratory syndrome (MERS), another coronavirus, also showed that both the NTD and CTD of the MERS-CoV N-protein harbored binding activity to its RNA packaging signal PS258 (nt 19712 to 19969) [26].
In conclusion, here we describe the development of an in vivo cell-based assay that detects the interaction between N-protein and packaging signal RNA sequences and demonstrate that N-protein itself can bind the RNA signal sequence specifically in living cells. This assay will not only be a powerful tool for future studies of in vivo interactions between viral RNA packaging signals and their putative binding proteins, but also will provide an easy and rapid protocol for screening of antiviral molecules against this specific interaction.
Author contributions
J.W. conceived the designed all experiments and generated figures. J.W. and M.L. contributed to construction of experimental plasmids. M.L. and T.K. contributed to protein detection analysis. J.W. and E.Y.L. wrote the manuscript. Y.E.C. supervised all aspects of the work.
Declaration of competing interest
The authors declare that there is no conflict of interest in this work.
Acknowledgments
This work was supported by the Brain Korea 21 PLUS Project for Medical Science, Yonsei University. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2017R1D1A1B03030315). In addition, this research was supported by a grant from The Health Fellowship Foundation. E.Y.L. was funded by the T32GM007367, Medical Scientist Training Program training grant at Columbia University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2019.09.115.
Transparency document related to this article can be found online at https://doi:10.1016/j.bbrc.2019.09.115.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Transparency document
References
- 1.Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., Rappuoli R. SARS--beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein--forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McBride R., Fielding B.C. The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis. Viruses. 2012;4:2902–2923. doi: 10.3390/v4112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao H.I., Olson C.A., Hwang S., Deng H., Wong E., Baric R.S., Roberts R.W., Sun R. mRNA display design of fibronectin-based intrabodies that detect and inhibit severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Biol. Chem. 2009;284:17512–17520. doi: 10.1074/jbc.M901547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fosmire J.A., Hwang K., Makino S. Identification and characterization of a coronavirus packaging signal. J. Virol. 1992;66:3522–3530. doi: 10.1128/jvi.66.6.3522-3530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo L., Masters P.S. Functional analysis of the murine coronavirus genomic RNA packaging signal. J. Virol. 2013;87:5182–5192. doi: 10.1128/JVI.00100-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang C.K., Hsu Y.L., Chang Y.H., Chao F.A., Wu M.C., Huang Y.S., Hu C.K., Huang T.H. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 2009;83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narayanan K., Chen C.J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 2003;77:2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuo L., Koetzner C.A., Hurst K.R., Masters P.S. Recognition of the murine coronavirus genomic RNA packaging signal depends on the second RNA-binding domain of the nucleocapsid protein. J. Virol. 2014;88:4451–4465. doi: 10.1128/JVI.03866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S.C., van den Born E., van den Worm S.H., Pleij C.W., Snijder E.J., Olsthoorn R.C. New structure model for the packaging signal in the genome of group IIa coronaviruses. J. Virol. 2007;81:6771–6774. doi: 10.1128/JVI.02231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narayanan K., Makino S. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 2001;75:9059–9067. doi: 10.1128/JVI.75.19.9059-9067.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escors D., Izeta A., Capiscol C., Enjuanes L. Transmissible gastroenteritis coronavirus packaging signal is located at the 5’ end of the virus genome. J. Virol. 2003;77:7890–7902. doi: 10.1128/JVI.77.14.7890-7902.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morales L., Mateos-Gomez P.A., Capiscol C., del Palacio L., Enjuanes L., Sola I. Transmissible gastroenteritis coronavirus genome packaging signal is located at the 5’ end of the genome and promotes viral RNA incorporation into virions in a replication-independent process. J. Virol. 2013;87:11579–11590. doi: 10.1128/JVI.01836-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh P.K., Chang S.C., Huang C.C., Lee T.T., Hsiao C.W., Kou Y.H., Chen I.Y., Chang C.K., Huang T.H., Chang M.F. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C.Y., Chang C.K., Chang Y.W., Sue S.C., Bai H.I., Riang L., Hsiao C.D., Huang T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saikatendu K.S., Joseph J.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J. Virol. 2007;81:3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan Y.W., Fang S., Fan H., Lescar J., Liu D.X. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu I.M., Gustafson C.L., Diao J., Burgner J.W., 2nd, Li Z., Zhang J., Chen J. Recombinant severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein forms a dimer through its C-terminal domain. J. Biol. Chem. 2005;280:23280–23286. doi: 10.1074/jbc.M501015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tylor S., Andonov A., Cutts T., Cao J., Grudesky E., Van Domselaar G., Li X., He R. The SR-rich motif in SARS-CoV nucleocapsid protein is important for virus replication. Can. J. Microbiol. 2009;55:254–260. doi: 10.1139/w08-139. [DOI] [PubMed] [Google Scholar]
- 20.Yu I.M., Oldham M.L., Zhang J., Chen J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J. Biol. Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang C.K., Jeyachandran S., Hu N.J., Liu C.L., Lin S.Y., Wang Y.S., Chang Y.M., Hou M.H. Structure-based virtual screening and experimental validation of the discovery of inhibitors targeted towards the human coronavirus nucleocapsid protein. Mol. Biosyst. 2016;12:59–66. doi: 10.1039/c5mb00582e. [DOI] [PubMed] [Google Scholar]
- 22.Huang C.Y., Hsu Y.L., Chiang W.L., Hou M.H. Elucidation of the stability and functional regions of the human coronavirus OC43 nucleocapsid protein. Protein Sci. 2009;18:2209–2218. doi: 10.1002/pro.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang S.I., Kim Y.H., Paik S.Y., You J.C. Development of a cell-based assay probing the specific interaction between the human immunodeficiency virus type 1 nucleocapsid and psi RNA in vivo. J. Virol. 2007;81:6151–6155. doi: 10.1128/JVI.00414-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo J., Yu K.L., Lee S.H., You J.C. Development of a functional cell-based assay that probes the specific interaction between influenza A virus NP and its packaging signal sequence RNA. Biochem. Biophys. Res. Commun. 2015;457:227–233. doi: 10.1016/j.bbrc.2014.12.092. [DOI] [PubMed] [Google Scholar]
- 25.Jain C., Belasco J.G. Rapid genetic analysis of RNA-protein interactions by translational repression in Escherichia coli. Methods Enzymol. 2000;318:309–332. doi: 10.1016/s0076-6879(00)18060-7. [DOI] [PubMed] [Google Scholar]
- 26.Hsin W.C., Chang C.H., Chang C.Y., Peng W.H., Chien C.L., Chang M.F., Chang S.C. Nucleocapsid protein-dependent assembly of the RNA packaging signal of Middle East respiratory syndrome coronavirus. J. Biomed. Sci. 2018;25:47. doi: 10.1186/s12929-018-0449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.