Abstract
Zebrafish endogenous retrovirus (ZFERV) is the unique endogenous retrovirus in zebrafish, as yet, containing intact open reading frames of its envelope protein gene in zebrafish genome. Similarly, several envelope proteins of endogenous retroviruses in human and other mammalian animal genomes (such as syncytin-1 and 2 in human, syncytin-A and B in mouse) were identified and shown to be functional in induction of cell–cell fusion involved in placental development. ZFERV envelope protein (Env) gene appears to be also functional in vivo because it is expressible. After sequence alignment, we found ZFERV Env shares similar structural profiles with syncytin and other type I viral envelopes, especially in the regions of N- and C-terminal heptad repeats (NHR and CHR) which were crucial for membrane fusion. We expressed the regions of N + C protein in the ZFERV Env (residues 459–567, including predicted NHR and CHR) to characterize the fusion core structure. We found N + C protein could form a stable coiled-coil trimer that consists of three helical NHR regions forming a central trimeric core, and three helical CHR regions packing into the grooves on the surface of the central core. The structural characterization of the fusion core revealed the possible mechanism of fusion mediated by ZFERV Env. These results gave comprehensive explanation of how the ancient virus infects the zebrafish and integrates into the genome million years ago, and showed a rational clue for discovery of physiological significance (e.g., medicate cell–cell fusion).
Keywords: Zebrafish endogenous retrovirus, Envelope protein, Fusion, Heptad repeats, Coiled-coil trimer
Graphical abstract
Highlights
-
•
ZFERV Env shares similar structural profiles with syncytin and other type I viral envelopes.
-
•
The fusion core of ZFERV Env forms stable coiled-coil trimer including three NHRs and three CHRs.
-
•
The structural mechanism of viral entry mediated by ZFERV Env is disclosed.
-
•
The results are helpful for further discovery of physiological function of ZFERV Env in zebrafish.
1. Introduction
Several ancient envelope genes derived from endogenous retroviruses have been identified in mammals, which are frozen in genomes and translated to functional proteins in physiology [1], [2], [3], [4]. In humans, syncytin-1 and syncytin-2, two envelope genes from human endogenous retroviruses (HERVs) have been existing in the genome at least 25 and 40 million years respectively [5], [6], [7]. They mediate cell–cell fusion in syncytiatrophoblast formation during placental development. Similar genes named as syncytin-A and syncytin-B from murine endogenous retroviruses were also found in mouse genome and proved to be also functional in mouse placental morphogenesis [2], [8]. Other HERV envelope genes such as EnvP (b) in human, syncytin-Ory1 in rabbit and syncytin-Car1 in carnivora had also retained its fusogenic capacity when tested in cell lines [3], [4], [9], [10], [11].
In previously study, other and our group found syncytin-1, syncytin-2, syncytin-A and other type I viral Envs (e.g., HIV-1 gp160 and influenza virus hemagglutinin) share similar core profiles structure [8], [12], [13]. They are synthesized and capped by an inactive precursor, proteolytic cleavage into surface subunit (SU) and transmembrane subunit (TM) of which normally required for fusogenic activity. The fusion is triggered by receptor binding and may also need low pH. In the post-fusion conformation, the Envs contain the fusion core structure termed six helix bundle composed of three helical N-terminal heptad repeat (NHR) regions forming a central trimeric core, and three helical C-terminal heptad repeat (CHR) regions packing into the grooves on the surface of the central core.
Zebrafish endogenous retrovirus (ZFERV) was integrated into zebrafish genome during evolution. It contains the intact open reading frames for the gag, pol, env genes and LTR sequences, and these sequences are located on chromosome 19. The gag and pol genes are contiguous and in the same reading frame. No other intact fish endogenous retrovirus has been identified yet and ZFERV may be limited to zebrafish [14]. In contrast the genes including gag, pol and env genes in HERVs have many copies on different chromosome and normally are not intact to be active. Their Envs encoded by env genes are similar to human and other animal endogenous retroviral Envs such as syncytin [15], [16]. As model organism, it is interesting to understand how the ancestral virus infects zebrafish and becomes part of the genome.
Since the enveloped viral infection is normally mediated by the viral Env, we analyzed the fusion core structure of the ZFERV Env and found its shares the similar profiles with other type I viral envelope proteins (e.g., syncytin), which gives a rational explanation on how the virus enters its host during the fusion step.
2. Materials and methods
2.1. Amino acids sequence analysis of syncytin
Alignment of Syncytin-1 (NP_055405), Syncytin-2 (NP_001074253), Syncytin-A (NP_001013773), ZFERV Env (AY075045), was analyzed by ClustalX and Bioedit 7. The 3D model of ZFERV Env was constructed by Rosetta 3.5 [17]. Then the manual model building was performed with Coot [18], [19]. And the quality of modeled 3D structure was checked using MolProbity [20].
2.2. NHR + CHR (N + C) construction
To express the predicted NHR and CHR as a single chain (N + C protein, residues 459–567) for characterization (Fig. S1), we amplified the region in the ZFERV Env from zebrafish genomic DNA (PCR primers: 5′GCA GGA TCC GTA GAC AGA ATA AAT TAC 3′ and 5′ GAT CTC GAG TCA TTC CCC AAA CAT ATC 3′) and cloned it into the BamHI/XhoI restriction sites of the GST fusion expression vector pGEX-6p-1 (GE Healthcare). GST tag can be removed by GST-fusion rhinovirus 3C protease [8].
2.3. Protein expression and purification
Escherichia coli strain BL21 (DE3) transformed with the recombinant pGEX-6p-1 plasmid containing N + C protein gene was grown at 37 °C in 2 × YT to an optical density of 0.8–1.0 (OD600). Then the cells was induced with 1 mM IPTG at 22 °C for 6 h. Bacterial cells were harvested and re-suspended in PBS (10 mM sodium phosphate, 150 mM NaCl, pH 7.4). The bacterial cells were lysed by sonication on ice with the final concentration of 1% Triton X-100. The lysate was subsequently clarified by centrifugation at 12,000 g for 40 min at 4 °C. The clarified supernatants were passed over glutathione-Sepharose 4B column (equalized by PBS before). The GST fusion protein-bound column was washed by PBS over 10 column volumes and eluted with glutathione elution buffer (10 mM reduced glutathione, 50 mM Tris-Cl, pH 8.0) for 1 column volume. Then the GST fusion protein was cleaved by GST-fusion rhinovirus 3C protease at 4 °C for 16 h in the cleavage buffer (50 mM Tris-Cl, pH 7.4; 150 mM NaCl; 1 mM DTT; 1 mM EDTA, pH 8.0). The free GST and the GST fusion rhinovirus 3C protease were removed by passing over the glutathione–Sepharose 4B column again. N + C protein was further purified through Hiload 16/60 Superdex G75 column (GE healthcare) running on ÄKTA explorer 100 chromatography system (GE healthcare). The purified N + C protein was concentrated to a proper concentration which was determined by the method of Bradford (Bio-Rad). Then the protein was stored at −70 °C for further analysis.
2.4. Size exclusion chromatography (SEC) analysis
The purified N + C protein was loaded into the Bio Suite TM 125-5 μm HR SEC column on HPLC system (Waters 1525) to assess oligomer formation. PBS (10 mM sodium phosphate, 150 mM NaCl, pH 7.4) was used as the mobile phase with a flow rate of 1 ml/min. The ultraviolet absorbance at 280 nm was recorded. A gel-filtration of standard consisting of bovine serum albumin (67 kDa), albumin egg (44 kDa), lactoglobulin (35 kDa), and chymotrypsin (25 kDa), Cytochrome C (12 kDa) (Sigma) was used to define the molecular weight of N + C protein.
2.5. Chemical cross-linking experiment
N + C protein in chemical cross-linking buffer (50 mM Hepes, 100 mM NaCl, pH 8.3) was cross-linked with ethylene glycol-bis-succinimidylsuccinate (EGS) (Sigma). The reaction was incubated at room temperature for 2 h and then terminated with 50 mM glycine in final concentration. Cross-linked products were electrophoresed on Tris-Tricine SDS-PAGE.
2.6. Circular dichroism spectroscopy
The secondary structure of N + C protein was detected by circular dichroism (CD) spectroscopy. The purified protein was dissolved in PBS at the final concentration of 0.6 mg/mL, and the CD spectra were recorded on a Jasco J-810 spectrophotometer (JASCO Corporation). The CD spectra were recorded at 25 °C and 90 °C in a 0.1 cm path length cuvette. Thermodynamic stability was measured by recording the CD signal at 222 nm in the temperature range from 25 °C to 90 °C.
2.7. Proteolytic digestion
The proteolysis reactions were performed in N + C protein with final protease K concentrations of 0 mM, 1 mM, 20 mM, and 50 mM respectively for 15 min at 4 °C. Samples were immediately subjected to Tris-Tricine SDS-PAGE analysis. Protease-resistant fragments were separated from the gel, then digested by trypsin and characterized by mass spectrometry (Voyager DE STR MALDITOF, Applied Biosystems).
3. Results
3.1. The ZFERV Env and type I viral Envs share similar structural profiling
By sequence alignment and analysis, we found that the ZFERV envelope protein and several envelope proteins from other ERVs (e.g., syncytin-1, syncytin-2 and syncytin-A) share similar structure features as typical type I viral envelope protein, especially in the core structure profiling (Fig. S2). It contains: a protease cleavage site, RNKR (399–402), which separates the two characteristic glycoprotein domains into surface subunit and transmembrane subunit; a fusion peptide (FP, 403–425) was located at the N-terminus containing hydrophobic, glycine-rich residues, which was essential for the initial penetration of the target cell membrane; a hydrophobic region following the fusion peptide including N-terminal heptad repeat (NHR, 459–511) and C-terminal heptad repeat (CHR, 521–565) linked by a linker with CX6CC motif. We found the sequences of NHR and CHR in ZFERV Env are highly homogenous with those in other syncytin proteins indicating they might form the fusion core.
3.2. Construction of 3D structures of NHR and CHR in ZFERV Env
To understand the detailed information of the fusion core, we did molecular modelling of NHR and CHR in ZFERV Env in silicon. The sequence of ZFERV Env was aligned using BLASTp [21]. The results showed that it was the member of HIV-1-like NHR and CHR superfamily while NHR region contained residues 468-505 and CHR region contained residues 535–543. The secondary structure comparison performed with Dali server indicated that the ZFERV Env structure was similar to that of the Env GP2 core domain from the CAS virus, a novel arena virus-like species and core domain of S2 from the human coronavirus NL63 Env (termed spike protein) [22]. The 3D model of ZFERV Env was constructed by Rosetta 3.5 [17] to further investigate how ZFERV Env forms homotrimer. This model fitted very well with an anti-parallel dimer of NHR + CHR complex in ZFERV Env, just like those in HIV-1 gp160 and coronavirus spike protein [23], [24], [25], and provided us putative detailed interactions between NHR and CHR in ZFERV Env.
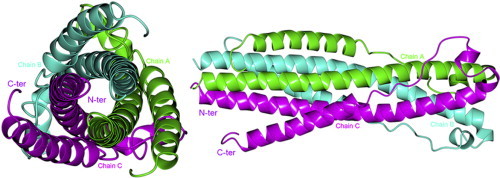
The overall structure of ZFERV Env contains fusion core structure termed six helix bundle composed of three helical NHRs forming a central trimeric core, and three helical CHRs peptides packing into the grooves on the surface of the central core (Fig. 1 A and B), as type I viral envelope proteins, HIV-1 gp160 [26], [27]. The interface of homotrimer that contains three homologous chains: Chain A (residues 468–543), Chain B and Chain C covers NHRs and CHRs. Several hydrophobic interactions occurre between NHRs and CHRs (Fig. 1C) and three helical NHRs form a hydrophobic core in the center of homotrimer. There might be also several hydrogen bonds existing between chain A, chain B and chain C (Fig. 1D and E). For example, in the N-terminal of NHR, R477 in chain A is hydrogen bonded to T476 in chain B, while R477 in chain B is hydrogen bonded to T476 in chain C, and R477 in chain C is hydrogen bonded to T476 in chain A. The homotrimer may be very stable because of the head-tail formation [28]. And in the C-terminal of NHR, Q486 and T490 in chain A are hydrogen bonded to S491 in chain C, S491 and N525 in chain C are hydrogen bonded to Q486 in chain A. Therefore, a network of hydrogen bonds between chain A, chain B and chain C is maintained to keep that the homotrimer is not easy to be apart.
Fig. 1.
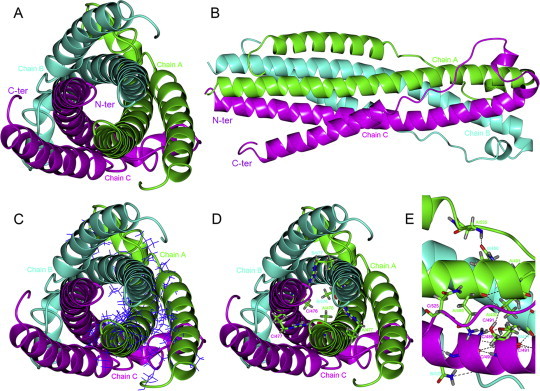
Computer modeling of N + C protein. The homotrimer chains (Chain A, Chain B and Chain C) are shown in ribbons and colored in green, cyan and magenta, respectively. The predicted structure of N + C protein (fusion core) which is a six helix bundle composed of three helical NHRs forming a central trimeric core, and three helical CHRs peptides packing into the grooves on the surface of the central core. N-terminus (N-ter) and C-terminus (C-ter) are labeled. (A): Top view of the overall structure. (B): Side view of the overall structure. (C): The hydrophobic amino acid residues of chain A are shown in bonds and colored in blue. (D): The hydrogen bonds existing in N-terminal of NHRs. These hydrogen-bond-forming residues are shown in cylinders and colored in atom type, these hydrogen bonds are marked by dotted lines. (E): Close-up view of the hydrogen bonds existing in C-terminal of NHRs. These hydrogen-bond-forming residues are shown in cylinders and colored in atom type, these hydrogen bonds are marked by dotted lines, too. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. N + C protein exists as a trimer
The oligomer formation of purified N + C protein in solution was analyzed by size exclusion chromatography and chemical cross-linking experiment. According to the standard curve, the elution time of N + C protein gave an apparent molecular mass of 42 kDa that is three folds of the theoretic molecular mass (12.3 kDa) indicating the formation of trimer (Fig. 2 A). In chemical cross-linking experiment, N + C protein existed as monomer, dimer, and trimer (Fig. 2B), while tetramer and larger oligomers were not observed in SDS-PAGE. The amount of trimer increased while the concentration of chemical cross-linking reagent increased. Taking together, these results showed that N + C protein existed as a trimer in native condition.
Fig. 2.
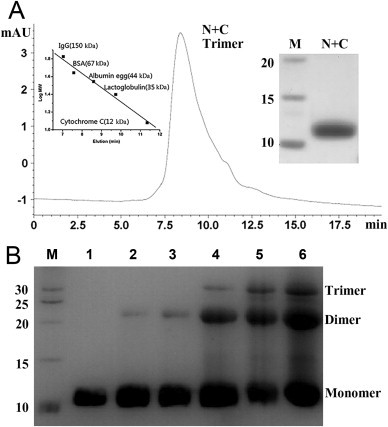
Oligomer formation of N + C protein. (A): Size exclusion chromatography of N + C protein in PBS. The left inset is a standard curve. The right insert is purified N + C protein in SDS-PAGE. Molecular mass standard was shown in lane M (kDa). The calculated mass from the peak was 42 kDa being close to the theoretical homotrimer mass of N + C protein (36.9 kDa). (B): Chemical cross-linking reaction of N + C protein. N + C protein cross-linked by EGS were separated in SDS-PAGE. Molecular mass standard was shown in lane M (kDa). lane 1, N + C protein without EGS, lanes 2–6, N + C protein with EGS at the final concentrations of 0.01 mM, 0.05 mM, 0.1 mM, 0.5 mM, 1 mM, respectively. Monomer, dimer, and trimer bands were found, while no bigger oligomer existed indicating the formation of trimer in N + C protein.
3.4. N + C protein is highly α-helix, thermo-stable, and denatured by low pH
The CD spectra showed that N + C protein had typical α-helix structure at 25 °C. (Fig. 3 A). The α-helical structure was slightly disrupted as the temperature increases (Fig. 3B) but still maintained α-helix structure at 90 °C (Fig. 3A). The curve still presented as a typical α-helical structure at 90 °C, indicating that it was stable and the melting temperature (Tm) was higher than 90 °C. These observations suggested that N + C protein trimer was highly α-helix, thermo-stable, which were similar to the characteristics of the fusion core structure of other identified viral Envs such as syncytin-1. The conformational change of N + C protein was tested by acid-induced denaturing (Fig. 3C). The α-helical structure of the protein was disrupted gradually as pH decreased, appearing to be in a folded state at pH 8.0 and in an unfolded state at pH 4.0.The “S” shaped curve displayed that this unfolding course accord with a two-state model. A neutral condition was enough to make the structure of N + C protein ordered and thermo-stable, which indicated that the ZFERV Env mediated fusion at neutral pH like HIV-1 gp160 but not at low pH like influenza virus hemagglutinin [29], [30].
Fig. 3.
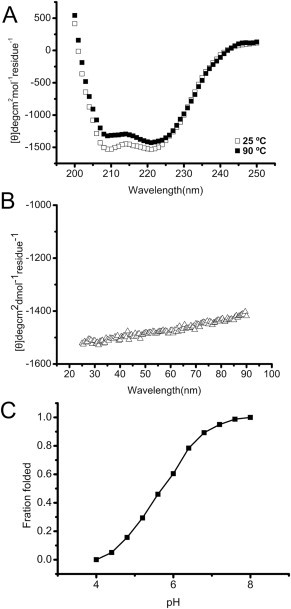
The CD spectra of N + C protein. (A): Secondary structure of N + C protein was recorded at 25 and 90 °C. It displays typical α-helical secondary structures with double minima of the ellipticities at 208 and 222 nm. (B): Thermal denaturing stability measurement by recording at 222 nm from 25 to 90 °C with the scanning rate of 1 °C/min. (C): The fraction folded (ff) of N + C protein by acid denatured at 25 °C. ff = ([θ] – [θM])/([θT] – [θM]). [θT] and [θM] were the mean residue ellipticities at 222 nm of the folded state at pH 8.0 and the unfolded states at pH 4.0, respectively.
3.5. N + C protein contains a protease-resistant core
N + C protein was firstly digested to two major bands and then a major band was left with gradient increased proteinase K concentration (Fig. 4 A). Subsequently, these bands were further digested by trypsin and analyzed by mass spectra. Two major fragments with molecular weights of 1319 Da and 2246 Da respectively were identified which can match two regions I462-R471 (calculated MW: 1319 Da) and D478-R498 (calculated MW: 2246 Da) in NHR according to deduction by molecular weight calculation and trypsin digestion site analysis (Fig. 4B). Therefore, after protease K treatment, the three inner NHRs were still retained due to the strong interactions while the three outer CHRs were completely digested, which was consistent with our mode (Fig. 1) and the crystal structure of syncytin-2 [12]. This phenomenon indicated the interaction between NHR and CHR in ZFERV Env might not be as strong as that in HIV-1 gp160 while might be similar to that in syncytin-2. In some type 1 viral Envs (e.g., HIV-1 gp160 and RSV F protein), the length of NHR is almost equal to that of CHR. In contrast, the length of CHR in other type 1 viral Envs (e.g., syncytin-2 and EBOV Gp2) is much shorter than that of NHR. The diversity of CHRs in different viruses might be related to the evolutional properties of the viruses with type 1 Envs, which needs to be further investigated.
Fig. 4.
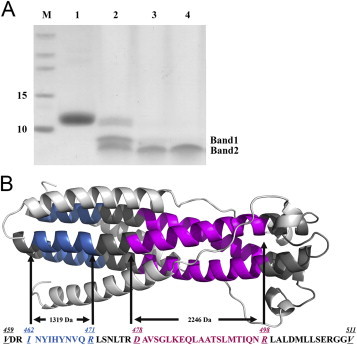
Protease-resistant core structure identification. (A): Limited protease digestion products were separated in Tris-Tricine SDS-PAGE. Lane 1, N + C protein without protease K; lane 2–4, N + C protein was digested by protease K with final concentrations of 1 mM, 20 mM and 50 mM respectively. Molecular mass standard was shown in lane M (kDa). There were two major digested bands in lane 2. With the increased concentration of protease K, only one band was left. (B): The protease-resistant core fragments in NHR: I462-R471 (INYIHYNVQR, M.W.: 1319 Da) is colored by blue; D478-R498 (DAVSGLKEQLAATSLMTIQNR, MW: 2246 Da) is colored by purple. Other residues in NHR were colored by gray; CX6CC motif and CHR regions are in green. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Although ERVs are found to exist in almost all vertebrate genomes, the interaction between the host factors (such as receptor and immune system) and ERV proteins (such as Env) is complex and largely unknown. Zebrafish is an excellent developmental model in vertebrates to study such questions because of the presence of ZFERV. ZFERV is the only intact fish endogenous retrovirus identified in zebrafish, which is present in lots varieties of tissues, including the brain, fin regenerates, heart, kidney, olfactory rosettes, and retina [14]. Although most ancient endogenous retrovirus in vertebrates contain many deletions and mutations in their genomes, the ZFERV Env gene transcript could correctly express and the protein might be still physiologically functional in vivo. However, there was not much information on the fusion function and mechanism of ZFERV Env. Here, we disclosed the molecule mechanism of how the ancestor virus enters the host that is mediated by ZFERV Env. Combined biophysical and biochemical methods were used to evaluate the structure of N + C protein in ZFERV Env. Our experiments implicate that N + C protein could form a stable triple-stranded coiled-coil that is the common structure found in other type I viral Envs including syncytin. These results show that ZFERV, frozen about 60–100 million years, still adopts similar mechanism with other viruses that use type I Envs when it enters the host cell suggesting a uniform viral entry structural conformation process during the long-term evolution.
Conflicts of interest
None.
Acknowledgements
We thank Dr. Lisa A. Steiner (Massachusetts Institute of Technology, Cambridge, MA) for kindly providing pCRII-TOPO-ZFERV plasmid expressing the envelope protein of ZFERV, and the Core Facility and Technical Support, Wuhan Institute of Virology, for the technical assistance, as well as the Wuhan Key Laboratory on Emerging Infectious Diseases and Biosafety, Wuhan Institute of Virology, for the support. This work was funded by the National Key Basic Research Program of China (Grant No.: 2011CB933600).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2015.03.081.
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2015.03.081.
Contributor Information
Rui Gong, Email: gongr@wh.iov.cn.
Gengfu Xiao, Email: xiaogf@wh.iov.cn.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Transparency document
References
- 1.Mi S., Lee X., Li X., Veldman G.M., Finnerty H., Racie L., LaVallie E., Tang X.Y., Edouard P., Howes S., Keith J.C., Jr., McCoy J.M. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 2.Dupressoir A., Marceau G., Vernochet C., Benit L., Kanellopoulos C., Sapin V., Heidmann T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. U.S.A. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heidmann O., Vernochet C., Dupressoir A., Heidmann T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new “syncytin” in a third order of mammals. Retrovirology. 2009;6:107. doi: 10.1186/1742-4690-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelis G., Heidmann O., Bernard-Stoecklin S., Reynaud K., Veron G., Mulot B., Dupressoir A., Heidmann T. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E432–E441. doi: 10.1073/pnas.1115346109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blond J.L., Beseme F., Duret L., Bouton O., Bedin F., Perron H., Mandrand B., Mallet F. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J. Virol. 1999;73:1175–1185. doi: 10.1128/jvi.73.2.1175-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaise S., de Parseval N., Benit L., Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaise S., Ruggieri A., Dewannieux M., Cosset F.L., Heidmann T. Identification of an envelope protein from the FRD family of human endogenous retroviruses (HERV-FRD) conferring infectivity and functional conservation among simians. J. Virol. 2003;78:1050–1054. doi: 10.1128/JVI.78.2.1050-1054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong R., Peng X., Kang S., Feng H., Huang J., Zhang W., Lin D., Tien P., Xiao G. Structural characterization of the fusion core in syncytin, envelope protein of human endogenous retrovirus family W. Biochem. Biophys. Res. Commun. 2005;331:1193–1200. doi: 10.1016/j.bbrc.2005.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vernochet C., Heidmann O., Dupressoir A., Cornelis G., Dessen P., Catzeflis F., Heidmann T. A syncytin-like endogenous retrovirus envelope gene of the guinea pig specifically expressed in the placenta junctional zone and conserved in Caviomorpha. Placenta. 2011;32:885–892. doi: 10.1016/j.placenta.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Dupressoir A., Lavialle C., Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta. 2012;33:663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Vargas A., Thiery M., Lafond J., Barbeau B. Transcriptional and functional studies of human endogenous retrovirus envelope EnvP(b) and EnvV genes in human trophoblasts. Virology. 2012;425:1–10. doi: 10.1016/j.virol.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 12.Renard M., Varela P.F., Letzelter C., Duquerroy S., Rey F.A., Heidmann T. Crystal structure of a pivotal domain of human syncytin-2, a 40 million years old endogenous retrovirus fusogenic envelope gene captured by primates. J. Mol. Biol. 2005;352:1029–1034. doi: 10.1016/j.jmb.2005.07.058. [DOI] [PubMed] [Google Scholar]
- 13.Peng X., Pan J., Gong R., Liu Y., Kang S., Feng H., Qiu G., Guo D., Tien P., Xiao G. Functional characterization of syncytin-A, a newly murine endogenous virus envelope protein. Implication for its fusion mechanism. J. Biol. Chem. 2007;282:381–389. doi: 10.1074/jbc.M606353200. [DOI] [PubMed] [Google Scholar]
- 14.Shen C.H., Steiner L.A. Genome structure and thymic expression of an endogenous retrovirus in zebrafish. J. Virol. 2003;78:899–911. doi: 10.1128/JVI.78.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villesen P., Aagaard L., Wiuf C., Pedersen F.S. Identification of endogenous retroviral reading frames in the human genome. Retrovirology. 2004;1:32. doi: 10.1186/1742-4690-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magiorkinis G., Blanco-Melo D., Belshaw R. The decline of human endogenous retroviruses: extinction and survival. Retrovirology. 2015;12:8. doi: 10.1186/s12977-015-0136-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R., Kaufman K., Renfrew P.D., Smith C.A., Sheffler W., Davis I.W., Cooper S., Treuille A., Mandell D.J., Richter F., Ban Y.E., Fleishman S.J., Corn J.E., Kim D.E., Lyskov S., Berrondo M., Mentzer S., Popovic Z., Havranek J.J., Karanicolas J., Das R., Meiler J., Kortemme T., Gray J.J., Kuhlman B., Baker D., Bradley P. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzym. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 19.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantalloube H., Labesse G., Chomilier J., Nahum C., Cho Y.Y., Chams V., Achour A., Lachgar A., Mbika J.P., Issing W. Automat and BLAST: comparison of two protein sequence similarity search programs. Comput. Appl. Biosci. : CABIOS. 1995;11:261–272. doi: 10.1093/bioinformatics/11.3.261. [DOI] [PubMed] [Google Scholar]
- 22.Holm L., Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weissenhorn W., Dessen A., Harrison S.C., Skehel J.J., Wiley D.C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 24.Chan D.C., Fass D., Berger J.M., Kim P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 25.Ingallinella P., Bianchi E., Finotto M., Cantoni G., Eckert D.M., Supekar V.M., Bruckmann C., Carfi A., Pessi A. Structural characterization of the fusion-active complex of severe acute respiratory syndrome (SARS) coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8709–8714. doi: 10.1073/pnas.0402753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Decroly E., Cornet B., Martin I., Ruysschaert J.M., Vandenbranden M. Secondary structure of gp160 and gp120 envelope glycoproteins of human immunodeficiency virus type 1: a Fourier transform infrared spectroscopic study. J. Virol. 1993;67:3552–3560. doi: 10.1128/jvi.67.6.3552-3560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poumbourios P., el Ahmar W., McPhee D.A., Kemp B.E. Determinants of human immunodeficiency virus type 1 envelope glycoprotein oligomeric structure. J. Virol. 1995;69:1209–1218. doi: 10.1128/jvi.69.2.1209-1218.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardarelli L., Lam R., Tuite A., Baker L.A., Sadowski P.D., Radford D.R., Rubinstein J.L., Battaile K.P., Chirgadze N., Maxwell K.L., Davidson A.R. The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins. J. Mol. Biol. 2010;395:754–768. doi: 10.1016/j.jmb.2009.10.067. [DOI] [PubMed] [Google Scholar]
- 29.Shi Y., Wu Y., Zhang W., Qi J., Gao G.F. Enabling the ‘host jump’: structural determinants of receptor-binding specificity in influenza A viruses. Nat. Rev. Microbiol. 2014;12:822–831. doi: 10.1038/nrmicro3362. [DOI] [PubMed] [Google Scholar]
- 30.Harrison J.S., Higgins C.D., O'Meara M.J., Koellhoffer J.F., Kuhlman B.A., Lai J.R. Role of electrostatic repulsion in controlling pH-dependent conformational changes of viral fusion proteins. Structure. 2013;21:1085–1096. doi: 10.1016/j.str.2013.05.009. (London, England : 1993) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


