Abstract
Anti-bacterial and anti-viral neuraminidase agents inhibit neuraminidase activity catalyzing the hydrolysis of terminal N-acetylneuraminic acid (Neu5Ac) from glycoconjugates and help to prevent the host pathogenesis that lead to fatal infectious diseases including influenza, bacteremia, sepsis, and cholera. Emerging antibiotic and drug resistances to commonly used anti-neuraminidase agents such as oseltamivir (Tamiflu) and zanamivir (Relenza) have highlighted the need to develop new anti-neuraminidase drugs. We obtained a serendipitous complex crystal of the catalytic domain of Clostridium perfringens neuraminidase (CpNanICD) with 2-(cyclohexylamino)ethanesulfonic acid (CHES) as a buffer. Here, we report the crystal structure of CpNanICD in complex with CHES at 1.24 Å resolution. Amphipathic CHES binds to the catalytic site of CpNanICD similar to the substrate (Neu5Ac) binding site. The 2-aminoethanesulfonic acid moiety and cyclohexyl groups of CHES interact with the cluster of three arginine residues and with the hydrophobic pocket of the CpNanICD catalytic site. In addition, a structural comparison with other bacterial and human neuraminidases suggests that CHES could serve as a scaffold for the development of new anti-neuraminidase agents targeting CpNanI.
Keywords: Clostridium perfringens, Neuraminidase, NanI, CHES, Anti-neuraminidase agents, Crystal structure
Abbreviations: CpNanI, Clostridium perfringens neuraminidase NanI; Neu5Ac, N-acetylneuraminic acid; CHES, 2-(cyclohexylamino)ethanesulfonic acid; SpNanB, Streptococcus pneumoniae NanB; RMSD, root mean square deviation
Graphical abstract
Highlights
-
•
We determined the crystal structure of CpNanI bound to CHES at 1.24 Å resolution.
-
•
CHES binds to the catalytic site of CpNanI similar to the substrate binding site.
-
•
We suggest strategies for modification of CHES for the development of anti-CpNanI agents.
1. Introduction
Neuraminidases are expressed in many bacteria including Clostridium perfringens (Cp), Streptococcus pneumonia (Sp), and Pseudomonas aeruginosa and in viruses including influenza A and B. Neuraminidases catalyze hydrolytic removal of terminal N-acetylneuraminic acid (Neu5Ac) from a number of glycoconjugates and play a pivotal role in human infection [1], [2], [3]. Viral neuraminidases cleave the terminal Neu5Ac from the cellular receptor to which the newly replicated viral particles are attached, and thus neuraminidases are involved in viral replication namely in the release from host cells [4]. Meanwhile, bacterial neuraminidases function at the initial stages of a pulmonary infection and participate in the biofilm formation, which plays important roles in bacterial growth and colonization [5], [6]. Thus, neuraminidases of pathogenic bacteria and viruses are considered promising drug targets for prevention of infections that cause fatal human diseases such as influenza, bacteremia, sepsis, cholera, food poisoning, and gas gangrene [6], [7], [8], [9], [10]. Several anti-neuraminidase agents such as substrate (Neu5Ac) mimics, cyclopentane, pyrrolidine-based derivatives, ethanesulfonic acid derivatives, and natural compounds are used for drug development [10], [11], [12], [13], [14], [15], [16], [17]. Commercial antibiotics and antiviral agents including oseltamivir (Tamiflu) and zanamivir (Relenza) have been developed to prevent bacterial and viral infections. Even though various commercial drugs are available on the market, emergence of resistance to antibiotics and antiviral drugs resistance mandates the development of new drugs [18], [19], [20].
2-(cyclohexylamino)ethanesulfonic acid (CHES) is commonly used as a buffering agent, and approximately 25 crystal structures complexed with CHES are available in Protein Data Bank (PDB) [21]. CHES binds to the active site of several enzymes including neuraminidase NanB from Streptococcus pneumonia (SpNanB, PDB code 2VW2), papain like protease from severe acute respiratory syndrome coronavirus (SARS-CoV) (PDB code 4M0W), sulfotransferase (Teg12, PDB code 3NIB), Sirtuin 5 (Sirt5, PDB code 3RIG), and haloalkaloic acid dehalogenase (HAD) phosphatase (PDB code 4YGR); the action inhibits substrate binding and enzymatic activity. These data suggest that CHES may serve a scaffold for new drug development for modulation on an enzyme's function. CHES contains a hydrophilic taurine (2-aminoethanesulfonic acid) moiety and a hydrophobic cyclohexyl groups. The amphipathic properties of CHES raise the possibility of CHES-based development of anti-neuraminidase agents, because the catalytic site of neuraminidases comprises hydrophilic residues (an arginine cluster with three arginine residues and a catalytic aspartate residue) and a hydrophobic pocket [22].
Clostridium perfringens, a major pathogen causing gas gangrene and enterotoxemia in humans, has three neuraminidases including NanH, NanI, and NanJ [23]. CpNanI, a secreted exo-sialidase, has been evaluated to be a potential drug target for inhibition of human infections caused by Clostridium perfringens. We obtained a serendipitous complex crystal of the catalytic domain of CpNanI (CpNanICD) with CHES serving as the buffer. Here we report the crystal structure of CpNanICD in complex with CHES (CpNanICD-CHES) at 1.24 Å resolution. On the basis of the structural comparison with other bacterial and human neuraminidases, we propose that CHES could serve a scaffold for the development of anti-neuraminidase agents targeting CpNanI.
2. Materials and methods
2.1. Protein expression and purification
Cloning, expression, and purification of CpNanICD (residues, 243–694) were reported previously [24]. Briefly, recombinant CpNanICD with a direct C-terminal hexahistidine (His6) tag fusion was expressed in Escherichia coli BL21 CodonPlus (DE3) cells. The protein was purified using a Ni-NTA affinity chromatography column with elution buffer consisting of 50 mM sodium phosphate (pH 7.0), 300 mM NaCl, and 300 mM imidazole. The protein was then subjected to size exclusion chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare Life Sciences) pre-equilibrated with gel-filtration buffer [25 mM CHES-HCl (pH 9.5) and 200 mM NaCl]. The protein was concentrated to 21.8 mg/ml using an Amicon Ultra-15 30 K (Millipore) and was stored at −80 °C.
2.2. Crystallization
Crystallization was performed by the hanging-drop vapor-diffusion method. Well-diffracted co-crystals were grown at 20 °C in 2 μl drops containing equal volumes of the protein solution [25 mM CHES-HCl (pH 9.5) and 200 mM NaCl] and mother liquor reservoir comprised of 20% (w/v) PEG 3350 and 0.2 M ammonium sulfate. The crystals were cryo-protected in cryoprotectant containing 20% (w/v) PEG 4000, 10% (v/v) glycerol and 0.2 M ammonium sulfate and were flash-frozen in liquid nitrogen for data collection.
2.3. Data collection and crystallographic analysis
The data set for the CpNanICD-CHES complex was collected using an ADSC Q315r detector at beamline 5C in the Pohang Accelerator Laboratory (PAL), Republic of Korea, using the X-ray beam at a single wavelength (0.9795 Å). Diffraction data were processed and scaled using the HKL2000 software [25]. The CpNanICD-CHES co-crystal was diffracted up to 1.24 Å resolution and belonged to space group P21 with cell parameters of a = 69.4, b = 98.0, c = 72.6 Å and β = 91.0°. Two molecules of the CpNanICD-CHES complex were present in an asymmetric unit and the Matthews coefficient (VM) was calculated to be 2.40 Å3Da−1, which corresponds to solvent content of 48.8%. The structure of the CpNanICD-CHES complex was solved by the molecular replacement method in the MOLREP software using the apo-structure of CpNanICD (PDB code 2VK5) as the search model [26]. Automated ligand building of CHES (ligand ID NHE) in the structure was carried out using the phenix.ligandfit [27]. Cycles of manual rebuilding were performed using Coot software, and subsequent refinement was carried out using Refmac5 and phenix.refine, with the final crystallographic R value of 14.4% (R free = 15.8%) [28], [29], [30]. The composite omit map (mF o-DF c) of CHES contoured at 1.5 σ was generated by phenix.mtz2map software [30]. The weak electron density regions (residues 691–694) were excluded from the final structure. The Ramachandran statistics were calculated using program MolProbity showing no residues with torsional angles in forbidden areas: 96.4% of the residues were in the favored regions and 3.6% of the residues were in allowed regions [31]. Data collection and refinement statistics are summarized in Table 1 . Atomic coordinates have been deposited in the Protein Data Bank under accession code 5TSP.
Table 1.
Data collection and refinement statistics.
| CpNanICD-CHES | |
|---|---|
| Data collection | |
| X-ray source | PAL-5C |
| Wavelength (Å) | 0.9795 |
| Space group | P21 |
| Unit-cell parameters (Å) | a = 69.4, b = 98.0, c = 72.6, α = 90.0, β = 91.0, γ = 90.0 |
| Resolution range (Å) | 50.0–1.24 (1.26–1.24) |
| No. of observed reflections | 1110058 |
| No. of unique reflections | 272513 |
| Completeness (%) | 99.5 (98.6) |
| Rmergea (%) | 4.7 (20.0) |
| Mean I/σ(I) | 12.0 (5.5) |
| Multiplicity | 4.1 (3.9) |
| Refinement statistics | |
| Resolution range (Å) | 35.2–1.24 |
| Rwork/Rfreeb (%) | 14.4/15.8 |
| No. of atoms | |
| Protein | 7233 |
| CHES | 26 |
| Ca2+ | 2 |
| Water | 1832 |
| Average B factors (Å2) | |
| Protein | 9.8 |
| CHES | 9.2 |
| Ca2+ | 6.8 |
| Water | 27.3 |
| R.m.s. deviation from ideal geometry | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.205 |
| Ramachandran plot | |
| Most favored regions (%) | 96.4 |
| Allowed regions (%) | 3.6 |
Values in parentheses are for the highest resolution shell.
Rmerge = ∑h ∑i |I(h)i−‹I(h)›|/∑h ∑iI(h)i, where I(h) is the intensity of reflection of h, ∑h is the sum over all reflections, and ∑i is the sum over i measurements of reflection h.
Rwork = Σhkl ||Fo|-|Fc||/Σhkl|Fo|; Rfree is the R value calculated for 5% of the data set not included in the refinement.
2.4. Homology modeling
Model structures of human neuraminidases Neu1, Neu3, and Neu4 were built using the structural template of human Neu2 (PDB code 1VCU) by MODELLER 9v7 software as described in our previous studies [24], [32].
3. Results and discussion
3.1. Overall structure of CpNanICD-CHES
The crystal structure of the CpNanICD-CHES was determined at 1.24 Å resolution with an R-factor of 14.4% (free R-factor of 15.8%) by molecular replacement using the CpNanICD-apo structure (PDB code 2VK5) as a template (Table 1). The crystal of CpNanICD-CHES contains two molecules in an asymmetric unit with a root mean square deviation (RMSD) of 0.27 Å for 437 Cα atoms. The overall structure of the CpNanICD-CHES complex shows not only the six-bladed β-propeller fold and Arg-Ile-Pro (RIP) and Asp-box motifs but also the catalytic site (including the arginine cluster, catalytic aspartate, glutamate, and tyrosine) as in other neuraminidases ranging from bacteria and viruses to humans (Figs. 1A and S1) [1], [16], [33].
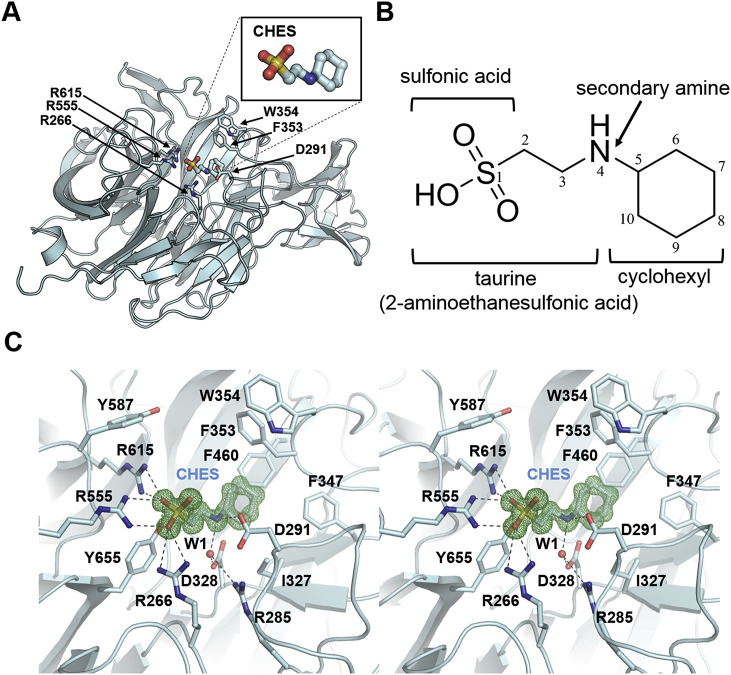
CHES comprises a hydrophilic taurine (2-aminoethanesulfonic acid) moiety and hydrophobic cyclohexyl groups and binds at the center of a catalytic site (Fig. 1 B and C). The sulfonic acid group of CHES forms direct hydrogen bonds with side chains of the arginine cluster (R266, R555, and R615). The secondary amine of CHES also interacts with D291 (thus forming a direct hydrogen bond) as well as with R285 and D328 through hydrogen bonding mediated by a water molecule (W1). The cyclohexyl group of CHES interacts with the residues in the hydrophobic pocket (β-carbon of D291, side chains of I327, F347, F353, W354, and F460) (Fig. 1C). Thus, functional groups of CHES including sulfonic acid, secondary amine, and cyclohexyl groups strongly contribute to the tight interaction with CpNanICD.
Fig. 1.
Structure of the catalytic domain (CpNanICD) of Clostridium perfringens in complex with 2-(cyclohexylamino)ethanesulfonic acid (CHES): CpNanICD-CHES. (A) A ribbon diagram of CpNanICD-CHES structure. CHES and residues are displayed as stick representation. (B) Chemical structure of CHES. (C) Stereoview of CHES binding mode in the CpNanICD catalytic site. A water molecule (W1) is shown as a red sphere. Hydrogen bonds are shown as dashed lines. The composite omit map (mFo-DFc) of CHES is contoured at 1.5 σ and displayed as mesh (green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
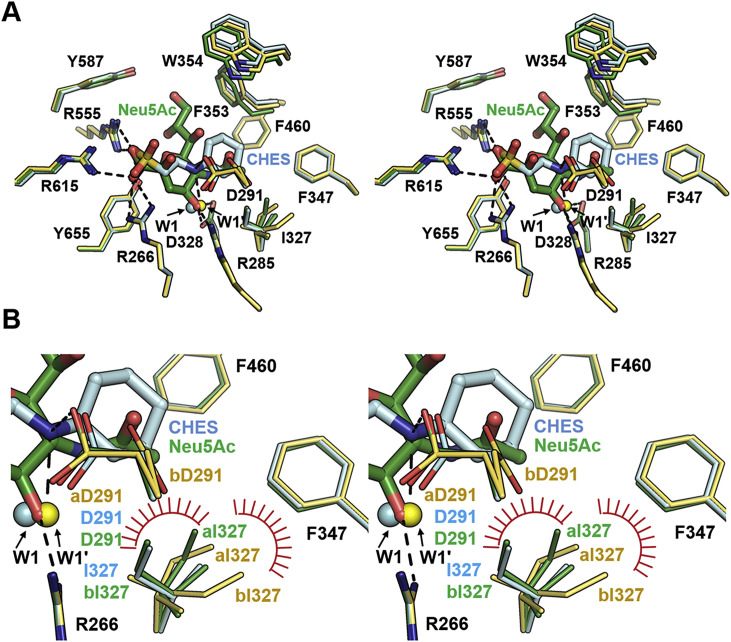
3.2. Structural comparison of apo, Neu5Ac, and CHES complexes of CpNanICD
The CpNanICD-CHES structure is similar to that of the apo-form and Neu5Ac-bound form, with RMSD of 0.21 and 0.19 Å among 393 Cα atoms, respectively (Fig. 2 ). In a detailed view, however, torsion angles of some of residues at the catalytic site are different among the three structures (Fig. 2B). Taking advantage of the high resolution structures, we can confidently analyze multiple conformations of some side chains. In the apo-form, D291, the acid/base catalyst, has a dual conformation. Meanwhile, in the Neu5Ac complex, the side chain of D291 adopts a single conformation forming a stable hydrogen bond with the 4-hydroxyl group of Neu5Ac. In case of the CHES complex, D291 forms a conformation similar to that of the substrate-bound structure interacting with the secondary amine of CHES (Fig. 2B).
Fig. 2.
Structural comparison among the catalytic domain (CpNanICD) of Clostridium perfringens NanI apo-form and Neu5Ac and 2-(cyclohexylamino)ethanesulfonic acid (CHES) complexes. (A) Stereoview of superimposed structures of apo-form (PDB code 2VK5, yellow), Neu5Ac complex (PDB code 2BF6, green), and the CHES complex (cyan). Neu5Ac and CHES in the catalytic site are highlighted in green and cyan, respectively. Water molecules (W1′ in the apo-form, W1 in the CHES complex) are presented as yellow and cyan spheres, respectively. Hydrogen bonds are shown as the dashed lines. (B) A zoomed in stereoview of D291 and I327 described in 2A. The residues for dual conformation were named aD291/bD291 and aI327/bI327, respectively. Hydrophobic interactions (I327 and F347 in the apo structure; Neu5Ac/CHES and I327 in the complex structures) are highlighted with the red eyelashes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Residue I327 behaves differently in the three structures. The side chain of I327 in CpNanI-apo has a dual conformation (aI327/bI327) and both conformations interact with F347 (Fig. 2B). In the Neu5Ac complex, the I327 side chain also has a dual conformation. Nevertheless, the positions are different from those in the apo-form. Although one of dual conformations (aI327) maintains a hydrophobic interaction with F347, the other conformation (bI327) forms a hydrophobic contact with 5′-acetamide moiety of Neu5Ac together with the β-carbon of D291. It is interesting to note that I327 in the CHES complex has only one conformation: the one interacting with the cyclohexyl group of CHES. In addition, water molecules (W1 in the CHES complex and W1′ in the apo-form) are located at a position similar to that of the 4-hydroxyl group of Neu5Ac and interact with R285 and D328 (Fig. 2A and B).
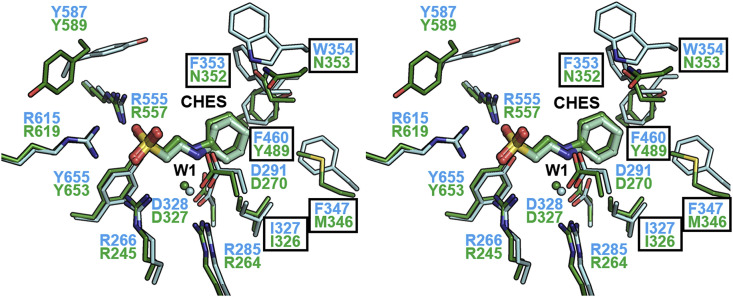
3.3. Structural comparison between the complexes of CpNanICD and SpNanBCD with CHES
To evaluate the structural differences between complexes, CpNanICD-CHES and SpNanBCD-CHES, we superimposed both complex structures. The CHES molecules located at the similar position in the catalytic sites of CpNanICD and SpNanBCD interact with the same residues including the arginine cluster (R266, R555, and R615 in CpNanICD versus R245, R557, and R619 in SpNanBCD) and the catalytic aspartate residue (D291 of CpNanICD versus D270 of SpNanBCD) (Fig. 3 ). Of note, the water molecule (W1) bridges the hydrogen bond between the secondary amine of CHES and R285/D328 in CpNanICD and R264/D327 in SpNanBCD, respectively, indicating that the water molecule (W1) plays an indispensable role in the CHES binding.
Fig. 3.
Structural comparison of complexes of the catalytic domain of the Clostridium perfringens NanI (CpNanICD) and the catalytic domain of Streptococcus pneumoniae NanB (SpNanBCD) with 2-(cyclohexylamino)ethanesulfonic acid (CHES). Neuraminidases, CHES, and water (W1) molecules in CpNanICD and SpNanBCD (PDB code 2VW2) are cyan and green, respectively. The residues of the hydrophobic pocket are highlighted with the black boxes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
One the other hand, residues in the hydrophobic pocket (F347, F353, W354, and F460 in CpNanICD; M346, N352, N353, and Y489 in SpNanBCD) are less conserved in CpNanI and SpNanB. Nevertheless, the hydrophilic residues (arginine cluster and catalytic aspartate) that interact with 2-aminoethanesulfonic acid of CHES are well conserved (Figs. 3 and S2). To compare the hydrophobicity levels of the cyclohexyl-binding pocket of CpNanICD and SpNanBCD, we calculated the hydrophobic pocket areas using software DeepView. We found that the hydrophobic contact area of CpNanICD (300 Å2, formed by β-carbon of D291, side chains of I327, F347, F353, W354, and F460) is two fold wider than that of SpNanBCD (162 Å2, formed by β-carbons of D270 and N352, side chains of I326, M346, N353, and Y489). Although cyclohexyl groups of CHES interact with a similar hydrophobic pocket in CpNanICD and in SpNanBCD, CHES may form stronger hydrophobic contacts with CpNanICD than with SpNanBCD because of the wider hydrophobic contact area [34].
3.4. Structural comparison of CpNanICD with human neuraminidases
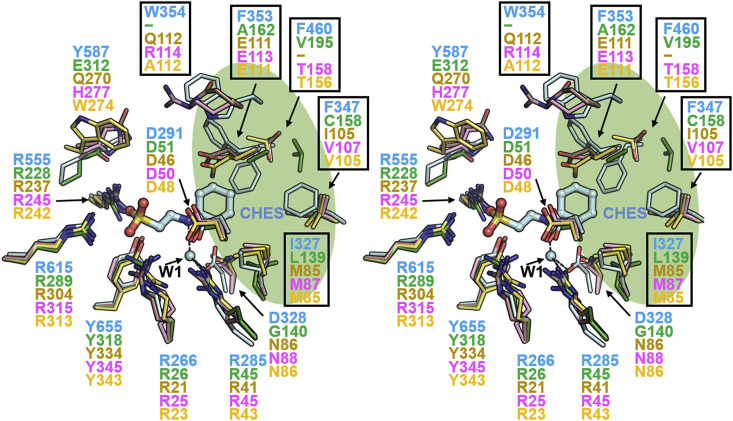
Among the four human neuraminidases (Neu1−Neu4), Neu2's structure was already reported (PDB code 1VCU) [35], and we modeled human Neu1, Neu3, and Neu4 reported in our previous studies (Fig. 4 ) [24]. In the CpNanICD-CHES structure, the hydrophobic pocket (residues I327, F347, F353, W354, and F460) contributes to the stable interaction with the cyclohexyl group of CHES. In human neuraminidases, however, hydrophobic pockets include polar residues (E111 and Q112 in Neu2; E113 and R114 in Neu3; E111 in Neu4). Thus, we predict that the hydrophobic interaction of CHES with human neuraminidases is less stable. In addition, CHES interacts with R285 and D328 of CpNanICD, and this interaction is mediated by one water molecule (W1). On the other hand, the residues corresponding to D328 of CpNanICD (G140 in Neu1, N86 in Neu2, N88 in Neu3, and N86 in Neu4) are not conserved, and the water-mediated hydrogen bonding with CHES may be different in human neuraminidases. Collectively, our data suggest that CHES interacts less favorably with human neuraminidases (Neu1–Neu4) because of a less hydrophobic interaction with the cyclohexyl group of CHES and the absence of water-mediated hydrogen bonding with the secondary amine of CHES.
Fig. 4.
Structural comparison of the catalytic domain of Clostridium perfringens NanI (CpNanICD) with human neuraminidases. Stereoview of the superimposed structure of CpNanICD with CHES (cyan) and catalytic domains of human Neu2 (PDB code 1VCU, brown), Neu1 (green), Neu3 (pink), and Neu4 (yellow). The structures of human Neu1, Neu3, and Neu4 are modeled using human Neu2 as a template (see Materials and Methods). CHES, residues, and the water molecule (W1) are displayed as stick models and a sphere, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. Strategies for modification of CHES for anti-neuraminidase candidates
We compared the CpNanICD-CHES structure with the structures of bacterial and viral neuraminidase complexed with oseltamivir and zanamivir (Fig. S3). After a structural comparison of human Neu1−Neu4 structures, we were able to suggest strategies for modification of CHES to facilitate for the development of new anti-neuraminidase agents targeting CpNanI. CHES contains a sulfonic acid group showing binding similar to that of a carboxyl group in Neu5Ac, oseltamivir, and zanamivir, where it interacts with the arginine cluster in the catalytic site (Figs. S3A and B). Nonetheless, there is a lack of hydrophilic functional groups such as the 6-glycerol side chain, the 4-hydroxyl group of Neu5Ac, the 4-amino group of oseltamivir, and 4-guanidino group of zanamivir. Thus, addition of a glycerol group at C3 of CHES and a hydrophilic group (such as hydroxyl, amino, or guanidine group) at nitrogen (N4) of CHES may replace the water molecule (W1) bridging the hydrogen bonds between CHES, R285 and D328 and may enhance the interaction of CHES with residues of CpNanICD (D291, R285, D328, E539, Y485, and R555) (Figs. 2 and S4). Moreover, introduction of additional moieties into the cyclohexyl ring of CHES was reported to increase the inhibitory effect on SpNanB [17]. Thus, we propose that the additional hydrophobic moieties such as linear hydrophobic groups (hydrophobic pentyloxy substituents) at the C3 position and cyclohexyl substituents (aryl groups of phenyl and benzyl, cycloalkane functional groups of cycloheptyl, and cyclooctyl) of CHES should enhance hydrophobic interactions as well as π-π stacking in the hydrophobic pocket of the bacterial neuraminidase catalytic sites. In addition, we predicted that addition of hydrophobic moieties in the cyclohexyl groups would also block the interaction with human neuraminidases.
The cyclohexyl group of CHES binds to a less conserved hydrophobic pocket; thus, it is possible to design a specific inhibitor of CpNanI by means of CHES's targeting this less conserved hydrophobic pocket of the catalytic site. On the basis of the structure-based comparative analyses of bacterial and human neuraminidases, we suggest strategies for modification of CHES for the development of specific anti-neuraminidase agents targeting CpNanI.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Acknowledgments
We thank the staff at beamline BL-5C of the Pohang Accelerator Laboratory (Pohang, South Korea), beamline NW12A at the Photon Factory (Tsukuba, Japan), and beamline BL26B1 at Spring-8 (Harima, Japan) for their kind help with data collection. This work was supported by the KRIBB Research Initiative Program and the National Research Foundation of Korea (NRF) grants (2015M2A2A4A03044653, 2013R1A2A2A01068440, and 2013M3A9A7046297).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2017.03.064.
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2017.03.064.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Transparency document
References
- 1.Taylor G. Sialidases: structures, biological significance and therapeutic potential. Curr. Opin. Struct. Biol. 1996;6:830–837. doi: 10.1016/s0959-440x(96)80014-5. [DOI] [PubMed] [Google Scholar]
- 2.Shinya K., Ebina M., Yamada S., Ono M., Kasai N., Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 3.Memoli M.J., Morens D.M., Taubenberger J.K. Pandemic and seasonal influenza: therapeutic challenges. Drug Discov. Today. 2008;13:590–595. doi: 10.1016/j.drudis.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J.-H., Resende R., Wennekes T., Chen H.-M., Bance N., Buchini S., Watts A.G., Pilling P., Streltsov V.A., Petric M., Liggins R., Barrett S., McKimm-Breschkin J.L., Niikura M., Withers S.G. Mechanism-based covalent neuraminidase inhibitors with broad-spectrum influenza antiviral activity. Science. 2013;340:71–75. doi: 10.1126/science.1232552. [DOI] [PubMed] [Google Scholar]
- 5.Soong G., Muir A., Gomez M.I., Waks J., Reddy B., Planet P., Singh P.K., Kanetko Y., Wolfgang M.C., Hsiao Y.-S., Tong L., Prince A. Bacterial sialidase facilitates mucosal infection by participating in biofilm production. J. Clin. Invest. 2006;116:2297–2305. doi: 10.1172/JCI27920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker D., Soong G., Planet P., Brower J., Ratner A.J., Prince A. The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect. Immun. 2009;77:3722–3730. doi: 10.1128/IAI.00228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corfield A.P. Bacterial sialidases–roles in pathogenicity and nutrition. Glycobiology. 1992;2:509–521. doi: 10.1093/glycob/2.6.509. [DOI] [PubMed] [Google Scholar]
- 8.Rood J.I. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 1998;52:333–360. doi: 10.1146/annurev.micro.52.1.333. [DOI] [PubMed] [Google Scholar]
- 9.Paulson J.C., Kawasaki N. Sialidase inhibitors DAMPen sepsis. Nat. Biotechnol. 2011;29:406–407. doi: 10.1038/nbt.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grienke U., Schmidtke M., von Grafenstein S., Kirchmair J., Liedl K.R., Rollinger J.M. Influenza sialidase: a druggable target for natural products. Nat. Prod. Rep. 2012;29:11–36. doi: 10.1039/c1np00053e. [DOI] [PubMed] [Google Scholar]
- 11.Hagiwara T., Kijima-Suda I., Ido T., Ohrui H., Tomita K. Inhibition of bacterial and viral sialidases by 3-fluoro-N-acetylneuraminic acid. Carbohydr. Res. 1994;263:167–174. doi: 10.1016/0008-6215(94)00133-2. [DOI] [PubMed] [Google Scholar]
- 12.Babu Y.S., Chand P., Bantia S., Kotian P., Dehghani A., El-Kattan Y., Lin T.H., Hutchison T.L., Elliott A.J., Parker C.D., Ananth S.L., Horn L.L., Laver G.W., Montgomery J.A. BCX-1812 (RWJ-270201): discovery of a novel, highly potent, orally active, and selective influenza neuraminidase inhibitor through structure-based drug design. J. Med. Chem. 2000;43:3482–3486. doi: 10.1021/jm0002679. [DOI] [PubMed] [Google Scholar]
- 13.Chand P., Kotian P.L., Dehghani A., El-Kattan Y., Lin T.-H., Hutchison T.L., Babu Y.S., Bantia S., Elliott A.J., Montgomery J.A. Systematic structure-based design and stereoselective synthesis of novel multisubstituted cyclopentane derivatives with potent antiinfluenza activity. J. Med. Chem. 2001;44:4379–4392. doi: 10.1021/jm010277p. [DOI] [PubMed] [Google Scholar]
- 14.Wang G.T., Chen Y., Wang S., Gentles R., Sowin T., Kati W., Muchmore S., Giranda W., Stewart K., Sham H., Kempf D., Laver W.G. Design, synthesis, and structural analysis of influenza neuraminidase inhibitors containing pyrrolidine cores. J. Med. Chem. 2001;44:1192–1201. doi: 10.1021/jm000468c. [DOI] [PubMed] [Google Scholar]
- 15.Stoll V., Stewart K.D., Maring C.J., Muchmore S., Giranda V., Gu Y.-G., Wang G., Chen Y., Sun M., Zhao C., Kennedy A.L., Madigan D.L., Xu Y., Saldivar A., Kati W., Laver G., Sowin T., Sham H.L., Greer J., Kempf D. Influenza neuraminidase inhibitors: structure-based design of a novel inhibitor series. Biochemistry. 2003;42:718–727. doi: 10.1021/bi0205449. [DOI] [PubMed] [Google Scholar]
- 16.Xu G., Potter J.A., Russell R.J., Oggioni M.R., Andrew P.W., Taylor G.L. Crystal structure of the NanB sialidase from Streptococcus pneumoniae. J. Mol. Biol. 2008;384:436–449. doi: 10.1016/j.jmb.2008.09.032. [DOI] [PubMed] [Google Scholar]
- 17.Brear P., Telford J., Taylor G.L., Westwood N.J. Synthesis and structural characterisation of selective non-carbohydrate-based inhibitors of bacterial sialidases. Chembiochem. 2012;13:2374–2383. doi: 10.1002/cbic.201200433. [DOI] [PubMed] [Google Scholar]
- 18.Ford S.M., Grabenstein J.D. Pandemics, avian influenza A (H5N1), and a strategy for pharmacists. Pharmacotherapy. 2006;26:312–322. doi: 10.1592/phco.26.3.312. [DOI] [PubMed] [Google Scholar]
- 19.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., Paterson D.L., Rice L.B., Stelling J., Struelens M.J., Vatopoulos A., Weber J.T., Monnet D.L. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 20.van der Vries E., Collins P.J., Vachieri S.G., Xiong X., Liu J., Walker P.A., Haire L.F., Hay A.J., Schutten M., Osterhaus A.D., Martin S.R., Boucher C.A., Skehel J.J., Gamblin S.J. H1N1 2009 pandemic influenza virus: resistance of the I223R neuraminidase mutant explained by kinetic and structural analysis. PLoS Pathog. 2012;8:e1002914. doi: 10.1371/journal.ppat.1002914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naumov P., Yasuda N., Rabeh W.M., Bernstein J. The elusive crystal structure of the neuraminidase inhibitor Tamiflu (oseltamivir phosphate): molecular details of action. Chem. Commun. (Camb). 2013;49:1948–1950. doi: 10.1039/c3cc38801h. [DOI] [PubMed] [Google Scholar]
- 23.Newstead S., Chien C.H., Taylor M., Taylor G. Crystallization and atomic resolution X-ray diffraction of the catalytic domain of the large sialidase, nanI, from Clostridium perfringens. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2063–2066. doi: 10.1107/S090744490402181X. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y., Ryu Y.B., Youn H.-S., Cho J.K., Kim Y.M., Lee W.S., Park K.H., Eom S.H. Structural basis of sialidase in complex with geranylated flavonoids as potent natural inhibitors. Acta Crystallogr. D. Biol. Crystallogr. 2014;70:1357–1365. doi: 10.1107/S1399004714002971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Vagin A., Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D. Biol. Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 27.Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., Zwart P.H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 33.Roggentin P., Rothe B., Kaper J.B., Galen J., Lawrisuk L., Vimr E.R., Schauer R. Conserved sequences in bacterial and viral sialidases. Glycoconj. J. 1989;6:349–353. doi: 10.1007/BF01047853. [DOI] [PubMed] [Google Scholar]
- 34.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 35.Chavas L.M., Tringali C., Fusi P., Venerando B., Tettamanti G., Kato R., Monti E., Wakatsuki S. Crystal structure of the human cytosolic sialidase Neu2. Evidence for the dynamic nature of substrate recognition. J. Biol. Chem. 2005;280:469–475. doi: 10.1074/jbc.M411506200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.