Abstract
An immune antibody phage-display library was constructed from B cells of SARS convalescent patients. More than 80 clones were selected from the library by using the whole inactivated SARS-CoV virions as target. One human scFv, B1, was characterized extensively. The B1 recognized SARS pseudovirus in vivo and competed with SARS sera for binding to SARS-CoV with high affinity (equilibrium dissociation constant, Kd = 105 nM). The B1 also has potent neutralizing activities against infection by pseudovirus expressing SARS-CoV S protein in vitro. Finally, we found that the B1 recognized an epitope on S2 protein, especially within amino acids 1023–1189 of S2 protein. This study not only first made a human neutralizing antibody, which recognized an epitope on S2 protein like natural antibody in sera, but also may help us to better understand the immunological characteristics of SARS protein and SARS vaccine design.
Keywords: SARS-CoV, Neutralizing antibody, S2 protein, scFv, Phage-display library
The worldwide outbreak of severe acute respiratory syndrome (SARS) as a newly emerging disease caused by SARS-associated coronavirus (SARS-CoV) represents the most recent threat to human health [1], [2], [3]. The SARS-CoV is highly contagious and the clinical course is severe with a high mortality. During the outbreak in 2003, there was no effective agent available for SARS prophylaxis and therapy [4], [5]. Although passive serotherapy using polyclonal immune sera from convalescent SARS patients has been shown to be an efficient treatment for SARS and potentially provides immediate protection against infection [6], [7], [8], human sera are very limited in sufficient amounts and have a possible contamination with infectious agents. Therefore, a viable strategy for SARS prophylaxis and treatment is to produce human antibody by engineering technology.
Viral envelope proteins initiate entry of viruses into host cells by binding to cell surface receptors followed by conformational changes leading to membrane fusion. It has been shown that spike (S) protein of SARS-CoV is not only responsible for binding to its receptor angiotensin-converting enzyme 2 (ACE2) that initiates entry into host cell [9], [10], [11], [12], but has also been identified as a dominant antigenic determinant eliciting neutralizing antibody during the infection of SARS-CoV [11], [12], [13], [14]. Several data have demonstrated that the antibodies selected by using S protein as antigen from naïve [15], semisynthetic antibody libraries [16], and memory repertoire [17] have potent neutralizing activity to SARS-CoV infection.
In the present study, we generated a SARS-CoV immune antibody library, and isolated a single chain Fv (scFv) with high specificity and affinity by selection using whole SARS-CoV virions, rather than a single recombinant S protein, as antigen. Our results show that the selected scFv with neutralizing activity recognizes an epitope on S2 protein, especially within amino acids 1023–1189 of S2 protein. This study would aid in making neutralizing antibody, which recognizes an epitope on S2 protein like natural antibody in sera, better understanding the immunological characteristics of SARS protein and the design of SARS vaccine.
Materials and methods
Cells, plasmids, and antibodies. Vero E6 was supplied by Institute of Microbiology and Epidemiology (Beijing, China). pET28a (+) vector was purchased from Novagen (Darmstadt, Germany). Phagemid pDNA5 [18] was provided by Dr. Andrew Bradbury from Los Alamos National Laboratory. pCDM8/S1-Fc vector [11] was provided by Dr. Hyeryun Choe from Harvard Medical School. The antibodies used were horseradish peroxidase (HRP)-conjugated anti-M13 from Amersham Pharmacia (England), HRP-conjugated anti-human Fc and FITC-conjugated anti-mouse IgG from Sigma (St. Louis, MO, USA). HRP-conjugated anti-mouse IgG was from Pierce (Rockford, IL, USA), while anti-His-Tag was from Novagen (Darmstadt, Germany).
Virion preparation. SARS-CoV BJ01 were inactivated in a biosafety level 3 laboratory and purified by differential centrifugation as previously described [19].
Construction of scFv library. Total RNA was prepared from peripheral blood lymphocytes of four convalescent SARS patients, followed by cDNA synthesis. The VH (heavy chain variable region) and VL genes (light chain variable region) were amplified using the primers listed in Table 1 and then cloned into phage-display vector pDAN5 [18]. The recombinant pDAN5 was transformed into Escherichia coli XL1-Blue by electroporation. The transformants were plated on dishes with 2× YT, containing 1% glucose, 100 μg/ml ampicillin, and 10 μg/ml tetracycline. All the colonies were collected by scraping, rescued by helper phage M13KO7 [20], and finally stored as a primary scFv library.
Table 1.
Primers for human VH and VL
| Human VH Back primers with XhoI | |
| VH1 Back XhoI | TTATCCTCGAGCGGTACCSAGGTSCAGCTGGTRCAGTCTGG |
| VH2 Back XhoI | TTATCCTCGAGCGGTACCCAGRTCACCTTGAAGGAGTCTG |
| VH3 Back XhoI | TTATCCTCGAGCGGTACCSAGGTGCAGCTGKTGSAG |
| VH4 Back XhoI | TTATCCTCGAGCGGTACCCAGSTRCAGCTRCAGSAGTS |
| Human VH For primers with NheI cite | |
| JH for NheI | GATTGGTTTGCCGCTAGCTGARGAGACRGTGACCRKKGT |
| Human Vκ Back primers with BssHII | |
| Vκ1 Back BssHII | AGCAAGCGGCGCGCATGCCGMCATCCRGDTGACCCAGTCTCC |
| Vκ2 Back BssHII | AGCAAGCGGCGCGCATGCCGATATTGTGMTGACBCAGWCTCC |
| Vκ3 Back BssHII | AGCAAGCGGCGCGCATGCCGAAATTGTGHTGACDCAGTCTCC |
| Vκ1 Back BssHII | AGCAAGCGGCGCGCATGCCGAAACKACACTCACGCAGTCTC |
| Human Vκ For primers with SalI | |
| Jκ1 for SalI | GAAGTTATGGTCGACCCTCCGGAACGTTTGATHTCCASYTTGGTCC |
| Jκ2 for SalI | GAAGTTATGGTCGACCCTCCGGAACGTTTAATCTCCAGTCGTGTCC |
| Human Vλ Back primers with BssHII | |
| Vλ1 Back BssHII | AGCAAGCGGCGCGCATGCCCAGYCWGYBYTGAYKCAGCC |
| Vλ2 Back BssHII | AGCAAGCGGCGCGCATGCCTCCTMTGWGCTGABDCAGS |
| Vλ3 Back BssHII | AGCAAGCGGCGCGCATGCCAATTTTATGCTGACTCAGCCCC |
| Human Vλ For primers with SalI | |
| Jλ for SalI | GAAGTTATGGTCGACCCTCCGGAACCTAGGACGGTSASCTTGGTCCC |
Note. A set of human antibody primers for amplification of the VH and VL genes from B cells of SRAS patients was optimized based on previous publications [20], [21], [27], [28], [29].
The italic indicates the flank sequence of the primers, the underline indicates the restriction enzyme site, and the bold indicates the primer sequence for amplifying the genes of human VH and VL.
Diversity assessment of the scFv library. A number of individual colonies from the primary library were randomly picked and their phagemids with scFv were prepared as described previously [21]. After amplifying with polymerase chain reaction (PCR), the individual scFv gene was digested by BstNI, and the fingerprinting of each colony was analyzed by agarose gels.
Selection of phage scFv to SARS-CoV. The immunotubes were coated with purified SARS-CoV virions in 0.05 M Na2CO3, pH 9.6, blocked 2 h with 3% BSA in phosphate-buffered saline (PBS), and incubated for 3 h at room temperature with 1012 phage scFv in 1 ml PBS containing 1% bovine serum albumin (BSA) and 0.1% Triton X-100. After intensive washes with PBST (0.1% Tween 20 in PBS), bound phage antibodies were eluted with 0.1 M glycine/HCl, pH 2.2, and immediately neutralized with 1.0 M Tris–Cl, pH 8.0. Eluted phage scFv were subjected to the next round of infection, rescue, and selection. After five rounds of panning, the higher binders to SARS-CoV were selected by enzyme-linked immunosorbent assay (ELISA).
ELISA. Ninety-six-well microtiter plates (Nunc, Rochester, NY) were coated overnight at 4 °C with inactivated SARS-CoV particles in 0.05 M Na2CO3, pH 9.6, blocked with 3% BSA in PBS, and incubated with an individual phage scFv in PBS containing 1% BSA. After five washes with PBST, the bound antibodies were detected by HRP-conjugated anti-M13 antibody followed by incubation with ortho-phenylenediamine (OPD) as substrate. The color reaction was measured at 490 nm in a BioRad ELISA reader (Hercules, CA, USA).
Expression and purification of scFv. In order to obtain a large amount of scFv, the genes of selected phage scFv were cloned into pET28a (+) vector and expressed with His-6-Tag in E. coli BL21 (DE3). The scFv in inclusion bodies was denatured with 8 M urea and purified by gel filtration on a column with Sephacryl S200 HR (10 × 110 mm) as previously described [22].
SDS–PAGE and Western blot. The protein samples were separated by electrophoresis in SDS–PAGE and then transferred onto membrane. The membranes were blocked with 5% non-fat milk in PBS and then incubated with a mouse anti-His-Tag antibody followed by a HRP-conjugated anti-mouse IgG. The specific protein bands were visualized by autoradiography on Kodak X-ray film.
Competitive inhibition assay. Sera from convalescent SARS patients were 1000 times diluted in PBS and then incubated 1 h at 37 °C with a serial twofold dilution of purified scFv B1, starting at a concentration of 24 μg/ml. The above mixture of sera with diluted scFv B1 was subjected to 96-well plate coated with SARS-CoV particles and incubated for another 1 h at 37 °C. HRP-conjugated anti-human Fc was added as secondary antibody. The following procedures were the same as described in ELISA.
Antibody affinity assay. The binding kinetics of scFv B1 to SARS-CoV was analyzed using a Variable Angle Spectroscopic Ellipsometer. Berifly, SARS-CoV virions were covalently immobilized to a protein chip and scFv B1 solution was added on the chip surface. Binding kinetic parameters were evaluated with antibody real-time binding curve [23], [24], [25].
Preparation of Luc/SARS S pseudovirus. The Luc/SARS S pseudovirus system was prepared based on the method described in [26]. Briefly, mammalian cell 293T was cotransfected by using three vectors that are mammalian expression vector pMT 21-S coding SARS-CoV S protein, pCMVR 8.2 coding for MuLV Gag and Pol, and pHR′-luc coding luciferase. After 48 h postinfection, Luc/SARS S pseudoviruses were packaged as virions in the infected cell and were collected from the culture supernatant.
In vitro neutralization assay. Luc/SARS S pseudovirus system was used as an infection model to evaluate the neutralization activity of scFv B1. First, the Luc/SARS S pseudoviruses were pre-incubated with a serial twofold dilution of purified scFv B1, starting at a concentration of 17 μg/ml. Then, Vero E6 cells were infected by the Luc/SARS S pseudovirus with or without scFv B1. After 3 h incubation at 37 °C, the culture medium was refreshed with DMEM containing 10% FBS and continuously cultured for another 48 h. The infected Vero E6 cells were lysed and assayed using a Luminometer DL Ready model TD2020 (Tartu, Estonia) for measuring the luciferase activity in the infected cells.
Immunofluorescence assay. Vero E6 cell monolayer infected with Luc/SARS S pseudovirus was fixed with the mixture of acetone and methanol. After blocking with goat sera, the slides were incubated with purified scFv B1, and then mouse anti-His-Tag, and finally FITC-labeled anti-mouse IgG. The scFv bound to SARS S pseudovirus in Vero E6 cells were visualized under a fluorescence microscope.
SARS antigen protein preparation. SARS S protein expressed with human Fc of IgG1 in CHO cell. Briefly, ectodomain (12–1194 amino acids) of S protein with human Fc of IgG1 was cloned into pMT 21, and a stably expressing cell line was selected after being transfected into CHO cell. After 48 h cultures, culture mediums were recovered and incubated with protein A–Sepharose beads for about 1 h at 4 °C. The beads were washed three times with PBS. Bound proteins were eluted in SDS–PAGE sample loading buffer at 100 °C for 5 min. Proteins were separated by 8% SDS–PAGE, identified by Western blot using HRP-goat anti-human Fc antibody, and visualized by autoradiography on X film.
The pCDM8-S1-Fc vector encoded a fusion of S1 protein (residues 12–672) and a human Fc of IgG1. The pCDM8-S1-Fc was transfected into 293T cells. After 48 h, the S1-Fc protein was produced by transfected 293T cells into culture supernatant and subjected to 8% SDS–PAGE and Western blot for analysis.
The coding sequences for 680–1189, 680–1029, 680–879, and 1023–1189 residues of S protein were PCR-amplified from the full-length S gene (a stop codon also was inserted just after the protein sequence by PCR) and cloned into expression vector pET28a (+) by restriction enzyme sites NcoI and EcoRI. After identification by sequences, protein was expressed by inducing with IPTG in E. coli BL21 (DE3). Protein samples were analyzed by using 15% SDS–PAGE and transformed into nitrocellulose membrane, which was blotted by scFv B1.
Results
SARS-CoV immune human scFv library
In order to make a large diversity of scFv library with a high affinity for SARS-CoV, we used pDNA5 as a phage-display vector and four SARS patient’s lymphocytes as a repertoire of antibody. All the patient’s sera showed high titer binding to SARS-CoV. Using PCR technique, four groups of VH and seven groups of VL (four for Vκ and three for Vλ) were amplified (Fig. 1 A) with a set of human antibody primers (Table 1) which is optimized based on previous publications [20], [21], [27], [28], [29]. The amplified VL and VH were, respectively, inserted into the pDNA5 vector and then electroporately transformed into E. coli to make a primary scFv library.
Fig. 1.
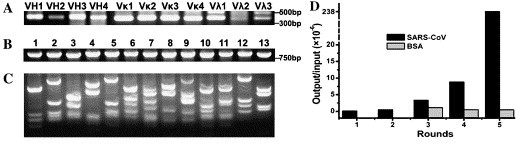
Construction of SARS immune scFv library. (A) Four groups of VH and seven groups of VL (four for Vκ and three for Vλ) were amplified by RT-PCR. Total RNA was prepared from peripheral blood lymphocytes of four convalescent SARS patients and followed by cDNA synthesis. The VH and VL genes were amplified using the primers listed in Table 1; (B) 13 scFv clones were randomly selected from the primary library and their genes were amplified by PCR; (C) fingerprinting of the 13 scFv genes digested 2 h at 60 °C by BstNI and then analyzed in 4% agarose gel, showing a diversity of the primary antibody library; (D) five panning rounds showing an enrichment of the scFv/phage to SARS-CoV. Purified SARS viral particles as antigen, five rounds of selection were performed according to standard procedure [20]. There is no significant change to immobilized control BSA during the last three round.
To assess the diversity of the scFv library, 13 colonies were randomly selected and their scFv genes were digested with BstNI since a number of BstNI sites randomly exist in the variable region of antibody. As shown in Fig. 1B, all the 13 clones contained a similar size of full-length scFv around 750 bp. However, each scFv clone showed a unique BstNI-digested fingerprinting pattern (Fig. 1C), indicating that individual clones in the primary library are different. This library was calculated to have a diversity of 1.85 × 106 members.
Selection of phage antibody binding to SARS-CoV
Bio-panning was performed with stringent conditions to enrich phage scFv for SARS-CoV. The phage scFv at 1012 pfu (input) was subjected to immunotubes coated with SARS-CoV virions. After 3 h incubation, the immunotubes were intensively washed to remove non-specific binders, and the bound phages (output) were calculated after each panning. The ratio of output/input was gradually increasing after the third and fourth rounds, and it was dramatically increasing after the fifth round. However, the phage scFv to immobilized control BSA did not show any significant changes (Fig. 1D). These results indicate that the phage scFv to SARS-CoV were specifically enriched after five round pannings.
From the fifth panning round, 96 phage clones were randomly selected for ELISA to evaluate their binding activity to SARS-CoV. We identified 86 of the 96 clones specifically binding to SARS-CoV, one of them, referred as B1, was a specific binder with consistently highest affinity. The B1 was identified as VH1 family and Vκ3 family based on its deduced amino acid sequence (Fig. 2 ).
Fig. 2.

Amino acid sequence of B1 scFv deduced from DNA sequence. Framework regions 1–4 (FW1–4) and complementarity determining regions 1–3 (CDR1–3) for both the VH and VL are shown, which was determined with the Vbase sequence directory. Based on its sequence, the scFv B1 is identified as VH1 family and Vκ3 family.
Expression and purification of B1 antibody
The B1 clone was expressed in the inclusion body of E. coli, and the refolding of scFv B1 was done during the gel filtration on a column with Sephacryl S200HR. The purity of B1 was analyzed as a single band of about 29 kDa by using SDS–PAGE (Fig. 3 A). This single band was specifically blotted by anti-His-Tag antibody (Fig. 3B), indicating that it was scFv with His-Tag.
Fig. 3.

Expression and purification of B1. (A) SDS–PAGE and (B) Western blot analysis of the scFv B1 before (lane 1) and after purification (lane 2). The scFv B1 was purified and refolded at the same time by gel filtration through a column with Sephacryl S200HR. The membrane with scFv B1 was blotted by using anti-His-6-Tag.
Specificity of B1 for SARS-CoV virions
To test whether soluble scFv B1 maintains the specific binding activity to SARS-CoV, we performed a competition assay. It was clearly shown that B1 significantly decreased the binding of SARS convalescent sera to SARS-CoV and the inhibition was getting stronger with increasing amounts of scFv B1, whereas the control scFv D4 did not affect sera activity (Fig. 4 ). The data suggest that B1 bears a similar antigen epitope as natural antibody in SARS convalescent sera. The binding kinetics of scFv B1 to SARS-CoV was analyzed using a Variable Angle Spectroscopic Ellipsometer. The equilibrium dissociation constant of scFv B1 was evaluated as K d = 105 nM (Table 2 ) with antibody real-time binding curve [23], [24], [25].
Fig. 4.
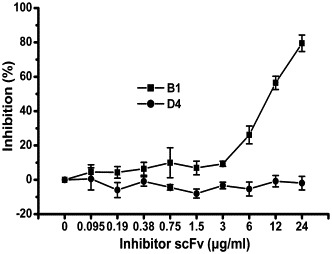
The specific binding of B1 to SARS-CoV. Competition ELISA shows the ability of B1 to abrogate the binding of SARS sera to SARS-CoV. Whereas the negative control (scFv D4) failed to bind to SARS-CoV; each point represents the mean of quadruple determinations; error bars, ±SD.
Table 2.
Kinetic rates and binding affinity of B1
| Kon (M−1 s−1) | Koff (s−1) | Ka (M−1) | Kd (M) |
|---|---|---|---|
| 5.22 × 103 | 5.5 × 10−4 | 9.5 × 106 | 1.05 × 10−7 |
B1 neutralizing Luc/SARS S pseudovirus infection
Luc/SARS S pseudovirus had been proven as an efficient in vitro model for the study of SARS-CoV infection [30], [31], [32]. In this model, luciferase activity reflects the infection and replication of pseudovirus in the host cells. Using this Luc/SARS S pseudovirus system, we tested the neutralizing activity of scFv B1 by using a luminometer. The results are shown in Fig. 5 , where B1 significantly inhibited the infection of SARS S pseudovirus into Vero E6 cells and the inhibition was effected in a dose-dependent manner, whereas a control scFv D4 did not cause any effect under the same conditions.
Fig. 5.

Neutralizing activity of scFv B1 was evaluated by a Lus/SARS pseudotyped virus system. Luc/SARS S pseudovirus had been proved as an efficient in vitro model for the study of SARS-CoV infection. In this model, luciferase activity that reflects the infection and replication of pseudovirus in the host cells was measured by a luminometer. B1 significantly inhibited the infection of SARS S pseudovirus into Vero E6 cells and the inhibition was effected in a dose-dependent manner, whereas a control D4 did not cause any effect under the same conditions. Each point represents the mean of triplicate determinations; error bars, ±SD. *Statistically significant differences (p ⩽ 0.01).
B1 recognizes S protein on SARS pseudovirus in vivo
We next determined whether the B1 recognizes S protein assembled on SARS pseudovirus in vivo. The Vero E6 cells were first infected with Luc/SARS S pseudovirus and then subjected to immunofluorescence assay. As shown in Fig. 6 , scFv B1, rather than scFv D4 as a negative control, recognized SARS pseudovirus in the infected Vero E6 cells (Figs. 6A and B), but not in mock-infected cells (Fig. 6C). These results are consistent with those of convalescent sera as positive control (data not shown), indicating that scFv B1 could bind to S protein assembled on viral particles in vivo.
Fig. 6.
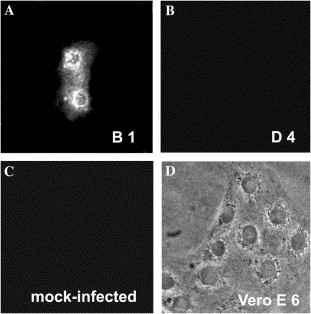
scFv B1 staining SARS pseudovirus in infected Vero E6 cell. (A) scFv B1 but not (B) scFv D4 recognizes SARS pseudovirus in the infected Vero E6 cells. (C) scFv B1 does not stain the mock-infected Vero E6 cells. (D) Pseudovirus-infected Vero E6 cells observed under a phase contrast microscope. Lus/SARS-CoV S protein mammalian expression vector pMT 21-S and pCMVR 8.2 coding for MuLV Gag and Pol, and pHR′-luc coding luciferase cotransfected into 293T cells. After 48 h culture, Vero E6 monolayer cells were fixed and blocked for incubation with purified scFv, and then mouse anti-His-Tag antibody, and finally FITC-labeled anti-mouse IgG. Binding to SARS S pseudovirus in Vero E6 cells was visualized under a fluorescence microscope.
B1 recognizes an epitope on S2 (1023–1189) protein
In order to identify the binding epitope, we tested the binding activity of scFv B1 with S-Fc and S1-Fc proteins using Western blot analysis. As shown in Figs. 7 A and B, the purified scFv B1 recognized the whole S protein, but it did not bind to the S1 protein. To further narrow the epitope of scFv B1, we constructed and expressed the recombinant S2 protein and its fragments. Using Western blot analysis, we found that purified scFv B1 recognized S2 protein (680–1189), but it did not bind to S2 (680–879) and S2 (680–1029) (Fig. 7D). The results suggested that the epitope of scFv B1 may be within amino acids 1029–1189 of the S2 protein. To confirm this, we made S2 (1023–1189) fragment and found that the scFv B1 recognized this fragment (Fig. 7D). Our data strongly demonstrated that the epitope of scFv B1 was located in the S2 protein, especially within amino acids 1023–1189 of the S2 protein.
Fig. 7.

Epitope mapping of scFv B1 by Western blot analysis. (A) scFv B1 recognizes S (12–1194)-Fc protein but does not bind to S1 (12–672)-Fc protein; (B) the proteins S (12–1194)-Fc and S1 (12–672)-Fc are recognized by anti-human Fc antibody as control; (C) SDS–PAGE analyzes the expression of S2 protein and its fragments; (D) Western blot shows the binding of scFv B1 to S2 (680–1189) and S2 (1023–1189), but does not recognize the S2(680–1029) and S2 (680–879).
Discussion
Antibodies have been used for a century for the prevention and treatment of infectious diseases. In viral disease, antibodies block viral entry into uninfected cells, promote antibody-directed cell-mediated cytotoxicity by natural killer cells, and neutralize virus alone or with the participation of complement. It has been shown that S protein is, as dimer and trimer, located on the surface of SARS virion, which is important for binding to receptor molecules that initiate entry into host cells [9], [19].
In order to obtain a neutralizing antibody, which mimics natural antibody elicited by viral pathogen, we constructed an immune scFv library, which was derived from B cells of four SARS patients with high titer to SARS virion. This B cell repertoire contains various antibodies with high specificity and matured affinity for SARS-CoV. Compared with naïve or synthetic antibody library, the immune library is more likely to obtain nature-like antibody with high specificity and affinity. Another feature to obtain nature-like antibody is in using complete SARS-CoV virions, rather than a single recombinant protein, as antigen for selections. The reason is that viral particle antigen bears natural and intact conformational immunogenic determinant. Thus, it is more feasible to select neutralizing antibody by panning against the whole SARS-CoV virion rather than a single recombinant S protein.
The selected scFv B1 with high affinity showed a specific binding to SARS-CoV particles in vitro and SARS pseudovirus in vivo. Moreover, the epitope recognized by scFv B1 is not on our suspected fragment according to a previous work [16], [17], [33], [34], but on the S2 protein, especially within amino acids 1023–1189 of the S2 protein, which is consistent with a recently published work on rabbits [35]. The competition assay showed that the B1 was able to abrogate the binding of SARS immune sera to SARS-CoV, suggesting that B1 bears a similar antigen epitope as native SARS antibodies in the sera. The neutralizing ability of B1 may be effected via its blocking the epitope on SARS virions.
S1 protein and S2 protein have been verified as important epitopes that elicit neutralization antibody in a previous study on SARS-CoV and other coronaviruses [36], [37]. However, neutralizing human antibody against S2 protein of SARS-CoV has not been reported, which could be significant in developing a cocktail of human SARS-CoV neutralizing monoclonal antibodies for SARS prophylaxis and treatment.
In summary, we describe here an effective method to obtain an antibody, which mimics natural antibody in immune sera, by using SARS virions, rather than a single recombinant protein, as antigen for selection from an immune antibody library. This study would have significance not only for developing neutralizing antibody, but also for a better understanding of the immunogenic characteristics of SARS protein and the design of SARS vaccine.
Acknowledgments
This work is supported by a CAS grant and the National 973 Grant specific for SARS prevention and therapy. We thank Dr. Andrew Bradbury for kindly providing phagemid pDNA 5 vectors, and Dr. Hyeryun Choe for kindly providing pCDM8/S1-Fc vector. And thanks to Dr. Zhanghui Wang for his assistance in the calculation of antibody affinity.
References
- 1.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. SARS Working Group, a novel coronavirus associated with severe acute respiratory syndrome. New Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Holmes K.V. SARS coronavirus: a new challenge for prevention and therapy. J. Clin. Invest. 2003;111:1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearson H., Clarke T., Abbott A., Knight J., Cyranoski D. SARS: what have we learned? Nature. 2003;424:121–126. doi: 10.1038/424121a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng Y., Wong R., Soo Y.O.Y., Wong W.S., Lee C.K., Ng M.H.L., Chan P., Wong K.C., Leung C.B., Cheng G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. 2005;24:44–46. doi: 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soo Y.O., Cheng Y., Wong R., Hui D.S., Lee C.K., Tsang K.K., Ng M.H., Chan P., Cheng G., Sung J.J. Retrospective comparison of convalescent plasma with continuing high-dose methylprednisolone treatment in SARS patients. Clin. Microbiol. Infect. 2004;10:676–678. doi: 10.1111/j.1469-0691.2004.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li G., Chen X., Xu A. Profile of specific antibodies to the SARS-associated coronavirus. New Engl. J. Med. 2003;349:508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 9.Xiao X., Feng Y., Chakraborti S., Dimitrov D.S. Oligomerization of the SARS-CoV S glycoprotein: dimerization of the N-terminus and trimerization of the ectodomain. Biochem. Biophys. Res. Commun. 2004;322:93–99. doi: 10.1016/j.bbrc.2004.07.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song H.C., Seo M.Y., Stadler K., Yoo B.J., Choo Q.L., Coates S.R., Uematsu Y., Harada T., Greer C.E., Polo J.M., Pileri P., Eickmann M., Rappuoli R., Abrignani S., Houghton M., Han J.H. Synthesis and characterization of a native, oligomeric form of recombinant severe acute respiratory syndrome coronavirus spike glycoprotein. J. Virol. 2004;78:10328–10335. doi: 10.1128/JVI.78.19.10328-10335.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho T., Wu S., Cheng S., Wei Y., Huang S., Hsiang C. Antigenicity and receptor-binding ability of recombinant SARS coronavirus spike protein. Biochem. Biophys. Res. Commun. 2004;313:938–947. doi: 10.1016/j.bbrc.2003.11.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y., Zhou Y., Wu H., Luo B., Chen J., Li W., Jiang S. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J. Immunol. 2004;173:4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- 15.Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAB to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. USA. 2004;101:2536–2641. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van den Brink E.N., Ter Meulen J., Cox F., Jongeneelen M.A., Thijsse A., Throsby M., Marissen W.E., Rood P.M., Bakker A.B., Gelderblom H.R., Martina B.E., Osterhaus A.D., Preiser W., Doerr H.W., de Kruif J., Goudsmit J. Molecular and biological characterization of human monoclonal antibodies binding to the spike and nucleocapsid proteins of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:1635–1644. doi: 10.1128/JVI.79.3.1635-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Traggian E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R., Murphy B.R., Rappuoli R., Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sblattero D., Bradbury A. Exploiting recombination in single bacteria to make large phage antibody libraries nature biotechnology. Nat. Biotechnol. 2000;18:75–80. doi: 10.1038/71958. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y., Yan X., Cao W., Wang C., Feng J., Duan J., Xie S. Probing the structure of the SARS coronavirus using scanning electron microscopy. Antivir. Ther. 2004;9:287–289. [PubMed] [Google Scholar]
- 20.O’Philippa M., BrienRobert A. Vol. 178. Humana Press; Totowa, New Jersey: 2002. (Methods in molecular biology: antibody phage display (methods and protocols)). pp. 59–73 and 133–147. [Google Scholar]
- 21.Borrebaeck C.A.K. Oxford university press; New York: 1995. Antibody engineering. pp. 53–87. [Google Scholar]
- 22.Zhang ZH., Lin Y., Yan X. Expression and refolding of an anti-tumor antibody AA98 fragment VH/κ. Prog. Biochem. Biophys. 2002;29:645–650. [Google Scholar]
- 23.Elwing H. Protein absorption and ellipsometry in biomaterial research. Biomaterial. 1998;19:397–406. doi: 10.1016/s0142-9612(97)00112-9. [DOI] [PubMed] [Google Scholar]
- 24.Wang ZH., Jin G. A label-free multisensing immunosensor based on imaging ellipsometry. Anal. Chem. 2003;75:6119–6123. doi: 10.1021/ac0347258. [DOI] [PubMed] [Google Scholar]
- 25.Malmborg A.C., Michaelsson A., Ohlin M., Jansson B., Borrebaeck A.K. Real time analysis of antibody–antigen reaction kinetics. Scand. J. Immunol. 1992;35:643–650. doi: 10.1111/j.1365-3083.1992.tb02970.x. [DOI] [PubMed] [Google Scholar]
- 26.Dehart J.L., Andersen J.L., Zimmerman E.S., Ardon O., An D.S., Blackett J., Kim B., Planelles V. The ataxia telangiectasia-mutated and Rad3-related protein is dispensable for retroviral integration. J. Virol. 2005;79:1389–1396. doi: 10.1128/JVI.79.3.1389-1396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong Zh., Wang Y. second ed. Peking University Medical Press; Beijing: 2002. Antibody engineering. pp. 308–315. [Google Scholar]
- 28.Sblattero D., Bradbury A. A definitive set of oligonucleotide primers for amplifying human V regions International School for Advanced Studies. Immunotechnology. 1998;3:271–278. doi: 10.1016/s1380-2933(97)10004-5. [DOI] [PubMed] [Google Scholar]
- 29.Knappik A., Liming Ge. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 30.Han D.P., Kim H.G., Kim Y.B., Poon L.L., Cho M.W. Development of a safe neutralization assay for SARS-CoV and characterization of S-glycoprotein. Virology. 2004;326:140–149. doi: 10.1016/j.virol.2004.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giroglou T., Cinatl J., Jr., Rabenau H., Drosten C., Schwalbe H., Doerr H.W., von Laer D. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J. Virol. 2004;78:9007–9015. doi: 10.1128/JVI.78.17.9007-9015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choy W.Y., Lin S.G., Chan P.K., Tam J.S., Lo Y.M., Chu I.M., Tsai S.N., Zhong M.Q., Fung K.P., Waye M.M., Tsui S.K., Ng K.O., Shan Z.X., Yang M., Wu Y.L., Lin Z.Y., Ngai S.M. Synthetic peptide studies on the severe acute respiratory syndrome (SARS) coronavirus spike glycoprotein: perspective for SARS vaccine development. Clin. Chem. 2004;50:1036–1042. doi: 10.1373/clinchem.2003.029801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H., Wang G., Li J., Nie Y., Shi X., Lian G., Wang W., Yin X., Zhao Y., Qu X., Ding M., Deng H. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J. Virol. 2004;78:6938–6945. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keng C.T., Zhang A., Shen S., Lip K.M., Fielding B.C., Tan T.H., Chou C.F., Loh C.B., Wang S., Fu J., Yang X., Lim S.G., Hong W., Tan Y.J. Amino acids 1055 to 1192 in the S2 region of severe acute respiratory syndrome coronavirus s protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. J. Virol. 2005;79:3289–3296. doi: 10.1128/JVI.79.6.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y.D., Li Y., Xu G.B., Dong X.Y., Yang X.A., Feng Z.R., Tian C., Chen W.F. Detection of antibodies against SARS-CoV in serum from SARS-infected donors with ELISA and Western blot. Clin. Immunol. 2004;113:145–150. doi: 10.1016/j.clim.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu M.W., Scott J.K., Fournier A., Talbot P.J. Characterization of murine coronavirus neutralization epitopes with phage-displayed peptides. Virology. 2000;271(1):182–196. doi: 10.1006/viro.2000.0310. [DOI] [PMC free article] [PubMed] [Google Scholar]


