Abstract
Recently, we reported a novel testis-specific sperm associated antigen 9 (SPAG9) protein, a new member of the JNK-interacting protein family, having a functional role in sperm–egg fusion [N. Jagadish, R. Rana, R. Selvi, D. Mishra, M. Garg, S. Yadav, J.C. Herr, K. Okumura, A. Hasegawa, K. Koyama, A. Suri, Biochem. J. 389 (2005) 73–82]. NCBI Blast searches revealed SPAG9 nucleotide sequence similarities with ESTs of various cancerous tissues. In the present study, we compared the efficiency of two independent SPAG9 specific small interfering RNA (siRNA) constructs, BS/U6/spag9 and BS/U6/spag9-I, to ablate the SPAG9 expression in mammalian cells. A positive correlation between the ratio of target gene versus siRNA and the suppression of SPAG9 expression was observed. Further, the cotransfection of BS/U6/spag9 with pcDNA-SPAG9 and pFlag-CMV2-JNK-3 resulted in specific suppression of SPAG9 without affecting JNK-3 expression. The present investigation will eventually extend the application of SPAG9 siRNA in in vivo targeting experiments that aim to define the SPAG9 functional genomics in tumor and reproductive biology.
Keywords: Green fluorescent protein, JNK-interacting protein, Short hairpin RNA, Sperm–egg interaction, Gene silencing
Sperm associated antigen 9 (SPAG9), a new member of JNK-interacting protein (JIP) family [1], involved in molecular interactions during sperm–egg fusion and MAPK signaling pathway [1], [2], [3], is conserved in human [4], baboon [5], and macaque [6]. MAPK interaction studies demonstrated that SPAG9 functions as a scaffolding protein exhibiting higher binding affinity to JNK3 and JNK2 compared to JNK1 [1], [2]. This interaction is interesting in view of the important regulatory role played by MAPKs in cell survival, proliferation, apoptosis, and tumor development [7]. The role of SPAG9 in MAPK signaling and its expression in various cancers such as esophageal adenocarcinoma [8] and Dermatofibrosarcoma protuberans (DFSP) [9] suggest its important function in cancer biology. In addition, SPAG9 sequence analysis revealed nucleotide sequence similarities with ESTs of various cancerous tissues and cancer cell lines. Recently, sperm protein 17 (Sp17) [10] and sperm c lysozyme-like protein 1 (SLLP1) expression [11] was also reported in various cancer tissues. Therefore, it may be interesting to study the siRNA-mediated inhibition of SPAG9 expression to address the possible eventual role of SPAG9 in tumor and reproductive biology.
RNA interference (RNAi) is known as a powerful tool for post-transcriptional gene silencing and expected to be involved in gene therapy strategies [12], [13], [14]. Double-stranded RNA (dsRNA), when introduced to cells, interferes with the expression of homologous genes, disrupting their normal function. In mammals, transient delivery of synthetic short interfering RNAs (siRNAs), which resemble the processed form of standard double-stranded RNAi trigger, is effective in silencing mammalian genes. Issues related to transfer efficiency and duration of the silencing effect, however, restrict the spectrum of applications of siRNAs in mammals. These shortcomings of siRNAs have been solved by the cellular expression of short hairpin RNAs (shRNAi) from DNA vectors. shRNAi are indistinguishable from siRNAs in terms of efficacy and mechanism but can be produced within cells from mammalian expression vectors. In this way, shRNAi expression makes possible the creation of continuous cell lines and transgenic animals in which suppression of a target gene is stably maintained by RNAi [15].
Although a few sperm proteins involved in sperm–egg interaction have been described [16], the precise roles of these proteins have not been established. Though mature sperms are transcriptionally inactive as a consequence of nuclear condensation, the SPAG9 protein is incorporated into spermatozoa and is involved in sperm–egg interaction [1]. In this study, we present data showing the relative effectiveness of two independent SPAG9-specific small interfering RNA constructs in the ablation of SPAG9 expression. The present investigation is beneficial in terms of providing preliminary data and proof of concept for future lentiviral vector-based siRNA approaches in vivo to study the effect of phenotypic changes in sperm as well as for the functional genomics of SPAG9.
Materials and methods
Cloning of chimeric construct expressing SPAG9 fused with green fluorescent (GFP) reporter protein. To generate the chimeric construct of SPAG9, a cDNA encoding complete open reading frame (ORF) of SPAG9 (comparable to 111–2410 bp of the published SPAG9, amino acid residues from 1 to 766) was amplified using forward 5′-ATGTCCATAATTATATGGAACATTTA-3′ and reverse 5′-TAAGTTGATGACCCATTATTAACCA-3′ primers, and cloned in pEGFPN2 vector. Briefly, the PCR product (which contains XhoI site at the 5′ end and a BamHI site at the 3′ end of the sense strand) was digested with XhoI and BamHI. The XhoI/BamHI fragment was inserted in-frame into XhoI/BamHI-digested pEGFPN2 plasmid (Clontech, USA) containing multiple cloning sites to obtain SPAG9-GFP. The nucleotide sequence of the constructs was confirmed by automated DNA sequencing.
Construction of small interfering RNA plasmid vector. The SPAG9-specific siRNA constructs were designed to be homologous to SPAG9 mRNA (GenBank Accession No. X91879). We selected two siRNA constructs: (1) BS/U6/spag9; the two complementary oligonucleotides (5′-GGGCCCAGATCTCAGTGGATATAAA TTCAAGAGA TTTATATCCACTGAGATCTTTTTTGAATTC-3′ and 5′-GAATTCAAAAAAGATCTCAGTGGATATAAA TCTCTTGAA TTTATATCCACTGAGATCTGGGCCC-3′), (2) BS/U6/spag9-I; the two complementary oligonucleotides (5′-GGGCCCACAGCTCATAGTAGAATTA TTCAAGAGA TAATTCTACTATGAGCTGTTTTTTGAATTC-3′ and 5′-GAATTCAAAAAACAGCTCATAGTAGAATTA TCTCTTGAA TAATTCTACTATGAGCTGTGGGCCC-3′, the 19-nucleotide sense or antisense strands are in bold letters and stem loop sequences are in italics), annealed to generate double-stranded DNAs, and ligated into the linearized empty vector pBS/U6.The siRNA plasmids encoded the hairpin RNAs that specifically targeted SPAG9 mRNA and had no significant homology with other known genes. The nucleotide sequences of SPAG9 siRNAs (BS/U6/spag9 and BS/U6/spag9-I) were verified by automated DNA sequencing.
The plasmids BS/U6 and BS/U6/gfp RNAi were a gift from Dr. Yang Shi (Department of Pathology, Harvard Medical School, Boston, USA).
Cell culture and transfections. COS-1 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Life Technologies) in a humidified incubator (5% CO2) at 37 °C. Cells grown on coverslips in 35 mm petri plates were transfected by Lipofectamine procedure (Life Technologies) and harvested 48 h after transfection. For dose-dependent experiment, 0.5 μg of chimeric construct (SPAG9-GFP) was cotransfected with siRNAs using different concentrations ranging from 0.5 to 6 μg. To evaluate the specificity of BS/U6/spag9, 0.5 μg pcDNA-SPAG9 and pFlag-CMV2-JNK-3, respectively, were cotransfected with 6 μg BS/U6/spag9. A constant concentration of 10 μg was used for all the transfection studies employing empty vector (BS/U6) or BS/U6/gfp.
The MAPK expression vector, pFlag-CMV2-JNK-3, was a gift from Dr. Katsuji Yoshioka (Department of Molecular Pathology, Cancer Research Institute, Kanazawa University, Japan).
Immunofluorescence microscopy. Cells were harvested 48 h post-transfection and were processed for immunofluorescence assay as described earlier [1]. Briefly, the rat anti-SPAG9 and anti-FLAG monoclonal antibodies M5 (Sigma) were used as primary antibodies to probe SPAG9 and JNK-3, respectively. After washing, cells were probed with secondary antibodies goat anti-rat Texas red and anti-mouse FITC conjugate for SPAG9 and JNK-3, respectively, and were analyzed by fluorescence microscopy under ECLIPSE, E 400 Nikon microscope (Nikon, Fukok, Japan).
Western blotting. After 48 h transfection, cells were harvested, lysed in lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100), and centrifuged at 2000g for 10 min [1]. The supernatant was used for protein determination by the Bradford procedure (Bio-Rad) and Western blotting. SPAG9, chimeric (SPAG9-GFP), JNK-3, and GFP proteins were resolved on 10% and 15% SDS–polyacrylamide gels, respectively, transferred onto nitrocellulose membranes, and incubated with the appropriate antibodies. Expression of GFP protein was probed with anti-GFP HRP-conjugated antibody (Santa Cruz Biotechnology, CA, USA). Expressions of SPAG9 and SPAG9-GFP protein in cell lysates were probed with primary antibody using rat anti-SPAG9 antibody and subsequently with goat anti-rat IgG HRPO (Jackson Immunoresearch, West Grove, PA) as secondary antibody. JNK-3 protein was probed with anti-FLAG monoclonal antibody Bio-M5 (Sigma). The detection was performed with 0.05% 3,3′-diaminobenzidene (Sigma).
Flow cytometry analysis. Cells transfected with SPAG9-GFP in the presence or absence of BS/U6/spag9 (using a range of concentrations from 0.5 to 6 μg) were harvested by trypsinization after 48 h transfection and washed twice with PBS. After the final wash, cells were resuspended in PBS and analyzed by flow cytometer (BD-LSR model, Becton–Dickinson, San Jose, CA). Data acquisition and analysis were done using WinMDI (version 2.8) software.
Results
To evaluate siRNA approach for SPAG9 gene silencing, we identified two independent sets of siRNA target sequences within ORF region of SPAG9 according to the criteria used in earlier studies [17]. Briefly, we inserted DNA fragments that acted as template for the synthesis of small RNAs under the control of mouse U6 promoter that directs the synthesis of a Pol III-specific RNA transcripts. The resulting RNA is composed of a single RNA that forms a stem–loop structure in which the sense and antisense strands form the stem of the hairpin (Fig. 1 A). Termination of transcription at a stretch of thymidine bases is predicted to generate a 2–4 bp uridine-nucleotide overhang at the 3′ end, identical with the overhang that is normally produced by the Dicer enzyme.
Fig. 1.
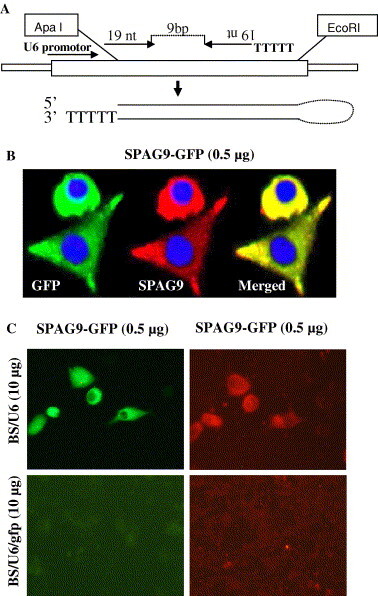
A siRNA synthesized from DNA template in vivo inhibited expression of transfected gene. (A) Strategy for generating siRNA from DNA template in vivo under the control of U6 promoter. The individual motif is 19-nt long and corresponds to the coding region of the gene of interest. The two motifs that form the inverted repeats are separated by a 9 bp. The transcriptional terminational signal of five thymidines (T) is added at the 3′ end of the inverted repeat. The resulting siRNA is predicted to fold back to form a hairpin dsRNA as shown (drawing not to scale). (B) Expression of chimeric SPAG9-GFP fusion protein in COS-1 cells. Fluorescence microscopy of COS-1 cells transfected with SPAG9-GFP: expression of chimeric GFP (the green) fused with SPAG9 (the red) in the same cell. The green and red fluorescence images were merged using Image Pro-Plus, version 5.1. DAPI for nuclear staining. (C) Inhibitory effect of gfp siRNA on SPAG9-GFP expression in COS-1 (original magnification 1000×). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
To investigate siRNA-mediated gene silencing, we first analyzed the effectiveness of gfp siRNA (BS/U6/gfp) in suppressing chimeric (SPAG9 fused with GFP) expression. COS-1 cells transfected with SPAG9-GFP revealed strong fluorescence after 48 h of transfection demonstrating GFP (green, Fig. 1B) as well as SPAG9 (red, Fig. 1B) expression in the same cells. Further, to demonstrate the inhibitory effect of BS/U6/gfp, SPAG9-GFP was cotransfected with the empty vector BS/U6 or BS/U6/gfp, respectively, with a ratio of 1:20 (target versus effector plasmids). The data indicated that empty vector BS/U6 had no effect on SPAG9-GFP expression (Fig. 1C), whereas BS/U6/gfp greatly reduced the expression of SPAG9-GFP to near background levels as shown in Fig. 1C.
Subsequently, two independent sets of SPAG9-specific siRNAs were evaluated for their effectiveness in inhibiting chimeric SPAG9 expression. A constant dose of 0.5 μg SPAG9-GFP was cotransfected with increasing concentrations of siRNAs ranging from 0.5 to 6.0 μg (Fig. 2 ). No appreciable difference in fluorescence intensity of the cells treated with 0.5 and 1.5 μg BS/U6/spag9 was observed under fluorescence microscope (Fig. 2A). In comparison, a higher dose of 3.0 μg BS/U6/spag9 resulted in diffused fluorescence in the treated cells indicating the inhibition of SPAG9-GFP expression (Fig. 2A). The strongest inhibition of SPAG9-GFP expression was observed with a concentration of 6 μg BS/U6/spag9 as demonstrated by abrogation of reporter (GFP) gene expression in the transfected cells (Fig. 2A). In these experiments, treatment with BS/U6/spag9 showed a greater impact than BS/U6/spag9-I on ablation of SPAG9 expression (data not shown). Hence, the subsequent experiments were restricted to siRNA BS/U6/spag9.
Fig. 2.
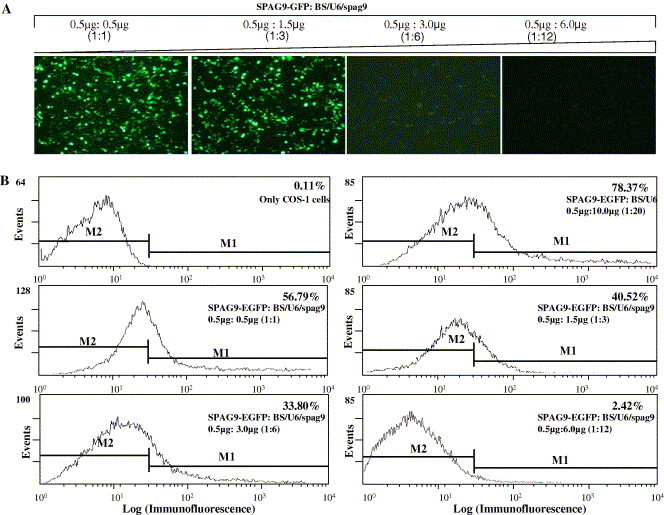
Experimental verification for screening effective siRNA-mediated gene silencing. COS-1 cells were cotransfected with SPAG9-GFP and increasing concentrations of BS/U6/spag9. At 48 h post-transfection, gene silencing was measured by quantitation of reporter gene (GFP) expression. (A) Fluorescence microscopy analysis of COS-1 cells expressing SPAG9-GFP in the presence of different concentrations of BS/U6/spag9 (original magnification 200×). (B) Quantitation of siRNA-mediated target gene silencing using fluorescence activated cell sorter analysis. M1 indicates the gating of GFP-positive cells, M2 the gating of GFP-negative cells. As apparent from the figures, the percentile of GFP expressing cells decreased with increase in the concentration of BS/U6/spag9 as indicated by the marker 1 (M1). The 6.0 μg BS/U6/spag9 concentration resulted in only 2.42% GFP expressing cells indicating the suppression of SPAG9-GFP expression using shRNAi approach.
A more accurate assessment of dose-dependent inhibition of chimeric SPAG9 expression was analyzed by fluorescence activated cell sorter (FACs) analysis. COS-1 cells were transfected with 0.5 μg SPAG9-GFP and increasing concentrations of BS/U6/spag9 ranging from 0.5 to 6.0 μg. The percentile fluorescence intensity in the treated cells is indicated by marker1 as compared to the control (only COS-1 cells), which is indicated by marker 2. The expression of SPAG9-GFP in the presence of empty vector BS/U6 revealed a fluorescence intensity of 78.37% as compared to 0.11% in the control cells (Fig. 2B). Further the fluorescence intensity of SPAG9-GFP was measured in the presence of different concentrations of BS/U6/spag9. The results indicated a positive correlation between target gene (SPAG9-GFP) versus effector siRNA (BS/U6/spag9) ratio and the suppression of SPAG9-GFP expression as shown in Fig. 2B. The fluorescence intensity decreased drastically to 2.42% in the cells treated with 6.0 μg of BS/U6/spag9 (Fig. 2B) as compared to 78.37% fluorescence intensity of SPAG9-GFP alone, indicating the effective and dose-dependent suppression of SPAG9-GFP expression using RNAi approach.
Our initial experiments established that BS/U6/spag9 is capable of inducing siRNA-mediated gene silencing, therefore we intended to assess the specificity of BS/U6/spag9. An independent experiment involving cotransfection of pcDNA-SPAG9 and pEGFPN2 with BS/U6 or BS/U6/gfp or BS/U6/spag9, respectively, was performed into COS-1 cells. Results indicated that BS/U6/gfp had no effect on SPAG9 expression (Fig. 3 C), whereas a reduced expression was observed in GFP expressing cells (Fig. 3D). In contrast, a reduction in SPAG9 (Fig. 3E), but not in GFP (Fig. 3F), expression by employing BS/U6/spag9 revealed the specific gene silencing of SPAG9 by BS/U6/spag9 and not by BS/U6/gfp. Yet another evidence for specific inhibition of target gene by BS/U6/spag9 was carried out using mammalian JNK-3 (c-Jun NH2-terminal Kinase-3) gene from MAPK signal transduction pathway. The cotransfection experiment using pcDNA-SPAG9 and pFLAG-CMV2-JNK3 with empty vector BS/U6 or BS/U6/spag9 or BS/U6/gfp revealed that BS/U6 or BS/U6/gfp did not inhibit the expression of either SPAG9 (Figs. 4 A and C) or JNK-3 protein (Figs. 4B and D). However, BS/U6/spag9 resulted in remarkable reduction in the expression of SPAG9 (Fig. 4E) without affecting JNK-3 expression (Fig. 4F). The results thereby demonstrate that the BS/U6/spag9-mediated specific gene silencing of SPAG9 without any effect on the expression of unrelated genes. Therefore, as previously published [18], our results confirm the efficiency and specificity of the siRNA silencing strategy.
Fig. 3.
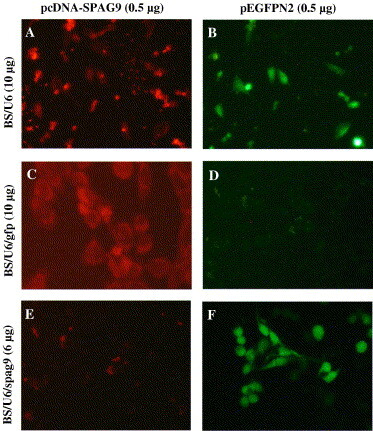
Fluorescence analysis of COS-1 cells cotransfected with plasmids expressing GFP and SPAG9 with BS/U6 (A,B) or BS/U6/gfp (C,D) or BS/U6/spag9 (E,F), respectively. Cells were analyzed after 48 h of transfection for SPAG9 (A) and GFP expression (B). BS/U6/gfp did not alter SPAG9 expression (C) BS/U6/gfp inhibited GFP expression (D). BS/U6/spag9 drastically reduced the number of SPAG9 expressing cells (E) without altering GFP expression (F) (original magnification 1000×).
Fig. 4.
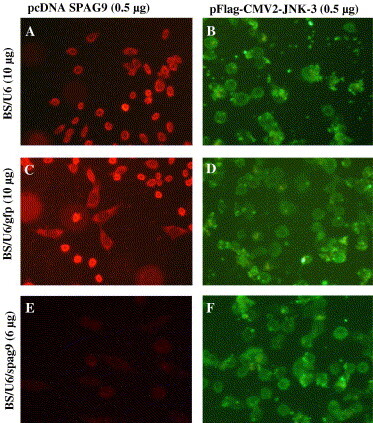
Specificity of siRNA-mediated target gene silencing. COS-1 cells were cotransfected with plasmids expressing SPAG9 and JNK-3 with BS/U6 (A,B) or BS/U6/gfp (C,D) or BS/U6/spag9 (E,F), respectively. Representative photomicrographs of cells expressing SPAG9 (A, C, and E; SPAG9 expression probed with anti-SPAG9 antibody) and JNK-3 (B, D, and F; JNK-3 expression probed with anti-FLAG monoclonal antibody). SPAG9 expression was drastically reduced in the BS/U6/spag9 treated cells (E), whereas normal JNK-3 expression was observed in all the treated cells (B, D, and F) (original magnification 1000×).
The inhibition of SPAG9-GFP expression at the protein level was also analyzed by Western blotting using anti-SPAG9 antibody. In a dose-dependent experiment, cells cotransfected with SPAG9-GFP and BS/U6/spag9 demonstrated a gradual decrease in the expression of SPAG9-GFP with the increasing concentrations of BS/U6/spag9 ranging from 0.5 to 6.0 μg (Fig. 5 A). Intriguingly, 6.0 μg pBS/U6/spag9 resulted in remarkable suppression of the SPAG9-GFP expression (Fig. 5A). In contrast, the treatment with BS/U6/spag9-I did not reveal decrease in the expression of SPAG9-GFP with increasing concentrations (Fig. 5A). To further strengthen the results, the expression of SPAG9-GFP was evaluated in the presence of BS/U6/gfp or BS/U6/spag9, respectively. No detectable expression of SPAG9-GFP was observed in either BS/U6/spag9 or BS/U6/gfp treated cells (Fig. 5B) as compared to the cells treated with empty vector BS/U6 (Fig. 5B).
Fig. 5.
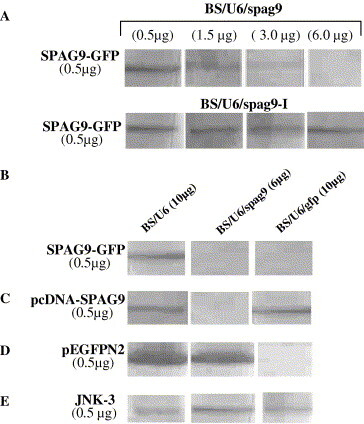
siRNA-mediated gene silencing in COS-1 cells. (A) Western blot analysis of COS-1 cells cotransfected with SPAG9-GFP with increasing concentrations of BS/U6/spag9 shows knockdown of SPAG9-GFP expression in a dose-dependent manner. In contrast, BS/U6/spag9-I treatment revealed no inhibition in SPAG9-GFP expression. (B) COS-1 cells cotransfected with SPAG9-GFP and BS/U6 or BS/U6/gfp or BS/U6/spag9 show no detectable chimeric protein bands in the presence of either BS/U6/gfp or BS/U6/spag9. (C) Western blot analysis of SPAG9 expression in cells cotransfected with either BS/U6 or BS/U6/spag9 or BS/U6/gfp. An appreciable inhibition of SPAG9 expression was observed with BS/U6/spag9, indicating that siRNA-mediated inhibition is gene-specific. (D) Cells transfected with pEGFPN2 together with BS/U6 or BS/U6/spag9 or BS/U6/gfp showed specific inhibition by BS/U6/gfp, whereas no effect was observed in the cells treated with BS/U6 and BS/U6/spag9. (E) Analysis of unrelated gene JNK-3 expression in the cells cotransfected with BS/U6 or BS/U6/spag9 or BS/U6/gfp revealed no effect on JNK-3 expression.
In addition, the specific knockdown of SPAG9 expression was investigated by cotransfecting 0.5 μg pcDNA-SPAG9 with BS/U6/spag9 (6 μg) or BS/U6/gfp (10 μg) or empty vector BS/U6 (10 μg) into COS-1 cells. In Western blot analysis, a drastic knockdown of SPAG9 expression was observed in BS/U6/spag9 treated cells (Fig. 5C), whereas the cells treated with BS/U6 or BS/U6/gfp revealed no inhibition in SPAG9 expression (Fig. 5C). Further, BS/U6/spag9 was examined for non-specific gene silencing using unrelated genes expressing GFP and JNK-3. The expression of GFP and JNK-3 was analyzed after transfecting COS-1 cells with pEGFPN2 and pFLAG-CMV2-JNK-3 plasmids with BS/U6/spag9 (6 μg) or BS/U6/gfp (10 μg) or empty vector BS/U6 (10 μg). The expression of GFP was remarkably reduced in BS/U6/gfp treated cells (Fig. 5D) when compared to cells treated with BS/U6 (Fig. 5D) or BS/U6/spag9 (Fig. 5D). However, there was no effect on JNK-3 expression in the presence of either BS/U6/gfp or BS/U6/spag9 (Fig. 5E). The overall results indicate that out of two independent sets of SPAG9 siRNAs, BS/U6/spag9 is capable of sequence-specific gene silencing in a dose-dependent manner without altering the expression of unrelated genes.
Discussion
The present investigation reports for the first time gene silencing of a sperm-specific protein using siRNA approach. SPAG9, a new member of JNK-interacting protein family, is a novel testis-specific protein exclusively expressed in testis [1], [19]. The amino acid sequence analysis revealed conservation of SPAG9 in human and non-human primates predicting functional interactions of proteins, as well as the evolutionary relationships between genomes [2], [4], [5], [6], [20]. MAPK interaction studies demonstrated that SPAG9 functions as a scaffolding protein exhibiting higher binding affinity to JNK3 and JNK2 compared to JNK1 [1], [2]. The interaction is important in view of the pleiotropic endpoints of JNK signaling pathways leading to cell proliferation, differentiation, apoptosis, immune cell function, and embryonic morphogenesis [21]. In most cases, the functional consequences of interaction between JNKs and scaffolding proteins are not yet fully understood; however, they are likely to play an important role in regulating JNK signaling pathways towards a particular physiological event. A recent study demonstrated that IB1/JIP1 facilitates the signal transduction in pancreatic β-cell lines mediated by the interacting proteins. IB1/JIP-1 interacts with JNK through the JNK binding domain (JBD), a domain able to prevent apoptosis of pancreatic β-cell lines induced by IL-1β and thus act as a crucial regulator of survival in insulin-secreting cells [22]. Similarly, SPAG9 interacts with JNKs through its JBD, which exhibits a significant sequence identity to JBD of JIP1, JIP2, and JIP3. The earlier study involving SPAG9 mutant lacking JBD (SPAG9ΔLZΔT) failed to show any interaction with JNK pathway, suggesting that JNK binding domain of SPAG9 is involved in JNK interaction [1]. Moreover, the expression of SPAG9 in various cancers such as esophageal adenocarcinoma [8] and Dermatofibrosarcoma protuberans (DFSP) [9] thereby suggests the possible involvement of SPAG9 interactions with JNKs in signal transduction pathways leading to cellular proliferation and tumor growth. Recently, sperm protein 17 (Sp17) [10] and sperm c lysozyme-like protein 1 (SLLP1) expression [11] was also reported in various cancer tissues. Though at present the exact nature of involvement of SPAG9 in tumorigenesis is not known, further investigations are warranted. The present study provides more evidence for the existence of RNAi induced gene silencing in mammalian cells and also suggests possibilities for using RNAi as an effective tool to determine the functional genomics of SPAG9 in tumor and reproductive biology.
Initially, we demonstrated the siRNA-mediated gene silencing of chimeric SPAG9-GFP using BS/U6/gfp, a construct, which has been already used and established for its siRNA function in mammalian cells by Sui and group [23]. A recent study reported the marked inhibition in the expression of chimera EGFP-tagged glucocerebrosidase (GBA) using GFP-directed siRNAs to evident the suppression of target (EGFP) as well as fused (GBA) protein expression [24]. Similarly, a successful knockdown of SPAG9-GFP expression was observed using GFP-directed siRNA, as indicated by immunofluorescence and Western blot analysis.
We further verified the gene silencing of SPAG9-GFP by BS/U6/spag9 using fluorescence activated cell sorter analysis. A dose-dependent inhibition of chimeric SPAG9 expression was successfully reported by the abrogation of reporter gene, i.e., GFP expression. The data from Western blot analysis also indicated that 6.0 μg BS/U6/spag9 was the most effective concentration of SPAG9 siRNA to mediate a marked inhibition of SPAG9-GFP expression. The results are in agreement with those of several other groups, that have developed vector-based siRNA expression systems that can induce dose-dependent RNAi effect in living cells such as inhibition of male-specific gynecophoral canal protein (SjGCP) [25], Bcl-2 expression [26], and SARS-CoV gene expression [27].
One more important aspect of siRNA studies is to explore the extent of siRNA specificity towards the gene of interest. siRNA-mediated gene silencing is generally believed to be highly sequence-specific. Tuschl and co-workers [18] demonstrated that even a single base mismatch between siRNA and its mRNA target abolished gene silencing. The caveats in specificity of gene silencing by RNAi mean there is an absolute requirement to test the selectivity of a siRNA before embarking on phenotype analysis. However, since RNAi, in contrast to the traditional knockout approach, does not completely eliminate the protein of interest, it was critical to evaluate the efficiency of siRNAs at the protein level. Therefore, we compared the silencing effect of two independent SPAG9 siRNAs (BS/U6/spag9 and BS/U6/spag9-I) on SPAG9 protein expression. The immunofluorescence and Western blotting results revealed that BS/U6/spag9 was fully competent to knockdown the SPAG9 expression. In contrast, SPAG9 expression was resistant to siRNA induced ablation using BS/U6/spag9-I. In addition, we evaluated the effect of BS/U6/spag9 on unrelated GFP and JNK3 proteins using SPAG9 as a positive control for determining the specificity of BS/U6/spag9. The expression of JNK-3 and GFP remained unaffected by BS/U6/spag9, suggesting that BS/U6/spag9 functions specifically to inhibit target gene and has no RNAi effect on unrelated gene expression.
The advent of RNAi has introduced a new tool for deciphering gene function by inducing post-transcriptional gene silencing. We hereby provide strong evidence that a DNA vector-based RNAi approach functions effectively in silencing SPAG9 gene expression in mammalian cells. The present study is beneficial in terms of providing proof of concept for further employment of a lentiviral vector-based siRNA approach in generating transgenic mice to study the functional genomics of SPAG9. Thus, the ease and convenience of deleting gene function combined with the ability to make transgenic animals using lentiviral vector-based RNAi would give a useful insight into exploring the biological function of SPAG9 in reproductive biology. Moreover, exploiting siRNA against SPAG9, which exhibits aberrant expression in various cancerous tissues, may give useful insight into the role of SPAG9 in cellular proliferation and programmed cell death and therefore warrants further investigations.
Acknowledgments
We thank Professor S.K. Basu, Director, National Institute of Immunology, for constant encouragement for this work. This work was supported by grants from the Department of Biotechnology, Government of India, Mellon foundation, and CONRAD, USA, Indo-US programme on CRHR.
References
- 1.Jagadish N., Rana R., Selvi R., Mishra D., Garg M., Yadav S., Herr J.C., Okumura K., Hasegawa A., Koyama K., Suri A. Characterization of a novel human sperm associated antigen 9 (SPAG9) having structural homology with c-Jun NH2-terminal Kinase interacting protein. Biochem. J. 2005;389:73–82. doi: 10.1042/BJ20041577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jagadish N., Rana R., Mishra D., Kumar M., Ramasamy S., Suri A. Sperm associated antigen 9 (SPAG9): a new member of c-Jun NH2-terminal kinase (JNK) interacting protein exclusively expressed in testis, Keio. J. Med. 2005;54:66–71. doi: 10.2302/kjm.54.66. [DOI] [PubMed] [Google Scholar]
- 3.N. Jagadish, R. Rana, D. Mishra, M. Garg, R. Selvi, A. Suri, Characterization of immune response in mice to plasmid DNA encoding human sperm associated antigen 9 (SPAG9), Vaccine (2005) (Epub ahead of print). [DOI] [PubMed]
- 4.Shankar S., Mohapatra B., Suri A. Cloning of a novel human testis mRNA specifically expressed in testicular haploid germ cells, having unique palindromic sequences and encoding a leucine zipper dimerization motif. Biochem. Biophys. Res. Commun. 1998;243:561–565. doi: 10.1006/bbrc.1997.7943. [DOI] [PubMed] [Google Scholar]
- 5.Shankar S., Mohapatra B., Verma S., Selvi R., Jagadish N., Suri A. Isolation and characterization of a haploid germ cell specific sperm associated antigen 9 (SPAG9) from the Baboon. Mol. Reprod. Dev. 2004;69:186–193. doi: 10.1002/mrd.20164. [DOI] [PubMed] [Google Scholar]
- 6.Jagadish N., Rana R., Selvi R., Mishra D., Suri A. Molecular cloning and characterization of a haploid germ cell specific sperm associated antigen 9 (SPAG9) from the Macaque. Mol. Reprod. Dev. 2005;71:58–66. doi: 10.1002/mrd.20245. [DOI] [PubMed] [Google Scholar]
- 7.Qi M., Elion E.A. MAP kinase pathways. J. Cell Sci. 2005;118:3569–3572. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- 8.Helm J., Enkemann S.A., Coppola D., Barthel J.S., Kelley S.T., Yeatman T.J. Dedifferentiation precedes invasion in the progression from Barrett’s metaplasia to esophageal adenocarcinoma. Clin. Cancer Res. 2005;11:2478–2485. doi: 10.1158/1078-0432.CCR-04-1280. [DOI] [PubMed] [Google Scholar]
- 9.Linn S.C., West R.B., Pollack J.R., Zhu S., Hernandez-Boussard T., Nielsen T.O., Rubin B.P., Patel R., Goldblum J.R., Siegmund D., Botstein D., Brown P.O., Gilks C.B., van de Rijn M. Gene expression patterns and gene copy number changes in dermatofibrosarcoma protuberans. Am. J. Pathol. 2003;163:2383–2395. doi: 10.1016/S0002-9440(10)63593-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiriva-Internati M., Grizzi F., Franceschini B., Hermonat P.L., Bright R.K., Bumm K., Dioguardi N., Kast W.M. Is sperm protein 17 a useful target for tumor immunotherapy? Blood. 2003;102:2308–2309. doi: 10.1182/blood-2003-05-1747. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z., Zhang Y., Mandal A., Zhang J., Giles F.J., Herr J.C., Lim S.H. The spermatozoa protein, SLLP1, is a novel cancer-testis antigen in hematologic malignancies. Clin. Cancer Res. 2004;10:6544–6550. doi: 10.1158/1078-0432.CCR-04-0911. [DOI] [PubMed] [Google Scholar]
- 12.Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 13.Hutvagner G., Zamore P.D. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 14.Dykxhoorn D.M., Novina C.D., Sharp P.A. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 15.Hannon G.J., Conklin D.S. RNA interference by short hairpin RNAs expressed in vertebrate cells. Methods Mol. Biol. 2004;257:255–266. doi: 10.1385/1-59259-750-5:255. [DOI] [PubMed] [Google Scholar]
- 16.Suri A. Sperm-based contraceptive vaccines: current status, merits and development. Exp. Rev. Mol. Med. 2005;7:1–16. doi: 10.1017/S1462399405009877. [DOI] [PubMed] [Google Scholar]
- 17.Elbashir S.M., Harborth J., Weber K., Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- 18.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 19.Jagadish N., Rana R., Mishra D., Garg M., Hasegawa A., Koyama K., Suri A. Immunogenicity and contraceptive potential of recombinant human sperm associated antigen (SPAG9) J. Reprod. Immunol. 2005;67:69–76. doi: 10.1016/j.jri.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Suri A. Contraceptive vaccines targeting sperm. Exp. Opin. Biol. Ther. 2005;5(3):381–392. doi: 10.1517/14712598.5.3.381. [DOI] [PubMed] [Google Scholar]
- 21.Davis R.J. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 22.Haefliger J.A., Tawadros T., Meylan L., Gurun S.L., Roehrich M.E., Martin D., Thorens B., Waeber G. The scaffolding protein IB1/JIP-1 is a critical mediator of cytokine-induced apoptosis in pancreatic β cells. J. Cell Sci. 2003;116:1463–1469. doi: 10.1242/jcs.00356. [DOI] [PubMed] [Google Scholar]
- 23.Sui G., Soohoo C., Affar E.B., Gay F., Shi Y., Forrester W.C., Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell T.N., Choy F.Y.M. Knockdown of chimeric glucocerebrosidase by green fluorescent protein-directed small interfering RNA. Genet. Mol. Res. 2004;3:282–287. [PubMed] [Google Scholar]
- 25.Cheng G., Lin J., Shi Y., Jin Y., Fu Z., Jin Y., Zhou Y., Cai Y. Dose-dependent inhibition of gynecophoral canal protein gene expression in vitro in the schistosome (Schistosoma japonicum) by RNA interference. Acta Biochim. Biophys. Sin. 2005;37:386–390. doi: 10.1111/j.1745-7270.2005.00058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao L., XU D., Wen L., Zhang X., Shao Y., Zhao X. Inhibition of STAT3 expression by siRNA suppresses growth and induces apoptosis in laryngeal cancer cells. Acta Pharmacol. Sin. 2005;26:377–383. doi: 10.1111/j.1745-7254.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 27.Peng T., Jun Z., Ni T., Bing-qiang Z., Tong-chuan H., Ai-long H. Potent and specific inhibition of SARS-CoV antigen expression by RNA interference. Chin. Med. J. 2005;118:714–719. [PubMed] [Google Scholar]


