Abstract
Enterovirus 71 (EV71) is associated with the severe hand foot and mouth disease (HFMD) outcomes, however the host-virus interaction mechanism and the pathogenesis remain poorly understood. Long non-coding RNAs (lncRNAs) are involved in variety physiological and pathological processes, but the functions of lncRNAs in EV71 infection remain elusive. Here we profiled the expression of lncRNAs in peripheral blood mononuclear cells (PBMCs) from EV71-infected mild patients, severe patients as well as the healthy controls, and identified 8541 lncRNAs were differentially expressed. Focused on the dynamic changed lncRNAs, we performed systematic bioinformatics analysis with Series Test of Cluster (STC) algorithm, Gene Ontology (GO) analysis, pathway analysis and lncRNA-mRNA co-expression network analysis, and revealed the potential functions and related pathways of these lncRNAs were associated with immunity and inflammation during the clinical process of EV71-infected HFMD. Among the significant dynamic changed lncRNAs, ten lncRNAs were screened whose expression were further validated in EV71-infected mild patients, severe patients and healthy control. These results shed light on the potential roles of lncRNAs in EV71-infected HFMD, especially in distinguishing the mild and severe cases for early diagnose and treatment, moreover, provide deeper insight into the mechanism of EV71-induced immune and inflammatory responses, as well as the pathogenesis of the imbalanced inflammation in severe EV71 infection.
Keywords: Enterovirus 71, Long non-coding RNA, Hand foot and mouth disease, Inflammation, Innate immunity
Highlights
-
•
LncRNAs expression profile were analyzed with EV71-infected HFMD patients' PBMCs.
-
•
Dynamic changed lncRNAs were analyzed in different severity EV71-infected patients.
-
•
Most of the lncRNAs were related to innate immune and inflammatory responses.
-
•
The candidate lncRNAs may serve as potential markers for EV71-induced severe HFMD.
1. Introduction
Enterovirus 71 is well known as one of the major causative agents of Hand, foot and mouth disease (HFMD), which usually initiates self-limiting acute febrile disease followed by papulovesicular rashes on the buccal mucosa and palms, soles and buttocks [1]. However, EV71 infection is also associated with severe neurological manifestations, including aseptic meningitis, acute flaccid paralysis and brainstem encephalitis, resulting in autonomic dysregulation, fulminant pulmonary edema, myocardial dysfunction, shock and severe sequelae, even death [2]. Accumulating evidences propose that the mutant of the virus, as well as the age, immune status and genetic profiles of the host may be involved in the clinical phenotypes exhibited by EV71-infected individuals [3], nevertheless, what determines the clinical outcome of EV71-induced HFMD is poorly understood.
The long non-coding RNAs (lncRNAs) are non-protein-coding transcripts at least 200 nucleotides, which play versatile roles in diverse physiological and pathological processes [4]. In 2009, the potential function of lncRNAs in innate immunity were identified for the fist time [5]. Since then, increasing number of lncRNAs have been identified to participated in innate immune system and the related immune and inflammatory responses, such as lincRNA-COX2 [5], Lethe [6], PACER [7] and lnc-DC [8]. The innate immune system is the first defense line of the host to recognize and clear the invasion pathogens. The tightly regulation of innate immune system is closely associated with the elimination efficiency of pathogen infection. Although, many lncRNAs have been uncovered exhibiting flexible manners in regulation of innate immunity, the function of most of lncRNAs are still unknown and need to be discovery.
Application of genome-sequence and microarray technologies have facilitated the deciphering of dramatic changes in the host transcriptome upon virus infection, and the function of lncRNAs in host-virus interaction attract more attentions [9]. Virus-inducible non-coding RNA (VINC) is the first reported viral infection related lncRNA [10]. There were widespread differential regulation of lncRNAs during SARS-CoV infection [11]. Moreover, the lncRNAs have distinctive kinetic expression profiles in type I interferon receptor and STAT1 knockout mice infected with SARS-CoV, including unique signatures of lncRNAs expression associated with lethal infection [11]. The expression of lncRNA Tmevpg1 (also known as NeST) was increased and essential for the host persistence of Theiler's virus [12]. In EV71-infected Rhabdomyosarcoma (RD) cells, more than 4800 lncRNAs differentially expressed [13], however, whether lncRNAs play essential roles in host-EV71 interaction is still lack in vivo and clinical evidences, which is needed further exploration.
Herein, we identified there were thousands of lncRNAs differently expressed in EV71-infected HFMD mild and severe patients and the healthy controls, and focused on the dynamic changed lncRNAs to performed the systematic bioinformatics analysis with GO analysis, pathway analysis as well as the lncRNA-mRNA co-expression network analysis, to reveal the potential roles of lncRNAs in EV71-infection.
2. Materials and methods
2.1. Study design and populations
In this case-control study, peripheral blood samples were obtained from 42 HFMD patients and 20 healthy controls in Shenzhen Children's Hospital, Shenzhen Baoan District People's Hospital and Shajing Institution of Disease Prevention and Healthcare from 2015 to 2016. All of the patients were confirmed as EV71 infection, through EV71 isolation and sequence identification with clinical samples, such as stool, rectal and throat swabs. Meanwhile, the patients were diagnosed with HFMD according to the WHO Guide to Clinical Management and Public Health Response for HFMD, and were further divided into 20 mild and 22 severe patients. Healthy candidates were age sex matched with the patients, and were identified without EV71 infection and other infection disease. All of the participants were informed consent and the study was approved by the Ethics Committee of the Shenzhen Center for Disease Control and Prevention.
2.2. RNA isolation
The peripheral blood mononuclear cells (PBMCs) were isolated from the blood samples by Ficoll paque peremium (GE Healthcare Life Science) within 2 h. The total RNA was extracted from the PBMCs using Trizol reagent (Invitrogen Corporation) and was validated with Agilent Array platform for microarray assay or store at −80 °C.
2.3. Microarray analysis
The microarray analysis was performed with the Affymetrix Human Transcriptome Array 2.0 (HTA 2.0) (Affymetrix), which contains more than 40,000 non-coding and 245,000 coding transcripts in human genome. Each transcript cloud be identified precisely by specific exon or exon-exon splice junction probes. The transcripts with P < 0.05 were selected, after significant and false discovery rate (FDR) analysis [14], [15], [16]. The microarray assay and bioinformatics analysis were performed by Gminix Biotechnology Company (Shanghai, China).
2.4. STC analysis
STC (Series Test of Cluster) were employed to study the gene expression dynamics profiles and to determine the profiles containing significant higher number of genes, revealing the change rule of gene expression [17]. We selected differentially expressed genes with randomized variance model corrected ANOVA. Profiles that are significant have higher probability than expected by Fisher's exact test and multiple comparison tests [18], [19].
2.5. GO analysis and pathway analysis
Gene Ontology (GO) analysis was used to explore the function of differentially expressed genes, and to assign the genes to biological processes GO terms according to the annotations [20]. The pathway analysis was based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to determine the significant pathway of the differentially expressed genes [20]. The GO terms and the KEGG pathways that had P < 0.05 were chosen.
2.6. Construction of the lncRNA-mRNA co-expression network
The lncRNA-mRNA co-expression network was constructed to clarify the interaction between differentially expressed lncRNAs and mRNAs in Profile9. Each lncRNA-mRNA pair was analyzed with Pearson correlation to choose the significant pairs for co-expression network construction [21], [22]. The lncRNA-mRNA pairs which with correlation coefficient significant more than 0.95 were selected.
2.7. Quantitative real-time PCR (qRT-PCR)
The total RNA was extracted with Trizol reagent (Invitrogen Corporation). Reverse Transcription was performed using One Step RT-PCR Kit (Takara). A lightCycler (Roche) and SYBR Quantitative real-time PCR kit (Takara) were employed for Q-PCR as described previously [23]. U6 served as the endogenous control. The primer sequences were presented in Table S1.
2.8. Statistical analysis
All statistical analysis were performed using SPSS version 17.0 software. ANOVA was used for multiple comparisons. P-values <0.05 were considered statistically significant.
3. Results
3.1. Genome-wide lncRNAs change in mild and severe EV71-infected HFMD patients and healthy control
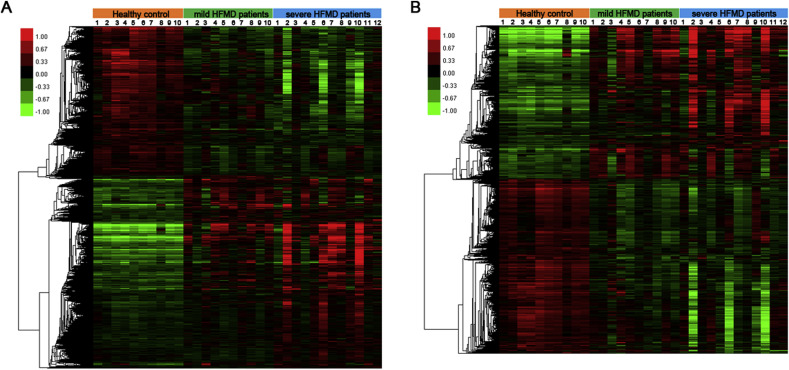
To determine the differential regulation of lncRNAs after EV71 infection, we analyzed the expression profiles of lncRNAs and mRNAs in peripheral blood mononuclear cells (PBMCs) derived from 10 mild HFMD patients, 12 severe HFMD patients and 10 healthy candidates, which were age (34.10 ± 17.10, 33.33 ± 13.81, 35.90 ± 11.41, P = 0.91) and gender (male, 5(10), 6(12), 5(10)) matched. A differential expression profile of each group was obtained with comparison between healthy control, mild patients and severe patients group, which showed that the expression of 8541 lncRNAs were significantly changed (P < 0.05) and 5955 mRNAs were significantly changed (P < 0.05) (Tables S2 and S3). The Hierarchical clustering analysis demonstrated general variations in lncRNAs and mRNAs expression in the PBMCs of EV71-infected HFMD patients (Fig. 1 A and B). These data indicate that the expression of large amount of lncRNAs are differently regulated in mild and severe EV71-infected HFMD patients.
Fig. 1.
Hierarchical clustering analysis of differentially expressed genes in healthy control, mild and severe HFMD patients. (A and B) Differentially expressed lncRNAs (A) and mRNAs (B) in healthy control (n = 10), mild HFMD patients (n = 10) and severe HFMD patients (n = 12). Red and green color indicates up-regulated and down-regulated transcripts, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Distinctive dynamic expression profiles of lncRNAs in mild and severe EV71-infected HFMD patients and healthy control
We utilized the STC algorithm to further investigate the expression change pattern of the lncRNAs in healthy control and patients with mild or severe clinical HFMD symptoms. The different expressed lncRNAs were placed into 16 model pattern profiles by STC analysis, and there were 8 significant profiles, respectively containing differential expressed lncRNAs with the similar trend change characteristics, the first 4 significant profiles were Profile9, profile11, Profile15 and Profile14 (Fig. 2 A). The Profile9 and Profile15 contained 1128 lncRNAs and 1362 lncRNAs respectively, which exhibited constantly expression up-regulation trend in healthy control, mild and severe HFMD patients (Table S4 and Fig. 2A and B). The Profile11 and Profile14 were constructed with 993 lncRNAs and 1153 lncRNAs respectively, which shown constantly down-regulated expression trend in healthy control, mild and severe HFMD patients (Table S4 and Fig. 2A and B). The different expressed mRNAs in each profiles were also shown (Fig. 2A).
Fig. 2.
STC analysis of distinctive dynamic expression profiles of lncRNAs and mRNAs in severe and mild EV71-infected HFMD patients and healthy control. (A) The differentially expressed lncRNAs and mRNAs are group into 16 model pattern profiles, and 8 significantly different profiles are identified and the change trend of these profile are shown in the red box. (B) In the first 4 significant profiles, the expression change of lncRNAs in severe and mild HFMD patients and healthy control are shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. GO analysis and KEGG pathway analysis
To further identify the functions of these dynamic expressed lncRNAs in Profile9, profile11, Profile15 and Profile14 upon EV71 infection, we took the mRNAs in the corresponding profile as input and performed the GO analysis and KEGG pathway analysis. In Profile9 and Profile15, the top 15 significant GO terms were most related to inflammatory response (GO:0006954) and innate immune response (GO:0045087) (Fig. 3 A and C and Table S5). Additionally, in Profile11 and Profile14, the top 15 significant GO terms were related to transcription and regulation of transcription (GO:0006351, 0006355 and 0045892) (Fig. 3B and D and Table S5).
Fig. 3.
GO enrichment analysis and KEGG pathway analysis of differentially expressed mRNAs. (A–D) The GO analysis of mRNAs in Profile 9 according to biological process, the horizontal axis represents –lg (P value) of the GO category. (E–H) The KEGG pathway analysis of mRNAs in Profile 9, the horizontal axis represents –lg (P value) of the pathway terms.
The KEGG pathways analysis revealed various of enrichment-related pathways, in which the lncRNAs and mRNAs derived from Profile9, Profile11, Profile 15 and Profile14 could be involved (Fig. 3E–H and Table S6). Notably, the pathways significantly enriched in Profile9 were most correlated with the pathogen infection and the innate immune signaling pathway, such as the MAPK signaling pathway (path id: 04010) and Toll-like receptor signaling pathway (path id: 04620) which play crucial roles in inflammatory response and innate immune response, moreover, the pathways significantly enriched in the other 3 profiles were also correlation with the pathogen infection and metabolic pathways.
3.4. LncRNA-mRNA co-expression network
According to the GO and pathway analysis, we took lncRNAs and mRNAs in the most significant STC Profile9 for the further investigation. For high-throughout analysis of the interaction between lncRNAs and mRNAs, we constructed the lncRNA-mRNA co-expression networks. The lncRNA-mRNA pairs were selected to construct the networks when the Pearson correlation coefficient were significant more than 0.95, and the regulation relationship between each pair of lncRNA-mRNA were also shown (Fig. 4 A and Table S7). The network was consist of 92 lncRNAs and 61 correlated mRNAs with 153 network nodes and 380 connection edges (Fig. 4A).
Fig. 4.
Co-expression network of lncRNAs-mRNAs and validation of candidate lncRNAs. (A) In Profile9, the network is consist of 92 lncRNAs and 61 correlated mRNAs with 153 network nodes and 380 connection edges, the purple node denote mRNA and the purple node around by yellow denote lncRNA. (B) Q-PCR analysis of 10 candidate lncRNAs in severe HFMD patients (n = 22), mild HFMD patients (n = 20) and healthy control (n = 20). Data are shown as mean ± S.D., **P < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. Confirmation of candidate lncRNAs expression level in mild and severe EV71-infected patients and healthy control
To identify the lncRNAs specifically involved in the EV71-induced immune and inflammation responses, we focused on the differentially expressed lncRNAs which took part in the co-expression network with immunity and inflammation related mRNAs in Profile9. There were 10 lncRNAs were screened out, n410673, n408005, n409323, TCONS_l2_00011393-XLOC_l2_006157, n406645, TCONS_00003079-XLOC_001808, TCONS_00022787-XLOC_011068, n410532, n345507 and n410510. We validated the expression level of the 10 lncRNAs in PBMCs of 20 healthy control, 20 mild and 22 severe HFMD patients which were age (35.40 ± 10.75, 33.15 ± 18.39, 31.11 ± 16.44, P = 0.67) and gender (male, 10 (20), 10 (20), 11 (22)) matched, and got the expression trends of these 10 lncRNAs as same as the lncRNA microarray (Fig. 4B).
4. Discussion
EV71 is the most causative virus leading to fatal HFMD in infants or children younger than 5 years old [24]. EV71-infected HFMD patients exhibit high level of cytokines and chemokines in serum or cerebrospinal fluid, especially the severe case always turn into dysregulation of immune response and inflammatory response, result in local or system complication even death [1]. The underlying mechanism about the abnormal regulation of EV71-induced immune and inflammatory state attract increasing concerns, and most of the researches are focus on the coding genes. Actually, non-coding RNAs such as microRNA and lncRNA have been revealed having variety of function in physiological and pathological processes. Although lncRNAs have been clarified functioning after many virus infection, while little was known in EV71-infected HFMD. In EV71-infected RD cells, 4800 lncRNAs were differentially expressed, 160 of the lncRNAs regulated enhancer-like lncRNA and mRNA pairs nearby the lncRNAs [13]. The study pointed out the potential roles of lncRNAs in the host response to EV71 infection, however, still need the clinical proof to confirm the putative function of EV71 infection-related lncRNAs. In our research, we investigated the differentially expressed lncRNAs in EV71-infected mild and severe HFMD patients and healthy control, and found 8541 lncRNAs were significantly changed, indicating that lncRNAs were involved in EV71 infection.
EV71 usually induces self-limiting HFMD, meanwhile, it is the most causative pathogen that lead to severe inflammation and neurological complications [25], however, what defines the clinical outcomes of EV71-induced HFMD remain elusive. In our research, we isolated and sequenced the EV71 RNA, and found the EV71-isolated from either mild or severe patients were all assigned to subgenotype C4a and with no sequence difference after sequence alignment (data not shown). These data indicated that the mild or severe clinical complications of the EV71-infected patients might not due to the virus mutant. The genes which dynamic change in response to the stimulations or accompany with the progression of diseases, may have crucial biological functions. Through STC analysis, the lncRNAs dynamic expression profiles were obtained at various time points after 2/3 partial hepatectomy, moreover, lncRNA-LALR1 was significantly increased in a time-dependent manner and was essential for the hepatocyte proliferation [26]. In the lung tissue of H1N1-infected mice, a total of 82 miRNA expression were significantly different from the control mice, and the dynamically expressed 17 miRNAs were confirmed more significant and related to the influenza A pathway through STC and further investigation [27]. We focused on the dynamic genetic profile after EV71 infection and screened the 8 significant dynamic expression profiles of lncRNAs using STC analysis. The first 4 significant profiles were Profile9, profile11, Profile15 and Profile14 each contained thousands of lncRNAs which constantly up or down regulated in healthy control, mild and severe HFMD patients. In the most significant Profile9, 10 lncRNAs were identified associated with the inflammation and innate immune response upon EV71 infection. The lncRNAs with dynamic expression change may be more correlated with the severity of the HFMD clinical symptoms, and could play crucial roles in the regulation of EV71-induced immune and inflammatory responses.
Since lncRNAs were reported having a diverse range of roles in innate immune system in 2009 [5], accumulating evidences reveal that lncRNAs are important in regulation of innate immune and inflammatory responses during host-virus interaction [9]. In lung tissue of SARS-CoV-infected mouse, the expression of lncRNAs widespread changed when compared with the control, and the similar changes were also proved during influenza virus infection, revealing that a common lncRNAs-based characterize of host response to respiratory viral infection [11]. After influenza virus or herpes simplex virus (HSV) infection, lncRNA NEAT1 (nuclear paraspeckle assembly transcript 1) expression was increased [28]. Then NEAT1 interacted with SFPQ (Splicing factor proline/glutamine-rich) and mediated SFPQ leaving from the promoter of IL-8, leading to activation of IL-8 in TLR3-p38 signaling pathway [28]. Moreover, upon HIV-1 infection, NEAT1 expression was up-regulated to inhibit HIV-1 replication [29]. On the other hand, viruses also can hijack lncRNAs for virus replication and viral protein synthesis benefiting for their own life process and suppressing the antiviral immune responses [30]. In response to the influenza A virus (IAV), virus inducible lincRNA (VIN) were up-regulated, which was also investigated in after H1N1, H3N2, H7N7 as well as vesicular stomatitis virus (VSV) [31]. The further function abolition of VIN leaded to the restriction of IAV replication and viral protein synthesis, highlighting the relevance of VIN in IAV virulence and virus pathogenesis [31]. We identified and confirmed 10 lncRNAs which were differentially expressed in EV71-infected mild patients, severe patients and healthy control. Among the 10 lncRNAs, the expression of n409323 was positive correlation with CARD8 expression level, and CARD8 may be involved in the negative regulation of IκB/NF-κB cascade upon EV71 infection, indicating that n409323 may play some roles in IκB/NF-κB signaling pathway regulation. Notably, the IκB/NF-κB cascade is one of most critical pathways for the production of cytokines such as IL-6 and TNF-α in the anti-viral innate immune responses [32]. The other screened lncRNAs were associated with innate immune and inflammatory responses according to the GO and pathway analysis, however, the function and the regulation mechanism of these lncRNAs still need further exploration.
Taken together, our research first exhibited the lncRNAs expression profile in EV71-infection clinical specimens, especially with the mild and severe EV71-infected HFMD patients. Moreover, we analyzed the lncRNAs dynamic differentially expressed profile revealing the potential crucial roles of lncRNAs in determinant of EV71-induced HFMD clinical manifestation progress. The screened 10 lncRNAs, that were correlated with the innate immune and inflammatory responses, suggested the pathogenesis of imbalanced inflammation during EV71 infection, and may severe as the candidate direction for intervention of severe EV71-infected cases.
Acknowledgments
We thank Gminix company (Shanghai, China) for bioinformatics support. This work was supported by Grants from the National Natural Science Foundation of China (81601386 and 81600652), Science and Technology Research Projects of Shenzhen (JCYJ20160428142411948 and JCYJ20160429190911895) and China Postdoctoral Science Foundation funded project (2016M590818).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bbrc.2017.09.141.
Transparency document related to this article can be found online at https://doi.org/10.1016/j.bbrc.2017.09.141.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Transparency document
References
- 1.Lei X.B., Cui S., Zhao Z.D., Wang J.W. Etiology, pathogenesis, antivirals and vaccines of hand,foot, and mouth disease. Natl. Sci. Rev. 2015;2:268–284. [Google Scholar]
- 2.Ho M., Chen E.R., Hsu K.H., Twu S.J., Chen K.T., Tsai S.F., Wang J.R., Shih S.R. An epidemic of enterovirus 71 infection in Taiwan. Taiwan enterovirus epidemic working group. N. Engl. J. Med. 1999;341:929–935. doi: 10.1056/NEJM199909233411301. [DOI] [PubMed] [Google Scholar]
- 3.Huang C.C., Liu C.C., Chang Y.C., Chen C.Y., Wang S.T., Yeh T.F. Neurologic complications in children with enterovirus 71 infection. N. Engl. J. Med. 1999;341:936–942. doi: 10.1056/NEJM199909233411302. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y.G., Satpathy A.T., Chang H.Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 2017;18:962–972. doi: 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P., Cabili M.N., Jaenisch R., Mikkelsen T.S., Jacks T., Hacohen N., Bernstein B.E., Kellis M., Regev A., Rinn J.L., Lander E.S. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rapicavoli N.A., Qu K., Zhang J., Mikhail M., Laberge R.M., Chang H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krawczyk M., Emerson B.M. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. Elife. 2014;3:e01776. doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S., Jiang Z., Xu J., Liu Q., Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 9.Atianand M.K., Caffrey D.R., Fitzgerald K.A. Immunobiology of long noncoding RNAs. Annu. Rev. Immunol. 2017;35:177–198. doi: 10.1146/annurev-immunol-041015-055459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saha S., Murthy S., Rangarajan P.N. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J. Gen. Virol. 2006;87:1991–1995. doi: 10.1099/vir.0.81768-0. [DOI] [PubMed] [Google Scholar]
- 11.Peng X., Gralinski L., Armour C.D., Ferris M.T., Thomas M.J., Proll S., Bradel-Tretheway B.G., Korth M.J., Castle J.C., Biery M.C., Bouzek H.K., Haynor D.R., Frieman M.B., Heise M., Raymond C.K., Baric R.S., Katze M.G. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1 doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vigneau S., Rohrlich P.S., Brahic M., Bureau J.F. Tmevpg1, a candidate gene for the control of Theiler's virus persistence, could be implicated in the regulation of gamma interferon. J. Virol. 2003;77:5632–5638. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin Z., Guan D., Fan Q., Su J., Zheng W., Ma W., Ke C. lncRNA expression signatures in response to enterovirus 71 infection. Biochem. Biophys. Res. Commun. 2013;430:629–633. doi: 10.1016/j.bbrc.2012.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright G.W., Simon R.M. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 15.Yang H., Crawford N., Lukes L., Finney R., Lancaster M., Hunter K.W. Metastasis predictive signature profiles pre-exist in normal tissues. Clin. Exp. Metastasis. 2005;22:593–603. doi: 10.1007/s10585-005-6244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke R., Ressom H.W., Wang A., Xuan J., Liu M.C., Gehan E.A., Wang Y. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nat. Rev. Cancer. 2008;8:37–49. doi: 10.1038/nrc2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao S., Mo D., Wang Q., Jia J., Qin L., Yu X., Niu Y., Zhao X., Liu X., Chen Y. Aberrant host immune response induced by highly virulent PRRSV identified by digital gene expression tag profiling. BMC Genomics. 2010;11:544. doi: 10.1186/1471-2164-11-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller L.D., Long P.M., Wong L., Mukherjee S., McShane L.M., Liu E.T. Optimal gene expression analysis by microarrays. Cancer Cell. 2002;2:353–361. doi: 10.1016/s1535-6108(02)00181-2. [DOI] [PubMed] [Google Scholar]
- 19.Ramoni M.F., Sebastiani P., Kohane I.S. Cluster analysis of gene expression dynamics. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9121–9126. doi: 10.1073/pnas.132656399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pujana M.A., Han J.D., Starita L.M., Stevens K.N., Tewari M., Ahn J.S., Rennert G., Moreno V., Kirchhoff T., Gold B., Assmann V., Elshamy W.M., Rual J.F., Levine D., Rozek L.S., Gelman R.S., Gunsalus K.C., Greenberg R.A., Sobhian B., Bertin N., Venkatesan K., Ayivi-Guedehoussou N., Sole X., Hernandez P., Lazaro C., Nathanson K.L., Weber B.L., Cusick M.E., Hill D.E., Offit K., Livingston D.M., Gruber S.B., Parvin J.D., Vidal M. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 22.Prieto C., Risueno A., Fontanillo C., De las Rivas J. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. PLoS One. 2008;3:e3911. doi: 10.1371/journal.pone.0003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng J., Liu X., Zhang P., Li D., Xu S., Zhou Q., Guo M., Huai W., Chen X., Wang Q., Li N., Cao X. Rb selectively inhibits innate IFN-beta production by enhancing deacetylation of IFN-beta promoter through HDAC1 and HDAC8. J. Autoimmun. 2016;73:42–53. doi: 10.1016/j.jaut.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Solomon T., Lewthwaite P., Perera D., Cardosa M.J., McMinn P., Ooi M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 2010;10:778–790. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- 25.Xing W., Liao Q., Viboud C., Zhang J., Sun J., Wu J.T., Chang Z., Liu F., Fang V.J., Zheng Y., Cowling B.J., Varma J.K., Farrar J.J., Leung G.M., Yu H. Hand, foot, and mouth disease in China, 2008-12: an epidemiological study. Lancet Infect. Dis. 2014;14:308–318. doi: 10.1016/S1473-3099(13)70342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu D., Yang F., Yuan J.H., Zhang L., Bi H.S., Zhou C.C., Liu F., Wang F., Sun S.H. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta-catenin signaling. Hepatology. 2013;58:739–751. doi: 10.1002/hep.26361. [DOI] [PubMed] [Google Scholar]
- 27.Bao Y., Gao Y., Jin Y., Cong W., Pan X., Cui X. MicroRNA expression profiles and networks in mouse lung infected with H1N1 influenza virus. Mol. Genet. Genomics. 2015;290:1885–1897. doi: 10.1007/s00438-015-1047-1. [DOI] [PubMed] [Google Scholar]
- 28.Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., Kai C., Yada T., Suzuki Y., Yamada T., Ozawa T., Kaneki K., Inoue T., Kobayashi M., Kodama T., Wada Y., Sekimizu K., Akimitsu N. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q., Chen C.Y., Yedavalli V.S., Jeang K.T. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio. 2013;4 doi: 10.1128/mBio.00596-12. e00596–00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Cao X. Long noncoding RNAs in innate immunity. Cell Mol. Immunol. 2016;13:138–147. doi: 10.1038/cmi.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winterling C., Koch M., Koeppel M., Garcia-Alcalde F., Karlas A., Meyer T.F. Evidence for a crucial role of a host non-coding RNA in influenza A virus replication. RNA Biol. 2014;11:66–75. doi: 10.4161/rna.27504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 2016;16:35–50. doi: 10.1038/nri.2015.8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.