Abstract
Eukaryotic cellular and most viral RNAs carry a 5′-terminal cap structure, a 5′-5′ triphosphate linkage between the 5′ end of the RNA and a guanosine nucleotide (cap-0). SARS coronavirus (SARS-CoV) nonstructural protein nsp16 functions as a methyltransferase, to methylate mRNA cap-0 structure at the ribose 2′-O position of the first nucleotide to form cap-1 structures. However, whether there is interplay between nsp16 and host proteins was not yet clear. In this report, we identified several potential cellular nsp16-interacting proteins from a human thymus cDNA library by yeast two-hybrid screening. VHL, one of these proteins, was proven to interact with nsp16 both in vitro and in vivo. Further studies showed that VHL can inhibit SARS-CoV replication by regulating nsp16 ubiquitination and promoting its degradation. Our results have revealed the role of cellular VHL in the regulation of SARS-CoV replication.
Keywords: VHL, SARS-CoV, nsp16, Replication, Ubiquitination
Highlights
-
•
Several host proteins were identified to interact with SARS-CoV nsp16 by yeast two-hybrid screening.
-
•
VHL was found to interact with SARS-CoV nsp16 and promotes nsp16 degradation.
-
•
VHL involves in regulating nsp16 ubiquination and stability, and modulating SARS-CoV replication.
1. Introduction
Coronaviruses are pathogenic agents of respiratory and enteral diseases in livestock, companion animals and humans. As members of Coronaviridae, the severe acute respiratory syndrome coronavirus (SARS-CoV) caused a worldwide SARS outbreak in 2003 and the newly identified Middle East respiratory syndrome coronavirus (MERS-CoV) has serious high virulence with a mortality rate of over 35% [1], [2]. The SARS-CoV has a positive-stranded RNA genome of 29 kb in length, containing 14 open reading frames (ORFs). The 5′ two-thirds of the genome includes 2 large overlapping ORFs (1a and 1b) which can be translated to two large poly-proteins. The precursor poly-proteins are then cleaved into 16 nonstructural proteins named as nsp1-16, which function to form a replication–transcription complex (RTC) in cells [3]. The remaining 3′ one-third of the genome is comprised of 12 ORFs, encoding the viral structural and accessory proteins that are translated from a set of nested subgenomic RNAs [4]. So far, the functions of nsp12-16 encoded by the ORF1b have been characterized based on previous experimental evidences. For example, nsp12 is an RNA-dependent RNA polymerase and nsp13 is an RNA helicase and 5′-triphosphatase [5], [6]. Nsp14 is identified as an exoribonuclease and N7- methyltransferase (N7-MTase) [7], [8]. Biochemical assays provide evidence that the nsp15 is an endoribonuclease [9]. While nsp16 is identified as a 2′-O-MTase [10]. As the components of SARS-CoV RTC, these proteins are mainly involved in virus genome replication and expression. Whether they also have interactions with cellular pathways remains unclear. In addition, the host partners involved in the lifecycle of SARS-CoV are poorly understood.
The von Hippel Lindau (VHL) protein is first identified as a component of an E3 ubiquitin ligase complex to target hypoxiainducible factor (HIF1α) for degradation under normal oxygen tension [11]. VHL loss or mutation induces tumorigenesis by causing HIF1α up-regulation in the cytoplasm and its translocation to the nucleus where it heterodimerizes with HIF1β to form an active transcription factor which can bind the promoters of several downstream genes. VHL interacts with Elongin C, Cul-2, Elongin B and Rbx1 to constitute the VBC ubiquitin ligase complex, targeting substrates for 26S proteasome-mediated proteolysis [11]. So far, other targets of VBC ubiquitin ligase complex are found, including RNA polymerase II seventh subunit [12], stimulated with retinoic acid 13 [13] and estrogen receptor-α [14]. VHL is also reported to stabilize p53 by removing the polyubiquitin chains on p53 [15]. The relationship between VHL and virus infection is also documented. The Adenovirus Ad5ΔE3 and human papilloma virus type 16 (HPV16) infections cause VHL protein degradation [16]. The Kaposi's sarcoma–associated herpesvirus (KSHV) infection induces VHL degradation in B lymphoma cells through the viral LANA protein [17]. In addition, VHL interacts with HIV-1 integrase and promotes its degradation. The regulation of HIV-1 integrase stability by VHL has a crucial influence on the viral replication cycle [18]. Although VHL has been shown to regulate virus replication, the more detailed mechanism of the regulation remain to be revealed.
In this report, several host proteins were identified to interact with SARS-CoV nsp16 by yeast two-hybrid screening, including VHL. We have confirmed the VHL-nsp16 interaction by GST pulldown, co-immunoprecipitation and co-localization assays. Further studies indicate that VHL is involved in regulating nsp16 ubiquination and stability, thus modulating SARS-CoV replication.
2. Materials and methods
2.1. Antibodies
The mouse anti-Flag (F4045) and anti-HA (H9658), the rabbit anti-Flag (F7425) antibodies were purchased from Sigma–Aldrich. The mouse anti-Myc (MA1-980), the HRP-labelled goat anti-mouse and goat anti-rabbit antibodies (32430 and 32460) were purchased from Thermo Fisher Scientific. The rabbit ant-VHL (#2738) antibody was purchased from Cell Signaling Technology. The FITC-labelled goat anti-rabbit IgG (CW0114), the rabbit anti-β-actin (CW0097), anti-GST (CW0085) and anti-His (CW0083) antibodies were purchased from Cwbio (Beijing, China).
2.2. Plasmids
The coding sequences of SARS-CoV nsp14 and nsp16 were PCR amplified from pET30a-nsp14 and pET30a-nsp16 respectively [8], [19]. Primers and PCR conditions employed to make the constructs are shown in Table S1. The SARS nsp16 and mouse hepatitis virus (MHV) nsp16 ORFs were PCR amplified and inserted into the EcoRI and BamHI sites of the pGBKT7 vector to generate pGBKT7-nsp16 (S) and pGBKT7-nsp16 (M). The SARS nsp16 ORF was cloned into the EcoRI and XhoI sites of pGST-4T-1 to form GST-nsp16. It also inserted into the HindIII and BamHI sites of the eGFP-C1 vector to obtain eGFP-nsp16. The SARS nsp14, nsp16 and human Dvl2 gene were cloned into pRK-2xFlag vector using the SalI and NotI sites to generate Flag-nsp14, Flag-nsp16 and Flag-Dvl2. The expression vectors His-VHL, HA-VHL, Flag-VHL, DsRed-VHL, Myc-VHL, PLKO.1, sh-VHL (1# and 2#), Myc-ubiquitin and HA-ubiquitin mutants were constructed previously [20].
2.3. Yeast two-bybrid screening, GST pull-down, fluorescence imaging, immunoprecipitation, western blot analysis, ubiquitination, CHX chase and Re-IP assays
Most of these assays were performed as described previously [20], [21]. For the Re-IP ubiquitination assays, the cell extracts were immunoprecipitated (IP) with an anti-Flag antibody and then the immunoprecipitates were denatured by boiling for 5 min with lysis buffer containing 1% SDS. Next, the denatured extracts were diluted with 10 volumes of lysis buffer and re-immunoprecipitated (Re-IP) with an anti-Flag antibody. The ubiquitination of proteins was analyzed by Western blotting with an anti-HA (WT or mutants of ub) antibody.
2.4. Luciferase reporter assays and statistical analysis
Vero cells (1 × 105) were transfected with the reporter SARS-rep-LUC as described previously [8]. All experiments were repeated at least three times. Statistical significance was determined with Student t test, with a p < 0.05 considered statistically significant.
3. Results
3.1. The identification of host proteins that interact with SARS-CoV nsp16
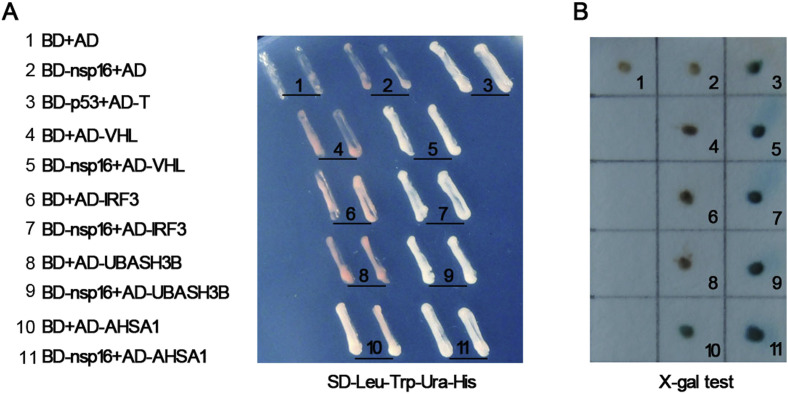
Previous reports have showed that the SARS-CoV nsp16 functions as a 2′-O-MTase involved in virus genome replication and immune escape [10]. In order to explore roles of nsp16 and its host partners which participate in the life cycle of SARS-CoV, we used the yeast two-hybrid system to screen a human thymus cDNA library for SARS-CoV nsp16 interacting proteins. From a total of 2 × 106 transformants, 14 potential positive clones were identified. Their sequences has been analyzed and the results were shown in Table 1 . Considering that the function of SARS-CoV nsp16 is similar to MHV nsp16, we also tested whether the MHV nsp16 interacts with these proteins. BD-nsp16 (MHV) and each of these prey plasmids were co-transformed into the yeast strain AH109. The result showed that four of these proteins (VHL, UBASH3B, AHSA1 or IRF3) could interact with MHV nsp16 as well (Fig. 1 A and B), suggesting that they could be associated with both SARS-CoV and MHV nsp16 protein.
Table 1.
Potential host cell interaction proteins of SARS nsp16.
| Number | Name | Abbreviation | Target region |
|---|---|---|---|
| 1 | ribosomal protein SA | RPSA | 117–295 |
| 2 | guanine nucleotide-binding protein, beta-2 subunit | GNB2 | 176–340 |
| 3 | cytochrome c oxidase subunit II | COX2 | 73–105 |
| 4 | SET domain, bifurcated 1 isoform 1 | ETDB1 | 1175–1291 |
| 5 | stabilin 1 precursor | STAB1 | 114–1182 |
| 6 | threonyl-tRNA synthetase-like 2 | ARSL2 | 98–401 |
| 7 | von Hippel-Lindau tumor suppressor isoform 1 | VHL | 44–213 |
| 8 | ubiquitin associated and SH3 domain containing, B | UBASH3B | 409–649 |
| 9 | activator of heat shock 90 kDa protein ATPase homolog 1 | AHSA1 | 248–338 |
| 10 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex | INCA1 | 1–172 |
| 11 | interferon regulatory factor 3 | IRF3 | 95–327 |
| 12 | metalloprotease 1 precursor | MP1 | 863–1037 |
| 13 | protein arginine methyltransferase 1 | PRMT1 | 337–363 |
| 14 | DNA directed RNA polymerase II polypeptide G | POLR2G | 1–172 |
Fig. 1.
The identification of nsp16 interaction proteins by yeast two-hybrid screening. (A) The AH109 competent cells were transformed with the binary plasmids and were plated twice on SD lacking Leu-Trp-Ura-His plate. BD-p53+AD-T shows positive control clone. (B) The single yeast colonies containing these plasmids were tested using the X-Gal assay. The blue color shows positive interaction of MHV nsp16 with candidate proteins. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. VHL interacts with nsp16
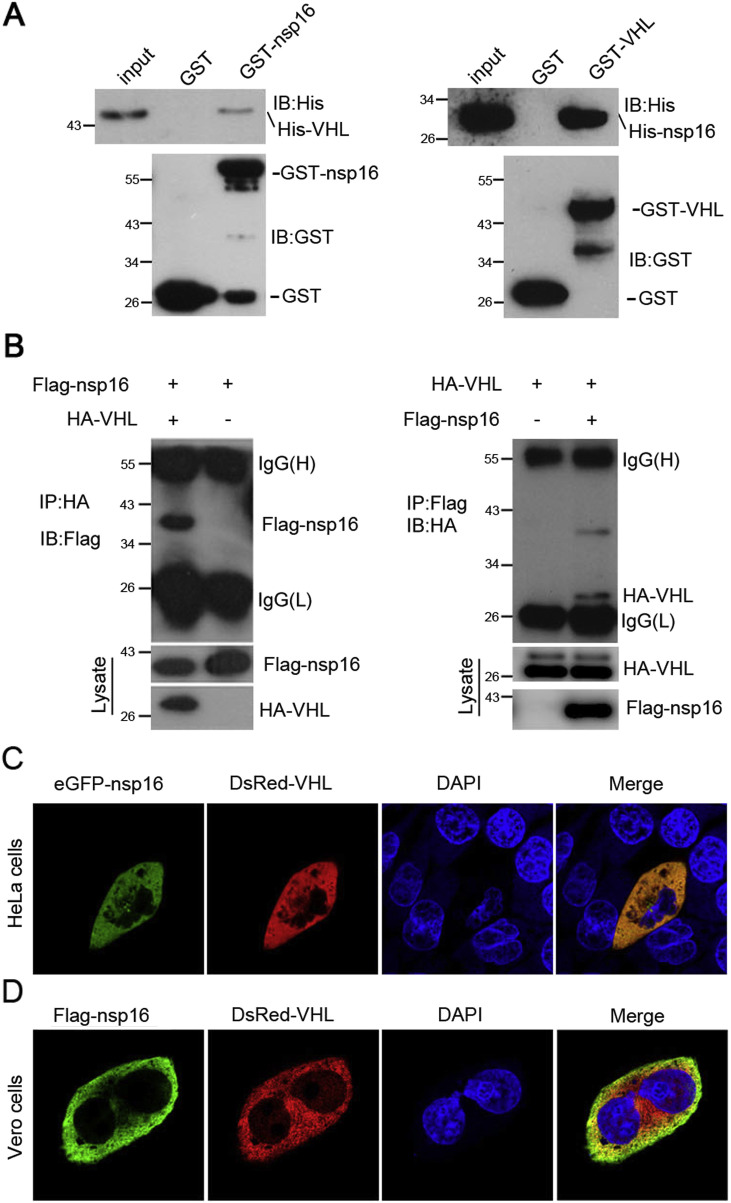
Among the four nsp16-interacting proteins, we selected VHL for further studies as VHL was previously shown to interact with the integrase of HIV-1 and promote its degradation to regulate the viral replication cycle [18]. To verify the interaction between SARS-CoV nsp16 and VHL, GST pull-down assay was employed. The results showed that GST-nsp16 bound to His-VHL in reciprocal pull-down assays, whereas GST control did not (Fig. 2 A). Further cellular reciprocal co-immunoprecipitation assays confirmed that nsp16 could associate with VHL within cells (Fig. 2B). Co-localization experiments showed that eGFP-nsp16 and Red-VHL have the same localization in HeLa cells. In addition, fluorescence imaging showed that Flag-nsp16 and Red-VHL have the same distribution in Vero cells. These results suggested that SARS nsp16 can interact with VHL in cells and the nsp16-VHL interplay may involve in SARS-CoV replication.
Fig. 2.
VHL interacts with SARS-CoV nsp16. (A) 6xHis-VHL or 6xHis-nsp16 proteins expressed in BL21 cells were incubated with glutathione beads and the purified GST, GST-nsp16 or GST-VHL proteins. The bound proteins were eluted and detected by Western blotting with an anti-His or anti-GST antibody. (B) Flag-nsp16 and HA-VHL were transfected into HEK293T cells. After 36 h, the cell extracts were immunoprecipitated (IP) with an anti-HA or anti-Flag antibody and analyzed by immunoblotting (IB) with an anti-Flag or anti-HA antibody (top panel). (C) The eGFP-nsp16 and Red-VHL plasmids were co-transfected into HeLa cells. After 24 h, the cells were fixed with 4% paraformaldehyde and the nuclei were stained with DAPI. The cells were examined by confocal microscopy. (D) After transfection, the Vero cells were fixed with 4% paraformaldehyde, labeled with rabbit anti-Flag antibody (1:100), then probed with FITC-tagged goat anti-rabbit IgG. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. VHL regulates SARS nsp16 protein stability, degradation and SARS replication
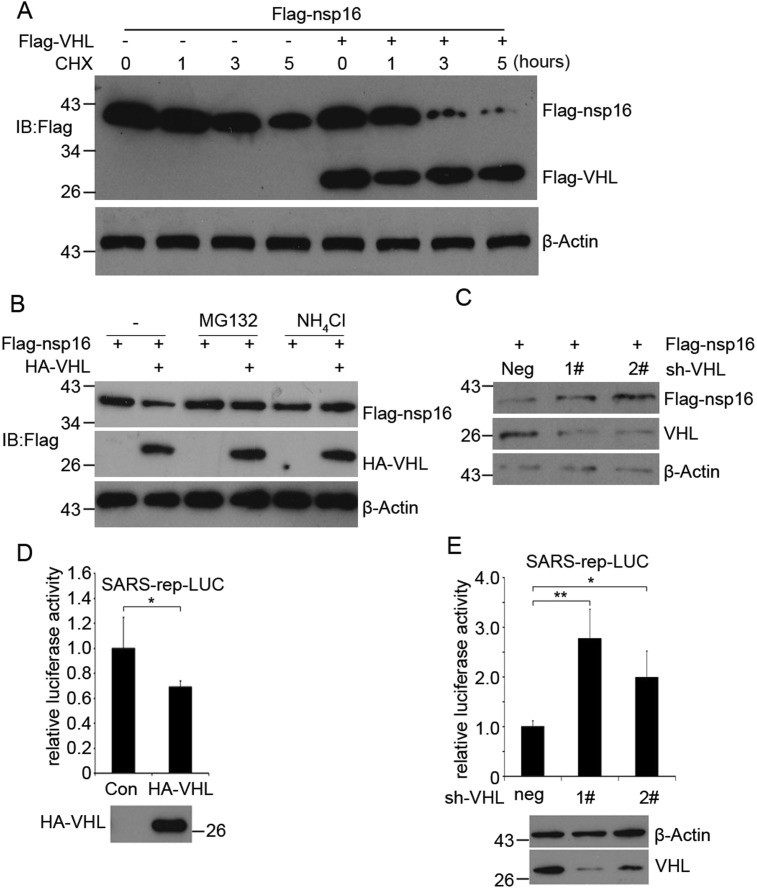
Because previous studies report that VHL inhibits protein synthesis [22], we decided to examine whether VHL regulates the protein stability of SARS-CoV nsp16. To answer this question, a CHX chase assay was performed. HEK293T cells were transfected with Flag-nsp16 and Flag-VHL, followed by treatment with the protein translation inhibitor CHX for 4 different time periods (0, 1, 3, and 5 h). We found that the half-life of nsp16 protein became shorter when it was co-transfected with VHL (Fig. 3 A). This result suggested that the stability of SARS nsp16 could be affected by VHL. In this experiment, we observed that VHL could promote nsp16 degradation (Fig. 3A lane 1 and 5). To study the mechanism of VHL-promoted nsp16 degradation, HEK293T cells were transfected with Flag-nsp16 and HA-VHL, and then the cells were treated with the 26S proteasome inhibitor MG132 or lysosome inhibitor NH4Cl. The results showed that both reagents could rescue the degradation of nsp16 protein induced by VHL (Fig. 3B), suggesting that VHL promotes SARS nsp16 degradation by the 26S proteasome and lysosome pathway. In addition, VHL knockdown increases the steady-state level of nsp16 protein in HEK293T cells (Fig. 3C).
Fig. 3.
VHL regulates SARS nsp16 protein stability, degradation, and SARS replication. (A) Flag-nsp16 and Flag-VHL plasmids were transfected into HEK293T cells. After 24 h, the cells were treated with CHX (100 μg/ml) for the different times. The cell lysates were analyzed by Western blotting with an anti-Flag or anti-β-actin antibody. (B) Flag-nsp16 and HA-VHL plasmids were transfected into HEK293T cells. The cells were treated with MG132 (20 μM) or NH4Cl (10 mM) for another 6 h after 24 h transfection. The cell lysates were analyzed by Western blotting with different antibodies. (C) HEK293T cells were transfected with Flag-nsp16 and control (PLKO.1) or VHL knockdown plasmids (sh-VHL-1#, sh-VHL-2#). After 24 h, the cells were treated with puromycin (1 μg/ml) for an additional 16 h. The cell lysates were analyzed by Western blotting with indicated antibodies. (D) The SARS-rep-LUC (0.1 μg) and HA-VHL (0.3 μg) were transfected into Vero cells. The pcDNA3.0 and pRL-TK were used to make an equal amount of total DNA and normalize the transfection efficiency. The reporter assays were processed 36 h after transfection. (E) Assays were performed as in (D) except for the transfection with VHL RNAi plasmids. *, p < 0.05, **, p < 0.01; the bar graphs show the mean value ± SD of triplicate samples.
In order to determine the effect of VHL on SARS replication, the SARS-CoV replicon that carries a luciferase reporter and Flag-VHL were transfected into Vero cells. The relative value of the reporter activity was decreased after the transfection of Flag-VHL (Fig. 3D), indicating that VHL impaired SARS RNA genome replication and/or transcription. Using this reporter system, knockdown of VHL can increase the level of SARS-CoV RNA genome replication and/or transcription (Fig. 3E).
3.4. VHL modulates SARS nsp16 ubiquitination
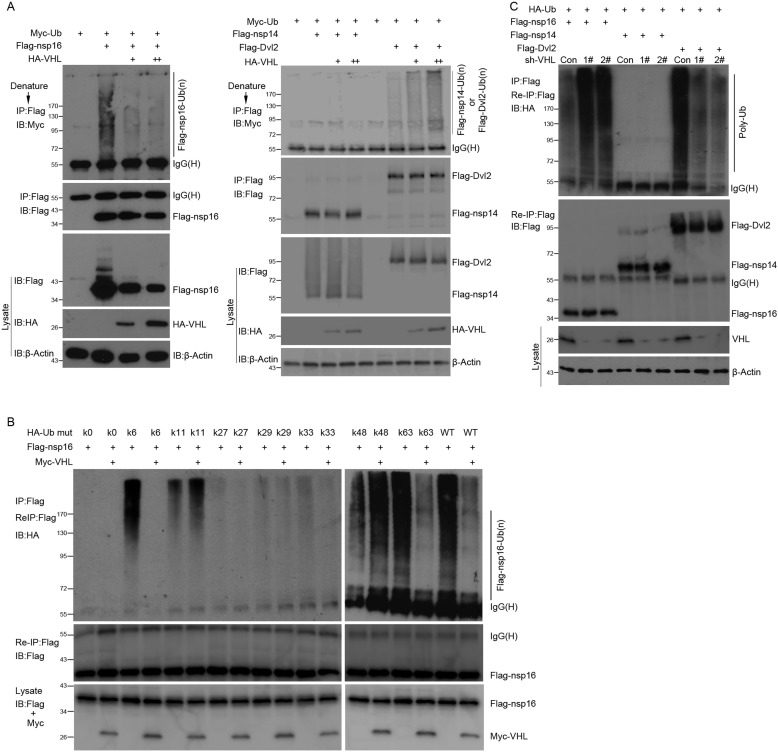
VHL is an E3 ubiquitin ligase that promotes many proteins ubiquitination. However, it is also involved in deubiquitinaion of other proteins such as p53 [15]. We examined whether VHL regulates SARS-CoV nsp16 ubiquitination by co-transfection Myc-ubiquitin, HA-VHL, and Flag-nsp16 or Flag-nsp14 as well as Dvl2 into HEK293T cells as indicated (Fig. 4 ). The results showed that overexpression of VHL inhibits SARS-CoV nsp16 ubiquitination (Fig. 4A, left panels); however, in control experiments, VHL promotes Dvl2 ubiquitination as reported previously [23] and has no effect on the ubiquitination of SARS-CoV nsp14 (Fig. 4A, right panels). We also determined the lysine specificity of PolyUb chain involved in the modification of SARS-CoV nsp16 and the effect of VHL on the ubiquitin modification with HA-tagged ubiquitin with all lysine mutated to arginine (K0), or containing only one lysine (K6, K11, K27, K29, K33, K48 or K63). Co-immunoprecipitation assays showed that the ubiquitination of SARS-CoV nsp16 mainly occurred at Ub-K6 and Ub-K63 (Fig. 4B, lane 3 and lane 15), and VHL inhibits the K6 and K63-linked poly-ubiquitination of SARS-CoV nsp16 (Fig. 4B, lane 4 and lane 16). In the same experiment, we observed that other ubiquitin site such as K11 was also involved in the nsp16 ubiquitination and VHL could promote the K48-linked ubiquitination of SARS-CoV nsp16 (Fig. 4B, lane 13 and lane 14). To determine whether VHL influences SARS-CoV nsp16 ubiquitination under physiological conditions, we used RNAi plasmids targeting the human VHL mRNA to reduce the VHL protein level. Co-immunoprecipitation assays showed knockdown of VHL potentiates the ubiquitination of SARS-CoV nsp16 and decrease the ubiquitination of Dvl2, with no effect on the ubiquitination of SARS nsp14 (Fig. 4C).
Fig. 4.
VHL modulates SARS-CoV nsp16 ubiquitination. (A) Left panels, HEK-293T cells were transfected with Myc-ubiquitin, Flag-nsp16 and an increasing amount of HA-VHL (1.0 and 2.0 μg). After 36 h transfection, the cells were collected and denatured in 1% SDS lysis buffer, and the cell lysate were diluted with lysis buffer until the concentration of SDS decreased to 0.1%. The cell extracts were used for co-immunoprecipitation with an anti-Flag antibody. The immunoprecipitates were detected by an anti-Myc or anti-Flag antibody. Right panels, assays were performed as in the left panels. SARS-CoV nsp14 as a negative control and Dvl2 as a positive control. (B) The HA-tagged ubiquitin mutants, Flag-nsp16 and Myc-VHL were transfected into HEK-293T cells. After 36 h, re-IP assays were performed with anti-Flag antibody. The immunoprecipitates were analyzed with an anti-HA or anti-Flag antibody. (C) The HA-ubiquitin, Flag-nsp16 or Flag-nsp14 or Flag-Dvl2, control (PLKO.1) or VHL RNAi plasmids (1# or 2#) were transfected into HEK-293T cells. After 24 h, the cells were treated with puromycin (1 μg/ml) for another 24 h and then lysed for re-IP assays with anti-Flag antibody. The immunoprecipitates were analysed with the anti-HA or anti-Flag antibody (upper panels).
4. Discussion
The SARS-CoV has a relatively large positive-stranded genome and forms a complicate RTC when replicating within cells. The RTC consists of 16 nonstructural proteins which can process viral genome or host RNA. SARS-CoV nsp16 acts as a 2′-O-MTase to selectively methylate cap structure at 2′-O position of nucleotide [7], [10], [19]. Host proteins are also involved in replication and transcription of viral genome. DDX1 was found to interact with SARS-CoV or avian infectious bronchitis virus (IBV) nsp14 to promote efficient coronavirus replication [24]. In order to find more host proteins that regulate SARS-CoV replication, a yeast two-hybrid screening with a human thymus cDNA library were performed. Using the nsp16 as a bait, 14 potential preys were identified. Four (VHL, UBASH3B, AHSA1 or IRF3) of these proteins were found to interact with both SARS-CoV nsp16 and MHV nsp16. Sequence analysis revealed that the two coronavirus nsp16 have more than 60% identity at the amino acid level and share the same stretches in the key functional region, implying a common mechanism for these two functionally equivalent proteins in coronaviruses.
In this study, the interaction between nsp16 and VHL was confirmed. However, the interaction during SARS-CoV infection was not addressed due to the unavailability of a valid anti-nsp16 antibody and the strict control of using wildtype SARS coronavirus. Previous research showed that VHL can interact with the integrase of HIV-1 and induce its degradation to regulate the viral replication cycle [18]. It might be possible that the interaction between nsp16 and VHL may also play a role during the replication of coronavirus genome RNA. Interestingly, Adenovirus Ad5ΔE3, HPV16 and KSHV infection can induce VHL degradation [16], [17], implying an interplay between hosts and virus regarding the important roles of VHL during virus infection and host immunity. Our data suggests that VHL impairs not only the virus genome replication and/or transcription but also nsp16 stability by promoting its degradation through both the 26S proteasome and lysosome pathways.
Ubiquitination of viral or host factors participates in almost every aspects of the viral life cycle [25]. For example, the influenza A virus (IAVs) matrix protein M1 was ubiquitinated by the E3 ubiquitin ligase Itch to release viral RNA and allow transport of vRNP to the nuclei [26], [27]. Hepatitis B viral X and core protein can be targeted by Siah-1 and Np95/ICBP90-like RING finger protein for ubiquitination and proteasome degradation [28], [29]. VHL is reported to promote or inhibit protein ubiquitination [11], [15]. In this study, we found that VHL could impede the SARS-CoV nsp16 ubiquitination linked through both K6 and K63 lysines. We also observed that VHL promotes the K48-linked ubiquitination of SARS-CoV nsp16. Ubiquitination linked through different ubiquitin lysine residues can target substrates for different cellular fates. In most cases, the K48-linked polyubiquitination leads to protein degradation and the K63-linked polyubiquitination provides long chains for protein interactions and stability [30]. However, ubiquitination linked through other lysines may also mediate protein stability [31]. We speculated that on one side VHL promotes SARS-CoV nsp16 degradation by enhancing its K48-linked polyubiquitination and impares SARS-CoV replication. On the other side, the SARS-CoV nsp16 could be stabilized through the K6 and K63-linked polyubiquitination in cells which can also be impeded by VHL to affect nsp16 stability. Previous researches have shown that the ubiquitin-proteasome system plays critical roles in RNA synthesis and protein expression during various stages of the coronavirus infection cycle [32]. Our results that VHL regulates the ubiquitination of nsp16 and inhibits SARS-CoV replication have provided evidence that ubiquitin modification with different linkage specificity also plays important roles within life cycle of coronavirus.
In conclusion, our results suggest that VHL may negatively regulates coronavirus replication by down-regulating nsp16, one of the viral replicase that functions as 2′-O-MTase. Through interacting with nsp16, VHL modulates its ubiquitination, therefore reducing its stability. Our results shed light on the function and mechanism of cellular proteins to defense against SARS-CoV infection. However, in vitro experiments indicates that the functional SARS-CoV genome replication/transcription complexes requires the involvement of cytoplasmic host factors [33]. In order to elucidate the mechanisms of coronavirus replication and its complicated interplay with host cells, more host partners involved in the coronavirus life cycle need to be identified and characterized in the future.
Conflict of interest
None.
Acknowledgments
This work was supported by China NSFC grants (#31170152 and 81130083) and China Postdoctoral Science Foundation (2014M560622).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2015.02.097.
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2015.02.097.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Transparency document
References
- 1.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 2.Memish Z.A., Zumla A.I., Al-Hakeem R.F., Al-Rabeeah A.A., Stephens G.M. Family cluster of Middle East respiratory syndrome coronavirus infections. N. Engl. J. Med. 2013;368:2487–2494. doi: 10.1056/NEJMoa1303729. [DOI] [PubMed] [Google Scholar]
- 3.Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., Canard B. SARS-CoV ORF1b-encoded nonstructural proteins 12–16: replicative enzymes as antiviral targets. Antivir. Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanner J.A., Watt R.M., Chai Y.B., Lu L.Y., Lin M.C., Peiris J.S., Poon L.L., Kung H.F., Huang J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5'–3' viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3'→5' exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decroly E., Imbert I., Coutard B., Bouvet M., Selisko B., Alvarez K., Gorbalenya A.E., Snijder E.J., Canard B. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2'O)-methyltransferase activity. J. Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaelin W.G., Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat. Rev. Cancer. 2008;8:865–873. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- 12.Na X., Duan H.O., Messing E.M., Schoen S.R., Ryan C.K., di Sant'Agnese P.A., Golemis E.A., Wu G. Identification of the RNA polymerase II subunit hsRPB7 as a novel target of the von Hippel-Lindau protein. EMBO J. 2003;22:4249–4259. doi: 10.1093/emboj/cdg410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanova A.V., Ivanov S.V., Danilkovitch-Miagkova A., Lerman M.I. Regulation of STRA13 by the von Hippel-Lindau tumor suppressor protein, hypoxia, and the UBC9/ubiquitin proteasome degradation pathway. J. Biol. Chem. 2001;276:15306–15315. doi: 10.1074/jbc.M010516200. [DOI] [PubMed] [Google Scholar]
- 14.Jung Y.S., Lee S.J., Yoon M.H., Ha N.C., Park B.J. Estrogen receptor alpha is a novel target of the Von Hippel-Lindau protein and is responsible for the proliferation of VHL-deficient cells under hypoxic conditions. Cell. Cycle. 2012;11:4462–4473. doi: 10.4161/cc.22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roe J.S., Kim H., Lee S.M., Kim S.T., Cho E.J., Youn H.D. p53 stabilization and transactivation by a von Hippel-Lindau protein. Mol. Cell. 2006;22:395–405. doi: 10.1016/j.molcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Pozzebon M.E., Varadaraj A., Mattoscio D., Jaffray E.G., Miccolo C., Galimberti V., Tommasino M., Hay R.T., Chiocca S. BC-box protein domain-related mechanism for VHL protein degradation. Proc. Natl. Acad. Sci. U. S. A. 2013;110:18168–18173. doi: 10.1073/pnas.1311382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai Q.L., Knight J.S., Verma S.C., Zald P., Robertson E.S. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2006;2:e116. doi: 10.1371/journal.ppat.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mousnier A., Kubat N., Massias-Simon A., Segeral E., Rain J.C., Benarous R., Emiliani S., Dargemont C. von Hippel Lindau binding protein 1-mediated degradation of integrase affects HIV-1 gene expression at a postintegration step. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13615–13620. doi: 10.1073/pnas.0705162104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P., Ahola T., Liang Y., Liu X., Guo D. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S., Chen K., Zhang Q., Cheng H., Zhou R. Regulation of the transcriptional activation of the androgen receptor by the UXT-binding protein VHL. Biochem. J. 2013;456:55–66. doi: 10.1042/BJ20121711. [DOI] [PubMed] [Google Scholar]
- 21.Chen S., Yu X., Lei Q., Ma L., Guo D. The SUMOylation of zinc-fingers and homeoboxes 1 (ZHX1) by Ubc9 regulates its stability and transcriptional repression activity. J. Cell. Biochem. 2013;114:2323–2333. doi: 10.1002/jcb.24579. [DOI] [PubMed] [Google Scholar]
- 22.Zhao W.T., Zhou C.F., Li X.B., Zhang Y.F., Fan L., Pelletier J., Fang J. The von Hippel-Lindau protein pVHL inhibits ribosome biogenesis and protein synthesis. J. Biol. Chem. 2013;288:16588–16597. doi: 10.1074/jbc.M113.455121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao C., Cao W., Bao L., Zuo W., Xie G., Cai T., Fu W., Zhang J., Wu W., Zhang X., Chen Y.G. Autophagy negatively regulates Wnt signalling by promoting dishevelled degradation. Nat. Cell. Biol. 2010;12:781–790. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 24.Xu L., Khadijah S., Fang S., Wang L., Tay F.P., Liu D.X. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J. Virol. 2010;84:8571–8583. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calistri A., Munegato D., Carli I., Parolin C., Palu G. The ubiquitin-conjugating system: multiple roles in viral replication and infection. Cells. 2014;3:386–417. doi: 10.3390/cells3020386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao S., Wu J., Liu R.Y., Li J., Song L., Teng Y., Sheng C., Liu D., Yao C., Chen H., Jiang W., Chen S., Huang W. Interaction of NS2 with AIMP2 facilitates the switch from ubiquitination to SUMOylation of M1 in influenza a virus-infected cells. J. Virol. 2015;89:300–311. doi: 10.1128/JVI.02170-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su W.C., Chen Y.C., Tseng C.H., Hsu P.W., Tung K.F., Jeng K.S., Lai M.M. Pooled RNAi screen identifies ubiquitin ligase itch as crucial for influenza A virus release from the endosome during virus entry. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17516–17521. doi: 10.1073/pnas.1312374110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao J., Wang C., Wang J., Yang X., Diao N., Li Q., Wang W., Xian L., Fang Z., Yu L. E3 ubiquitin ligase Siah-1 facilitates poly-ubiquitylation and proteasomal degradation of the hepatitis B viral X protein. FEBS Lett. 2011;585:2943–2950. doi: 10.1016/j.febslet.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Qian G., Jin F., Chang L., Yang Y., Peng H., Duan C. NIRF, a novel ubiquitin ligase, interacts with hepatitis B virus core protein and promotes its degradation. Biotechnol. Lett. 2012;34:29–36. doi: 10.1007/s10529-011-0751-0. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z.J. Ubiquitination in signaling to and activation of IKK. Immunol. Rev. 2012;246:95–106. doi: 10.1111/j.1600-065X.2012.01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dynek J.N., Goncharov T., Dueber E.C., Fedorova A.V., Izrael-Tomasevic A., Phu L., Helgason E., Fairbrother W.J., Deshayes K., Kirkpatrick D.S., Vucic D. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raaben M., Posthuma C.C., Verheije M.H., te Lintelo E.G., Kikkert M., Drijfhout J.W., Snijder E.J., Rottier P.J., de Haan C.A. The ubiquitin-proteasome system plays an important role during various stages of the coronavirus infection cycle. J. Virol. 2010;84:7869–7879. doi: 10.1128/JVI.00485-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Hemert M.J., van den Worm S.H., Knoops K., Mommaas A.M., Gorbalenya A.E., Snijder E.J. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4:e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.