Abstract
The severe acute respiratory syndrome (SARS) is a contagious disease that killed hundreds and sickened thousands of people worldwide between November 2002 and July 2003. The nucleocapsid (N) protein of the coronavirus responsible for this disease plays a critical role in viral assembly and maturation and is of particular interest because of its potential as an antiviral target or vaccine candidate. Refolding of SARS N-protein during production and purification showed the presence of two additional protein bands by SDS–PAGE. Mass spectroscopy (MALDI, SELDI, and LC/MS) confirmed that the bands are proteolytic products of N-protein and the cleavage sites are four SR motifs in the serine–arginine-rich region—sites not suggestive of any known protease. Furthermore, results of subsequent testing for contaminating protease(s) were negative: cleavage appears to be due to inherent instability and/or autolysis. The importance of N-protein proteolysis to viral life cycle and thus to possible treatment directions are discussed.
Keywords: Coronavirus, Nucleocapsid protein, N-protein, Proteolysis, SARS
Severe acute respiratory syndrome (SARS) is an atypical pneumonia first described in November 2002 [1]. The causative agent, the SARS coronavirus, belongs to the coronaviradae family of enveloped, positive-sense RNA viruses and posseses four structural proteins. The nucleocapsid (N) protein has been found to bind to a specific packaging-signal motif on the viral RNA; it is the interaction of this protein–RNA complex with the membrane (M), envelope (E) and spike (S) proteins that leads to budding through the cell membrane and virus maturation. The N-protein is of particular interest because of its potential as a vaccine candidate [2], [3], as a diagnostic marker for SARS [4], [5], and because it appears to play a critical role in the perturbation of several host cell processes during infection [6].
SARS N-protein is a 432 amino acid, 46 kDa protein with the high pI (10.1) and high content of basic amino acids characteristic of many DNA- or RNA-binding proteins. Some atypical characteristics of the N-protein include a low percentage of hydrophobic amino acids and an absence of cysteine residues [7]. The peculiar composition of the protein may be important for the RNA-binding properties, but the absence of strong intramolecular interactions also suggests that, in contrast to other viral structural proteins, the structure of SARS N-protein is unstable [8], [9], [10], [11], [12].
As with other viral N-proteins, the SARS N-protein exhibits extensive oligimerization that is presumably linked to SARS virus packaging and maturation. Of the potential intermolecular binding sites that have been identified [13], [14], [15], the first to be characterized was the highly hydrophilic serine and arginine rich region, 184ssrsssrsrgnsr196 [7], [16], [17], [18]. Deletion of this sequence abrogates N-protein self-association and prevents N-protein localization around the nucleus and thus the RNA-binding and packaging required for SARS virus maturation.
In the process of purifying the N-protein to study its potential in the production of vaccines against the virus, the appearance of two other protein/protein fragment bands on SDS–PAGE was detected. This paper describes the work to identify these proteins/protein fragments and to determine their source. A series of experiments were designed to isolate and characterize these fragments, pinpoint the cleavage site(s), and determine if the cleavage was due to a bacterial protease contaminant or to autolysis. It is anticipated that identification of the source/conditions for cleavage and subsequent inactivation of N-protein and concomitant disabling of the coronavirus’ ability to package RNA and/or interfere with host cell function, may lead to development of novel and efficacious treatments for SARS.
Materials and methods
SARS N-protein expression and purification. Escherichia coli M15 and BL21(DE3) cells, transformed with pQE-2/NP (pQE-2 expressing the N-protein), were used to produce protein as previously described [17], [19]. Purification was performed under denaturing conditions using a His-trap HP metal affinity column (GE Lifesciences).
The purified N-protein was transferred to dialysis tubing (7500 MWCO) and dialyzed into a urea-supplemented refolding buffer (10 mM Tris, 100 mM sodium phosphate, 150 mM NaCl, and 8 M urea, pH 8.0). Urea was then gradually removed by a stepwise replacement of the buffer with Tris/phosphate buffer (10 mM Tris, 100 mM sodium phosphate, and 150 mM NaCl, pH 8.0) containing decreasing concentrations of urea (8, 4, 2, 1, 0.5, and 0 M). In samples where the N-terminal (His)6-tag was removed, the pH of the buffer was adjusted to 7.0 by dialysis and Qiagen DAPase (dipeptidase) added to the protein solution.
N-protein solutions were analyzed on SDS–poly-acrylamide gel using the 12% cross-link method described by Laemmli [20] or the 20% Tricine method described by Schagger [21]. Gels were stained using Coomassie blue or Sypro Ruby Red.
Mass spectrometric analyses. SELDI-TOF/MS data were generated using a PBS-IIC instrument (Ciphergen, Fremont, CA) that was calibrated using All-in-One peptide standards (Ciphergen) adhered to a normal phase, NP20 protein array. One microgram of the SARS N-protein was applied to each of the remaining sample spots for analysis. SELDI-TOF spectra were generated by laser desorption/ionization using an average 130 laser shots with an intensity of 190–200 (arbitrary units) and detector sensitivity of eight.
MALDI-TOF mass spectrometry was performed on peptides after SDS–PAGE separation and in-gel tryptic digestion of proteins/peptide bands [22]. Peptide fragments were analyzed using a Micromass MALDI-LR instrument (Waters, Mississauga, ON) and analyzed using MassLYNX 3.5 software (Waters). Peptide fingerprint searches were performed using MASCOT (Matrix Science, Boston, MA) and the NCBI protein database.
N-protein fragments were purified on a Thermo Spectra System HPLC using a Vydac C8 reverse phase column. Proteins were eluted using 0.01% trifluoroacetic acid (TFA) and a 10–90% acetonitrile gradient and protein-containing fractions analyzed by LC/MS as previously described [23].
Protease assessment. Quanticleave protease assay using fluorescein isothiocyanate-(FITC)-conjugated casein as described by manufacturer (Pierce). Fluorescence was detected using 485/538 nm excitation/emission wavelengths in a Genios plate reader running XFluor 4 software (Tecan, Durham, NC).
Non-specific protease activity was tested by mixing a 10-fold excess (w/w relative to SARS N-protein) of either ovalbumin or RNAse A and co-refolded with the SARS N-protein. Ovalbumin and RNase A were prepared by cleaving and capping of disulfide bonds with iodoacetamide to prevent disulfide bond formation prior to refolding. The test proteins (N-protein and ovalbumin or RNAase) were then denatured by addition to 6 M guanidine buffer containing SARS N-protein and proteins simultaneously refolded. The presence or absence of cleavage peptides was determined using SDS–PAGE.
Fluorescent resonance energy transfer-labelled peptides (EDANS/DABCYL-conjugated 168LPKGFYAEGSRGGSQASS185- and 181SQASSRSSSRSRGNSRNSTP200-SARS N-protein peptides) were purchased from JPT Peptide Technologies (Acton, MA). EDANS/DABCYL-conjugated peptides were designed to correspond to putative cleavage sites of the SARS N-protein. Cleavage of either peptide would lead to separation of the EDANS reagent from the DABCYL and result in a 40-fold increase in fluorescent signal. An aliquot (100 μl) of each EDANS/DABCYL-conjugated peptide dissolved in PBS at pH 7.2 (0.5 mg/ml final concentration) was placed in a 96-well plate and 100 μl of either N-protein preparation (25 μg total protein) or trypsin (positive control) was added. Fluorescence (relative fluorescent units, RFU) was recorded using 360/465 nm excitation/emission wavelengths over 60 min at 25 °C.
Results and discussion
SARS N-protein production, purification, and refolding
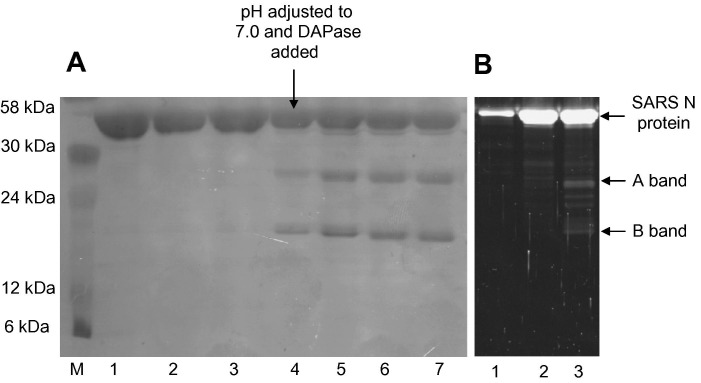
SARS N-protein was produced in both M15 and BL21(DE3) cells. Proteins were expressed as insoluble inclusion bodies and purified using metal affinity resin, then urea-denatured proteins were refolded by stepwise dialysis and the His-tag removed by DAPase digestion. SDS–PAGE confirmed that the recovered N-protein was purified to near homogeneity under denaturing conditions (Fig. 1 ). However, following the refolding, two additional bands (A and B bands) were observed at approximately 29 kDa (A band) and 25 kDa (B band). The masses were consistent with proteolyed N-protein (approximately 50 kDa) and similar to the SARS N-protein proteolysis products reported in the presence of caspases [24].
Fig. 1.
SARS N-protein refolding and proteolysis. (A) SARS N-protein produced in M15 cells and purified was refolded by gradual dialysis and the engineered (His)6-tag was removed using Qiagen DAPase as described in Materials and methods. Ten microliters of the protein mixtures were loaded onto 12% SDS–PAGE and stained using Coomassie blue. Lane M, molecular weight markers; lane 1, SARS N-protein in 8 M urea; lane 2, 0.5 M urea; lane 3, 0 M urea; lane 4, 0 M urea; lane 5, 10 min following DAPase addition; lane 6, 20 min following DAPase addition; lane 7, 30 min following DAPase addition. (B) SARS N-protein was produced and refolded without DAPase. Samples were separated on 12% SDS–PAGE and stained with Sypro Ruby Red lane 1, SARS N-protein in 2 M urea; lane 2, 1 M urea; and lane 3, 0.5 M urea.
To rule out the possibility that the A and B bands could be attributed either to N-protein cleavage by DAPase or a contaminant in DAPase, the purification was repeated using BL21(DE3) cells without DAPase addition. BL21(DE3) cells lack the lon and ompT protease genes and thus reduced recombinant protein proteolysis is expected. However, the A and B bands were still observed, confirming that the SARS N-protein cleavage was not due to the DAPase preparation and was independent of the cell line used (data not shown).
Characterization of protein fragments
To determine the source of the A and B bands, mass spectrometry techniques were used. The experiments used preparations of recombinant N-protein from which the His-tag had not been enzymatically removed; thus, calculation of the resultant protein/peptide masses must take into consideration the presence of the His-tag (an additional 11 amino acid sequence).
While SDS–PAGE showed two discrete bands of approximately 29 and 25 kDa, surface enhanced laser desorption ionization (SELDI) mass spectrometry (data not shown) showed that the bands comprised of four or five proteins/peptides with similar molecular masses: 24.0–26.0 (A band) and 21.5–23.0 kDa (B band). Each SELDI peak was separated from its nearest neighbor by approximately 200–250 Da (two amino acids). Thus, the A and B bands were tentatively identified as several site-specific hydrolyzed fragments of the full-length N-protein (47 kDa).
Preliminary identifications of the SARS N-protein fragments were confirmed by in-gel digestion of the A and B bands and MALDI-TOF/mass spectrometry. Detected peptide molecular weights were searched against the MASCOT database and both the A and B bands were identified as SARS N-protein fragments. MALDI-TOF/MS data was then compared to the expected molecular masses of tryptic peptides determined by an in silico digest of the N-protein sequence. Only peptides corresponding to the N-protein’s C-terminal region were detected in the A band, while only N-terminal peptides were detected in the B band. This result, combined with SELDI-TOF/MS data, indicated that the recombinant N-protein undergoes cleavage at multiple sites near the center of its amino acid sequence.
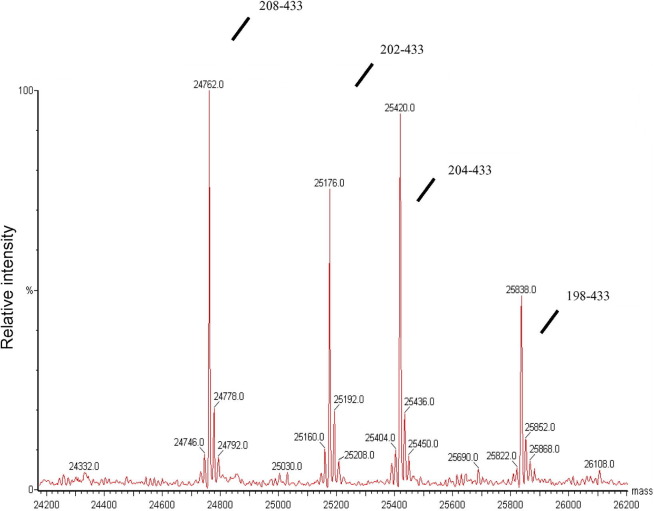
To determine the exact location of N-protein cleavage, N-protein samples were analyzed by LC/MS. Resultant masses were compared to the theoretical masses of all possible peptides derived from the N-protein’s amino acid sequence. The results showed that cleavage had occurred between residues 197/198, 201/202, 203/204, and 205/206 of the His-tagged N-protein (i.e., 184ssr/sssr/sr/gnsr/196 in the untagged protein sequence). The deconvoluted mass spectrum of the cleaved C-terminus protein fragment is shown in Fig. 2 and is consistent with the result of SELDI-TOF/MS that showed successive cleavage sites with gaps of approximately two amino acids.
Fig. 2.
LC/MS analysis of N-protein fragments purified by reverse phase HPLC Proteins were purified by reverse phase HPLC and analyzed by LC/MS as described in Methods. Protein fragment masses of 24,762, 25,176, 25,420, and 25,838 Da were detected.
Protease assessment
With the site of N-protein cleavage identified, experiments were performed to determine whether this cleavage was the result of protease contamination of the preparation, N-protein autocatalysis, or an inherent susceptibility to specific bond cleavage. Initially, the commercially available Quanticleave assay, which uses FITC-labelled casein, was used to test for non-specific proteolysis. When the FITC-labelled casein was added to preparations of the purified N-protein, no increase in fluorescence was detected.
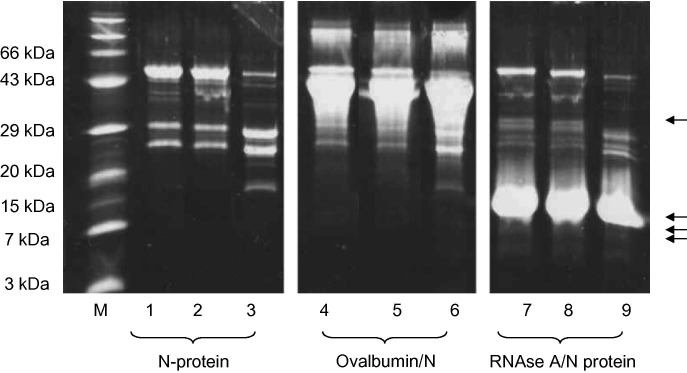
It was possible that a contaminating protease was present but undetectable by the Quanticleave assay, as casein contains several R residues but no SR motif. To further evaluate the possibility that contaminating proteases were responsible for N-protein hydrolysis, denatured SARS N-protein was refolded in the presence of excess denatured ovalbumin (possessing an SR motif at a.a. 104–105) or denatured RNAse A (possessing an SR motif at a.a 36–37). After refolding, the protein/protein fragment mixtures were separated by SDS–PAGE; while the SARS N-protein still showed the distinctive cleavage pattern upon refolding, neither ovalbumin nor RNAse A showed any proteolysis (Fig. 3 ). If a contaminating protease was responsible for cleavage of the SR motif, preferential digestion of the more concentrated (10-fold excess) ovalbumin or RNAse A is expected. The absence of cleavage of these two proteins suggests the absence of protease contamination in the N-protein sample.
Fig. 3.
SARS N-protein refolding with excess ovalbumin or RNAse A. SARS N-protein was refolded in the presence of a 10-fold excess of ovalbumin or RNAase. Samples were separated by 20% Tricine gel lane M, molecular size markers; lanes 1, 4, and 7, samples obtained after overnight dialysis into a final refolding buffer (0 M urea); lanes 2, 5, and 8, obtained following additional 2 h incubation at 4 °C; lanes 3, 6, and 9, obtained following 120 h incubation at 4 °C. Lanes 1–3 contained N-protein refolded in the absence of secondary protein. Lanes 4–6 contained SARS N-protein refolded with ovalbumin, lanes 7–9 contained SARS N-protein refolded with RNAase. Arrows indicate molecular weights of predicted ovalbumin (32 and 12 kDa) and RNAse A (9 and 8 kDa) cleavage products.
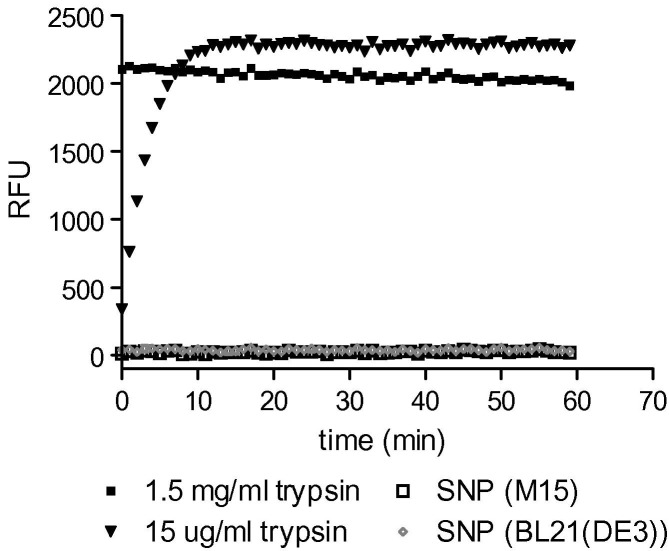
A specific proteolysis assay was then developed using peptides containing the SR-rich region corresponding to the SARS N-protein cleavage site. Peptides derived from the N-protein, were synthesised and conjugated with EDANS and DABCYL for FRET analysis. The FRET-labelled peptide substrates were incubated with N-protein, trypsin (positive control), or buffer (negative control). In this assay peptide cleavage would result in the liberation of EDANS and DABCYL and a concomitant increase in fluorescence [25]. In positive controls, incubation of both FRET substrates with trypsin resulted in increased fluorescence and demonstrated that the assay could detect proteases. However, in N-protein trials, no similar increase in fluorescence was seen (Fig. 4 ) indicating that contaminating proteases were unlikely unless in amounts below the detection limit of the assay (for trypsin, approximately 2.5 ng/ml). SARS N-protein present in the same mixture as these SR-containing substrates did, itself, undergo cleavage as evidenced by the appearance of A and B bands on SDS–PAGE (data not shown), indicating that if the N-protein is responsible for its own cleavage (autolysis), it is unable to cleave FRET-labelled peptides containing the cleavage site. These data can be explained if the sequences for protease binding and subsequent cleavage are located in different regions of the N-protein. Separation of binding and cleavage sites on a proteolytic substrate has been reported previously [26] but it is not common for a protease to bind a substrate at a sequence remote from the scissile bond. Taken together, the results from the FITC-labelled casein, the refolding, and the FRET (EDANS/DABCYL) detection experiments strongly suggest that the SR-specific cleavage of N-protein is not the result of a contaminating protease activity.
Fig. 4.
EDANS/DABCYL-conjugated SARS N-protein peptide assay. EDANS/DABCYL-conjugated 181SQASSRSSSRSRGNSRNSTP200 peptide was generated as artificial protease substrates. Increased fluorescence (measured in relative fluorescence units, RFU) indicated proteolysis. Ten micrograms and 100 ng trypsin were used as a positive controls along with SARS N-protein samples from either M15 or BL21(DE3) cells.
Cleavage of N-protein
This detection of SARS N-protein proteolysis is not the first observation of N-protein cleavage. Cleavage has also been reported in SARS coronavirus-infected Vero (monkey-derived) cells [24], [27]. Ying and co-workers [24] observed three N-protein-related bands with apparent molecular size of 27–31 kDa and four at 16–23 kDa in addition to bands for the full-length protein. This hydrolysis attributed to the action of endogenous caspase-3 as N-protein incubation with exogenous caspases, resulted in similar cleavage products. The authors did not note that the SARS N-protein sequence lacks the canonical—DXXD—caspase-3 cleavage site. More recently, Diemer et al. [27] showed that caspase-6 can cleave SARS N-protein following residue 400 or 403, giving fragments of 44 and 2 kDa. The cleavage was cell-type specific and only observed in Vero-E6 or human epithelial and not Caco or N2a (murine neuronal) lines. In the current study, SDS–PAGE results using E. coli-derived SARS N-protein showed similar cleavage to that detected by Ying et al., but in the absence of any caspases.
The SR-rich region thus appears to be readily cleaved in both E. coli and Vero cells and our attempts to identify a contaminating protease responsible for that cleavage, though not exhaustive, were unsuccessful. Guruprasad and co-workers [28] report that certain dipeptide sequences are statistically more likely to be found in proteins with shorter in vivo half-lives than in those with longer in vivo half-lives. Both SR and RS (arginine–serine) dipeptides (and the N-protein SR-rich region contains both) are among those found more frequently in “unstable” proteins. In vivo instability is usually linked to digestion by circulating or cellular proteases. Therefore, a database of proteases was searched to find an enzyme capable of the observed cleavages. Not only is the SR motif cleavage site is not a recognition site for any known E. coli proteases (consistent with our inability to detect protease contamination in our N-protein preparations), it is not recognized by any other protease in the database [29]. Several proteases do cleave following positively charged residues, including arginine (Arg-C and clostripain) or any arginine or lysine (trypsin); however, this cleavage is otherwise non-specific and does not require S in the P2 position.
The putative biological role(s) of this SR-rich region is not completely known, but it has been proposed that this region allows the SARS N-protein to form the dimers or tetramers [13], [17] thought to be essential in formation of the mature viral nucleocapsid and aiding in compacting the RNA/nucleocapsid complex [13], [14], [15], [30]. Deletion of the SR-rich region not only completely abolished the oligimerization of the N-protein but also resulted in a disordered distribution of N-proteins in mammalian cells [17]. Considering that the region responsible for self-association identified by these previous studies was identical to the cleavage region identified in this study, it is reasonable to speculate that the cleavage of this sequence may aid in the unpacking of viral RNA needed to allow this RNA to serve as a template for viral genome replication. If binding and subsequent lysis of the SR-rich amino acid region are inherent properties of the SARS N-protein, the SR-rich amino acid sequence presents a potential target for a SARS therapeutic and an agent that prevents its cleavage (perhaps a competitive inhibitor such as a mimetic of the SR-region) may prove effective in interrupting viral replication in SARS-infected individuals.
Acknowledgment
Thanks to Dr. Laura Stewart for critical input into the manuscript.
References
- 1.Drosten C., Gunther S., Preiser W., van der W.S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 2.Zhu M.S., Pan Y., Chen H.Q., Shen Y., Wang X.C., Sun Y.J., Tao K.H. Induction of SARS-nucleoprotein-specific immune response by use of DNA vaccine. Immunol. Lett. 2004;92:237–243. doi: 10.1016/j.imlet.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao J., Huang Q., Wang W., Zhang Y., Lv P., Gao X.M. Identification and characterization of dominant helper T-cell epitopes in the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:6079–6088. doi: 10.1128/JVI.02568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y.H., Li J., Liu X.E., Wang L., Li T., Zhou Y.H., Zhuang H. Detection of the nucleocapsid protein of severe acute respiratory syndrome coronavirus in serum: comparison with results of other viral markers. J. Virol. Methods. 2005;130:45–50. doi: 10.1016/j.jviromet.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li F.Q., Xiao H., Tam J.P., Liu D.X. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005;579:2387–2396. doi: 10.1016/j.febslet.2005.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Surjit M., Lal S.K. The SARS-CoV nucleocapsid protein: a protein with multifarious activities. Infect. Genet. Evol. 2007 doi: 10.1016/j.meegid.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J., Ji J., Ye J., Zhao X., Wen J., Li W., Hu J., Li D., Sun M., Zeng H., Hu Y., Tian X., Tan X., Xu N., Zeng C., Wang J., Bi S., Yang H. The structure analysis and antigenicity study of the N protein of SARS-CoV. Genomics Proteomics Bioinformatics. 2003;1:145–154. doi: 10.1016/S1672-0229(03)01018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Wu X., Wang Y., Li B., Zhou H., Yuan G., Fu Y., Luo Y. Low stability of nucleocapsid protein in SARS virus. Biochemistry. 2004;43:11103–11108. doi: 10.1021/bi049194b. [DOI] [PubMed] [Google Scholar]
- 9.Garzon M.T., Lidon-Moya M.C., Barrera F.N., Prieto A., Gomez J., Mateu M.G., Neira J.L. The dimerization domain of the HIV-1 capsid protein binds a capsid protein-derived peptide: a biophysical characterization. Protein Sci. 2004;13:1512–1523. doi: 10.1110/ps.03555304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ihnat P.M., Vellekamp G., Obenauer-Kutner L.J., Duan J., Han M.A., Witchey-Lakshmanan L.C., Grace M.J. Comparative thermal stabilities of recombinant adenoviruses and hexon protein. Biochim. Biophys. Acta. 2005;1726:138–151. doi: 10.1016/j.bbagen.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Lidon-Moya M.C., Barrera F.N., Bueno M., Perez-Jimenez R., Sancho J., Mateu M.G., Neira J.L. An extensive thermodynamic characterization of the dimerization domain of the HIV-1 capsid protein. Protein Sci. 2005;14:2387–2404. doi: 10.1110/ps.041324305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yelamos B., Nunez E., Gomez-Gutierrez J., Datta M., Pacheco B., Peterson D.L., Gavilanes F. Circular dichroism and fluorescence spectroscopic properties of the major core protein of feline immunodeficiency virus and its tryptophan mutants Assignment of the individual contribution of the aromatic sidechains. Eur. J. Biochem. 1999;266:1081–1089. doi: 10.1046/j.1432-1327.1999.00952.x. [DOI] [PubMed] [Google Scholar]
- 13.He R., Leeson A., Ballantine M., Andonov A., Baker L., Dobie F., Li Y., Bastien N., Feldmann H., Strocher U., Theriault S., Cutts T., Cao J., Booth T.F., Plummer F.A., Tyler S., Li X. Characterization of protein–protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Res. 2004;105:121–125. doi: 10.1016/j.virusres.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C.Y., Chang C.K., Chang Y.W., Sue S.C., Bai H.I., Riang L., Hsiao C.D., Huang T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neuman B.W., Adair B.D., Yoshioka C., Quispe J.D., Orca G., Kuhn P., Milligan R.A., Yeager M., Buchmeier M.J. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006;80:7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang X., Ye L.B., Zhang Y., Li B., Li S., Kong L., Wang Y., Zheng H., Wang W., Wu Z. Nucleocapsid amino acids 211 to 254, in particular, tetrad glutamines, are essential for the interaction between the nucleocapsid and membrane proteins of SARS-associated coronavirus. J. Microbiol. 2006;44:577–580. [PubMed] [Google Scholar]
- 17.He R., Dobie F., Ballantine M., Leeson A., Li Y., Bastien N., Cutts T., Andonov A., Cao J., Booth T.F., Plummer F.A., Tyler S., Baker L., Li X. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004;316:476–483. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu I.M., Gustafson C.L., Diao J., Burgner J.W., Li Z., Zhang J., Chen J. Recombinant severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein forms a dimer through its C-terminal domain. J. Biol. Chem. 2005;280:23280–23286. doi: 10.1074/jbc.M501015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang D., Pereboev A.V., Korokhov N., He R., Larocque L., Gravel C., Jaentschke B., Tocchi M., Casley W.L., Lemieux M., Curiel D.T., Chen W., Li X. Significant alterations of biodistribution and immune responses in Balb/c mice administered with adenovirus targeted to CD40(+) cells. Gene Ther. 2008;15:298–308. doi: 10.1038/sj.gt.3303085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Schagger H., von J.G. Tricine–sodium dodecyl sulfate–polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 22.Jiminez C.R., Huang L., Qiu Y., Burlingame A.L. In gel digestion of proteins for MALDI-MS fingerprint mapping. In: Taylor G.P., editor. Current Protocols in Protein Science. John Wiley and Sons, Inc.; Hoboken: 1998. pp. 16.4.1–16.4.5. [DOI] [PubMed] [Google Scholar]
- 23.Simons B.L., Kaplan H., Fournier S.M., Cyr T., Hefford M.A. A novel cross-linked RNase A dimer with enhanced enzymatic properties. Proteins. 2007;66:183–195. doi: 10.1002/prot.21144. [DOI] [PubMed] [Google Scholar]
- 24.Ying W., Hao Y., Zhang Y., Peng W., Qin E., Cai Y., Wei K., Wang J., Chang G., Sun W., Dai S., Li X., Zhu Y., Li J., Wu S., Guo L., Dai J., Wang J., Wan P., Chen T., Du C., Li D., Wan J., Kuai X., Li W., Shi R., Wei H., Cao C., Yu M., Liu H., Dong F., Wang D., Zhang X., Qian X., Zhu Q., He F. Proteomic analysis on structural proteins of severe acute respiratory syndrome coronavirus. Proteomics. 2004;4:492–504. doi: 10.1002/pmic.200300676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matayoshi E.D., Wang G.T., Krafft G.A., Erickson J. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science. 1990;247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 26.Breidenbach M.A., Brunger A.T. New insights into clostridial neurotoxin-SNARE interactions. Trends Mol. Med. 2005;11:377–381. doi: 10.1016/j.molmed.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 27.Diemer C., Schneider M., Seebach J., Quaas J., Frosner G., Schatzl H.M., Gilch S. Cell type-specific cleavage of nucleocapsid protein by effector caspases during SARS coronavirus infection. J. Mol. Biol. 2008;376:23–34. doi: 10.1016/j.jmb.2007.11.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guruprasad K., Reddy B.V., Pandit M.W. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990;4:155–161. doi: 10.1093/protein/4.2.155. [DOI] [PubMed] [Google Scholar]
- 29.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker J.M., editor. The Proteomics Protocols Handbook. Humana Press; Totowa: 2005. pp. 571–607. [Google Scholar]
- 30.Tan Y.W., Fang S., Fan H., Lescar J., Liu D.X. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]