Abstract
Caused by a new coronavirus, severe acute respiratory syndrome (SARS) is a highly contagious disease associated with significant fatality that emerged in 2003. The molecular cause of the unusually high human pathogenicity of the SARS coronavirus (SARS-CoV) is still unknown. In an effort to characterize molecular components of the virus that are absent in other coronaviruses, all of which are considerably less pathogenic for humans, we recombinantly produced the SARS-unique domain (SUD) within non-structural protein 3 (Nsp3) of SARS-CoV and characterized its nucleic-acid binding properties. Zone-interference gel electrophoresis and electrophoretic mobility shift assays revealed a specific affinity of SUD for oligo(G)-strings. A few such segments are present in the SARS-CoV genome, but also in mRNAs of host proteins involved in the regulation of signaling pathways. A putative role of SUD in virus-induced apoptosis or survival of host cells is discussed.
Keywords: Coronavirus, SARS, Nsp3, SUD, Non-structural protein, bbc3, Apoptosis, In-vitro translation, Oligo(G)-binding, Zone-interference gel electrophoresis, ZIGE
Severe acute respiratory syndrome (SARS) is a highly infectious disease that first surfaced in 2002/2003 and was associated with a fatality rate of about 10% [1]. A new coronavirus, SARS-CoV, was identified as the causative agent [1], [2], [3]. SARS-CoV is an enveloped virus with a large (29.7 kb) single-stranded RNA genome of positive sense that encodes several structural and auxiliary proteins as well as two large overlapping polyproteins, pp1a (∼486 kDa) and pp1ab (∼790 kDa) [4]. These polyproteins are processed by two proteases to yield 16 non-structural proteins (Nsp1–Nsp16), which are essential for RNA replication and processing.
Nsp3 is the largest among the non-structural proteins of SARS-CoV. It is believed to comprise at least six domains (Fig. 1 A): (1) an N-terminal acidic domain which is rich in glutamate; (2) the ‘X domain’ which binds poly(ADP-ribose) and has a fold similar to the macrodomain of histone 2A [5], [6]; (3) the SARS-unique domain (SUD) which has only been found in SARS-CoV so far [4], [7]; (4) a papain-like protease (PL2pro), which also exhibits deubiquitinating activity and is involved in suppression of the type-I interferon response of the host cell [8], [9], [10], [11], [12]; (5) an additional, non-canonical papain-like domain identified by bioinformatics (Ferron and Canard, personal communication); and (6) the ‘Y domain’, which contains an N-terminal transmembrane subdomain [13]. Except for the PL2pro, the functions of these domains are unclear. Some may play a role as part of the replicase/transcriptase complex of the virus, and others may interact with components of the host cell. A recent study of the interactome of the proteins of the SARS coronavirus indicated some interaction between an N-terminal fragment of Nsp3 with Nsp2 and with Orf3a. However, no results were obtained for the SARS-unique domain (SUD) because for cloning reasons, it was cleaved into two halves [14].
Fig. 1.
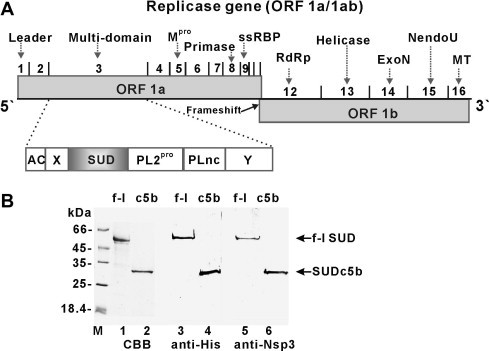
(A) Organization of SARS-CoV genome. The location of different Nsps in ORF1a/1ab and their function is indicated. (B) Expression of full-length SUD (f-l) and SUDc5b (c5b). SDS–PAGE (15%, lanes 1 and 2) and Western blot using anti-(His)4-antibody (lanes 3 and 4) and anti-Nsp3-antibody (lanes 5 and 6).
We believe that the SUD might be of particular interest for elucidating the basis for the high pathogenicity of SARS-CoV for humans. All other coronaviruses, including the recently discovered human coronaviruses NL63 [15] and HKU1 [16], do not have this domain, and since they are much less pathogenic for humans, it is conceivable that there is a correlation between the presence of the SUD and viral pathogenicity. We therefore decided to produce this domain using recombinant DNA technology and in-vitro protein synthesis in order to study its interactions with components of the virus and the host cell. In this communication, we show for the first time that full-length SUD as well as a truncated version bind oligo(G) stretches (“G-strings”) in nucleic acids.
Materials and methods
In-vitro protein synthesis. For expression of the SUD, a plasmid encoding the full-length domain with an N-terminal His-tag and a factor Xa recognition sequence was constructed by transfer of the NcoI-XhoI fragment from pETBlue-2-SUD (see below) into the dephosphorylated pIVEX1.3-WG vector (Roche) cleaved with the same restriction enzymes. After expression using the RTS wheat-germ continuous-flow cell-free system (Roche), the His-tagged protein was purified using immobilized metal-affinity chromatography (IMAC; HisTrap HP column), followed by elimination of imidazole through a HiTrap desalting column (GE HealthCare) equilibrated with 50 mM Tris–HCl, 300 mM NaCl, pH 7.5. Protein purity was estimated by 15% SDS–polyacrylamide gel electrophoresis (PAGE) under denaturing conditions. After transfer to a nitrocellulose membrane (Schleicher & Schuell), the protein was immunologically detected by anti-His (Novagen) or anti-Nsp3 (Rockland) antibodies.
Expression of full-length SUD in Escherichia coli. For the production of the full-length SUD in E. coli, the coding region corresponding to residues 349–726 of Nsp3 was PCR-amplified using sense (GGATTATCTTGATAACCTGAAGC) and anti-sense (GTCTTAACCTCCCGCAGGGATAA) primers and SARS-CoV cDNA (TOR2 strain; Accession No. AY274119; kind gift of Dr. J. Ziebuhr) as template. The PCR product was cloned into the EcoRV site of the plasmid pETBlue-2 (Novagen), resulting in a gene encoding a protein with three additional amino acids (M-A-M) at the N-terminus and a C-terminal extension of 56 residues including a C-terminal (His)6 tag. The fusion gene was expressed in E. coli Tuner DE3 (pLacI) cells (Novagen).
Expression of the SUDc5b fragment. Step-wise N- and C-terminal truncations led to a stable minimal version of the SUD protein consisting of residues 389–562 of Nsp3, named SUDc5b. The coding region for this protein was PCR-amplified from the plasmid encoding the full-length protein and ligated into the StuI site of the plasmid pQE30, providing an N-terminal His-tag and a short linker sequence. E. coli M15 (pRep4) was used as expression host for this construct. The coding regions of all expression plasmids were verified by DNA sequencing.
Purification of SUDc5b. The cells were resuspended in lysis buffer (10 mM imidazole, 300 mM NaCl, 25 mM NaH2PO4, pH 8.0) and disrupted with a sonicator. After centrifugation at 16,000g (20 min, 4 °C), Benzonase was added to the supernatant, followed by ultracentrifugation at 38,000g (1 h, 4 °C). SUDc5b was purified by IMAC and desalted as described above for the full-length protein. Further purification was achieved by ion-exchange chromatography using a gradient from 25 mM to 1 M NaCl. In all experiments, protein concentration was determined using the BCA Protein Assay Kit (Pierce) or by absorbance at 280 nm. The protein was stored at 4 °C.
Analytical size-exclusion chromatography. The apparent molecular mass of SUDc5b was determined by an analytical BioSep-SEC-S 3000 column (Phenomenex) connected to an HPLC DuoFlow (Bio-Rad). The column was calibrated in 25 mM Tris–HCl, 300 mM NaCl, 0.5 mM EDTA, pH 7.5, using the following standard proteins: cytochrome c (12.5 kDa), chymotrypsinogen (25.0 kDa), ovalbumin (42.75 kDa), bovine serum albumin (66.4 kDa), and glucose oxidase (160 kDa). The sample volume was 25 μl with a concentration of 2 mg/ml.
Dynamic light-scattering. Dynamic light-scattering (DLS) was performed using a Laser-Spectroscatter 201 (RiNA GmbH). After centrifugation for 1 h at 4 °C and 13,000g, sample (20 μl) was applied to a 10-mm quartz cuvette (Hellma).
Electrophoretic mobility shift analysis (EMSA). Radioactive RNA probes bbc3-G14 and bbc3-A14 (see Results and discussion), respectively, were transcribed by T7 RNA polymerase in the presence of [α-32P]-GTP (3000 Ci/mmol, Hartmann) from the following deoxyoligonucleotide duplexes containing the T7 promoter (underlined, only the sense strand 5′-to-3′ direction is indicated): TGTAATACGACTCACTATAGGGTGACACTGGGGGGGGGGGGGGCTCTCCTCTCGGTGCTCCTTCACTC and TGTAATACGACTCACTATAGGGTGACACTAAAAAAAAAAAAAACTCTCCTCTCGGTGCTCCTTCACTC. EMSA was performed with radioactive RNA probes as described [17] with the following modifications: the reaction mixture containing 32P-labeled RNA (1–3 × 104 cpm) and 1–5 μg of the protein tested in binding buffer (5 mM Hepes, pH 7.9, 25 mM KCl, 2 mM MgCl2, 1.75 mM ATP, 6 mM DTT, 0.05 mM phenylmethylsulfonyl fluoride, 166 μg/ml of E. coli tRNA, and 5% glycerol) was incubated for 20 min at 30 °C. For electrophoresis, non-denaturing polyacrylamide 4–12% gradient TBE gels (Invitrogen) were used. RNA–protein complexes were visualized using a PhosphorImager (Fujifilm BAS 1000) and the software PCBAS (Raytest).
Zone-interference gel electrophoresis. The zone-interference gel electrophoresis (ZIGE) device was adapted from [18] (see Fig. S1 in Supplementary Material). The agarose was dissolved in 1× TBE buffer. The protein was incubated at RT for 0.5 h with deoxyoligonucleotides of different lengths (6–14 and 24 in case of bbc3), with concentrations of 0–32 μM in a sample volume of 10 μl. The samples were mixed with dimethylsulfoxide (DMSO; final concentration of 10% (v/v)) and a trace of bromophenolblue (BPB). These protein-oligonucleotide samples were applied to the small slots (see Fig. S1). Oligonucleotide with the same concentration as in the small slots was also mixed with DMSO and BPB in 1× TBE buffer and applied to the long slots of the gel (total volume 100 μl). Electrophoresis was performed at 4 °C for 1 h with a constant current of 100 mA. Staining was performed as outlined in [18].
Results and discussion
The DNA sequence encoding the full-length SUD (residues 349–726 of Nsp3; see Fig. 1A), also referred to as Nsp3c, was successfully expressed with an N-terminal His-tag in E. coli (not shown) and in the in-vitro RTS wheat-germ system and purified to homogeneity (Fig. 1B, lane 1). Besides the full-length SUD, a soluble, more stable minimal version, SUDc5b, was constructed by systematic N- and C-terminal truncation and produced, with an N-terminal His-tag, in E. coli (Fig. 1B, lane 2). The identity of the proteins was verified by N-terminal sequencing (not shown) and reaction with anti-(His)4 and anti-Nsp3 (SARS-CoV) (Fig. 1B, lanes 3–6).
Independent of the expression system used, the full-length SUD displayed a strong and reproducible tendency to proteolytic (self-)degradation. This was reduced but still detectable in the preparations of the SUDc5b protein. Analytical size-exclusion chromatography and dynamic light-scattering measurements indicated that SUDc5b is a monomeric protein (31 kDa) (see Figs. S2 and S3A in Supplementary Material).
We were able to show that both full-length SUD and its truncated version, SUDc5b, bind single-stranded DNA and RNA. For short nucleotides (decamers), this interaction is highly specific for oligo-guanidine nucleotides. Both deoxyribo- and ribo-G10 are bound with comparable affinities (Fig. 2 A and B). K d values derived from zone-interference gel electrophoresis (ZIGE; [18]) are in the low micromolar range (in case of SUDc5b, 1.0 μM for (dG)10 and 0.93 μM for G10). The binding affinity is dependent on the length of the oligo(G) nucleotide. For SUDc5b, the highest affinity was found for (dG)12–14 (not shown). For full-length SUD, the K d value derived from ZIGE is about 2.1 μM (Fig. 2C).
Fig. 2.
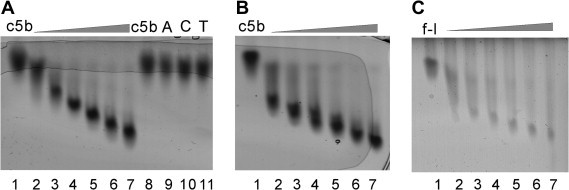
Zone-interference gel electrophoresis experiments (ZIGE) performed to evaluate the affinity of SUD for different nucleic-acid species. (A) SUDc5b in presence of increasing concentrations (1, 2, 4, 8, 16, and 32 μM, lanes 2–7) of (dG)10. Lanes labeled c5b, A, C, and T correspond to nucleic acid-free protein, mixture of SUDc5b with 16 μM of (dA)10, (dC)10, and (dT)10, respectively. (B) SUDc5b in presence of increasing concentrations (1, 2, 4, 8, 16, and 32 μM, lanes 2–7) of ribo-G10. (C) Full-length SUD (f-l) in presence of increasing concentrations (1, 2, 4, 8, 16, and 32 μM, lanes 2–7) of (dG)10.
A database and literature search for potential oligo(G) stretches that might be recognized by SUD within the host cell revealed interesting potential targets. Since SUD does not seem to differentiate between DNA and RNA, the availability of potential binding sites will depend on the sub-cellular localization of the protein. Oligo(dG) sequences, so called G-strings, have been identified as binding sites for transcription factors in eukaryotic cells [19], [20]. Since there is no evidence for a nuclear localization of Nsp3 in the host cell [8], [14], we focused on potential cytosolic interaction partners. Oligo(G) sequences are found in 3′-UTRs of several mRNAs including the mRNA for the human pro-apoptotic protein Bbc3 (Accession No. U82987; [21]), the transcript variant 1 of the mRNA for the human MAP kinase 1 (NM_002745), the mRNA for the human RAB6B protein, a member of the Ras oncogene family (NM_016577), and the mRNA for TAB3, a component of the NF-κB signaling pathway (NM_152787). These proteins are prime candidates for an interference of the virus with cellular signaling pathways. Changes in the stability and/or translation efficiency of these mRNAs due to the binding of a regulatory factor should result in an altered response of the infected cell to extracellular signals.
The truncated SUDc5b binds efficiently (K d = 0.45 μM) to an oligodeoxynucleotide (named bbc3-(dG)14) of 24 bases in length, with a central (dG)14 stretch, which we used as a model for the bbc3 mRNA. The replacement of the (dG)14 stretch by (dA)14 leads to a dramatically reduced binding affinity (Fig. 3 ). Similar results were obtained for full-length SUD (not shown).
Fig. 3.
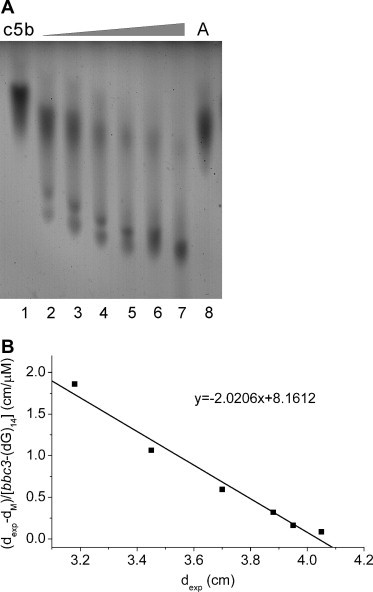
ZIGE results illustrating the complex formation of SUDc5b with bbc3-(dG)14. (A) SUDc5b in presence of increasing concentrations (1, 2, 4, 8, 16, and 32 μM, lanes 2–7) of bbc3-(dG)14. Lanes labeled c5b and A correspond to nucleic acid-free protein and mixture of SUDc5b with 32 μM of bbc3-(dA)14 (lane 8), respectively. The sequence of bbc3-(dG)14 is 5′-ACA CTG GGG GGG GGG GGG GCT CTC-3′, and the sequence of bbc3-(dA)14 is 5′-ACA CTA AAA AAA AAA AAA ACT CTC-3′. (B) The Kd value is derived from the slope of the graph [18].
To further assess the RNA-binding affinity of SUD and SUDc5b, a radioactive RNA probe (named bbc3-G14) containing 14 G bases and flanking regions (a total of 48 bases) from the bbc3 gene (nucleotides 983–1030) was transcribed by T7 RNA polymerase from a synthetic deoxynucleotide duplex template harbouring the T7 promoter [22]. As a control for the specificity of the RNA–protein interaction, an RNA probe (bbc3-A14) containing an A14-string instead of G14 was prepared in a similar manner. An electrophoretic mobility shift assay (EMSA) was used to probe the interaction between SUD proteins and RNAs. To prevent non-specific binding of radioactive RNA probes to the proteins, a 1000-fold molar excess of unlabeled tRNA was added to the reaction mixtures. As can be seen from Fig. 4 A, full-length SUD is able to retard the mobility of the radioactive bbc3-G14 RNA in a concentration-dependent manner (lanes 1–4). An anti-Nsp3 antibody that had been tested for specific interaction with SUD (see Fig. 1B, lanes 5 and 6), but not an irrelevant anti-T7 RNA polymerase antibody, supershifts the migration of the SUD–RNA complex (Fig. 4A, lanes 7 and 8, respectively), implying direct and highly specific interaction between SUD and bbc3-G14. Both full-length SUD (Fig. 4A, lanes 2–4 and 6) and SUDc5b (Fig. 4B, lane 1) are able to retard the electrophoretic mobility of free bbc3-G14 RNA due to RNA–protein complex formation. In contrast, bbc3-A14 RNA is unable to form a similar complex either with full-length SUD (Fig. 4B, lane 4) or with SUDc5b (Fig. 4B, lane 5), thus confirming the specificity of the RNA–protein interaction. Interestingly, the appearance of an additional, extremely slow-migrating band was observed with SUDc5b and bbc3-G14 RNA (Fig. 4B, lane 1, asterisk), but not with bbc3-A14 RNA (lane 5). This is possibly due to a multimerization of a specific RNA–protein complex. The ability of SUDc5b to form multimers in presence of (dG)8 was confirmed by dynamic light-scattering (Fig. S3B). No interaction of bovine serum albumin (BSA), included as a further control of specificity, with bbc3-G14 RNA (Fig. 4B, lane 2) or with bbc3-A14 RNA (Fig. 4B, lane 6) was detectable.
Fig. 4.
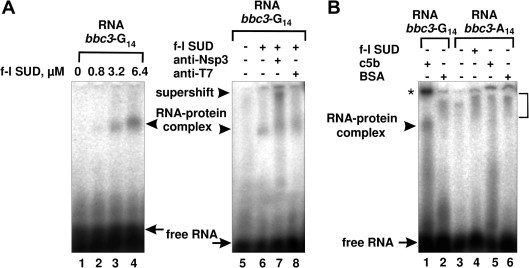
(A) Concentration-dependent binding of full-length SUD to [32P]-labeled (1.5 × 105 cpm) bbc3-G14 RNA representing a segment of bbc3 mRNA, and demonstration of specificity of interaction by the formation of a supershift. Indicated amounts of the full-length SUD (lanes 1–4) were incubated in binding mixtures before electrophoresis. In lanes 7 and 8, full-length SUD (6.4 μM) was incubated either with anti-Nsp3 or with anti-T7 RNA polymerase antibodies. (B) Specificity of interaction to bbc3-G14 RNA. Full-length SUD (lane 4), its truncated form c5b (lanes 1 and 5), or BSA (lanes 3 and 6) were incubated with radioactive bbc3-G14 RNA (lanes 1 and 2) or with control RNA, bbc3-A14 (lanes 3–6). Retarded and supershifted RNA–protein complexes are shown with arrowheads. Free RNAs are shown with arrows. Unspecific retardation of radioactivity, detected also in lanes with free RNAs and BSA, is marked with the bracket.
The observed affinity of SUD to the G-string of bbc3 RNA (bbc3-G14) is highly specific, since a 1000-fold molar excess of tRNA is not able to prevent the formation of the complex. Moreover, the exchange of G14 to A14 completely prevents the interaction.
Other possible interaction partners for SUD are the coronaviral RNAs within the infected cells. We found three potential binding sites in the genome of the TOR2 isolate of the SARS coronavirus. A single G6 motif (nucleotides 21,604–21,609) and two C6-motives (nucleotides 17,386–17,391 and 28,157–28,162) are present in the (+)-strand (Fig. S4A). SUDc5b also binds to G6-containing deoxyribonucleotides derived from these regions of the viral genome, again with an estimated affinity of approximately 1 μM (Fig. S4B and C). The binding affinity to the reversed complementary sequences representing the opposite strand of the genome is considerably lower (Fig. S4). However, in accordance with the preferred length of the oligo(G) stretch of 12–14 nucleotides, the highest binding affinity was found for the oligonucleotides derived from the human bbc3 mRNA.
As mentioned before, G11–15 motifs are present in the 3′-UTRs of human mRNAs encoding Bbc3, MAP kinase 1, RAB6B, and TAB3 proteins, all of which are components of cellular signaling pathways. It is conceivable that binding of SUD to these RNAs will result in an alteration of their stability, or translation efficiency, as it has been demonstrated for eukaryotic trans-acting factors such as HuR, CP1, and TIA-1 [23], [24], [25], [26], and thereby will interfere with cellular signal transduction. Bbc3 is a pro-apoptotic protein that is induced by a variety of apoptotic stimuli. It is subject to transcriptional regulation by multiple cell death-signaling pathways [21] and has been demonstrated to mediate apoptosis in neuroblastoma cells [27]. Blocking Bbc3 production at the translation level could override the transcriptional activation and render a cell less sensitive to apoptotic signals. Similarly, an interaction of SUD with the TAB3 mRNA may contribute to the recently described NF-κB activation and the increased CXCL10 (interferon γ-inducible protein 10) levels observed in the blood of SARS patients [28]. In addition to the spike, nucleocapsid, Orf3b, Orf6, Orf7a, and PL2pro proteins [12], [28], SUD could thus be another component of SARS-CoV involved in the modulation of host-cell signaling pathways.
Acknowledgments
This work was supported by the Sino-European Project on SARS Diagnostics and Antivirals (SEPSDA) of the European Commission (contract number SP22-CT-2004-003831), the Sino-German Center for Promotion of Research, Beijing, and the Schleswig-Holstein Innovation Fund. R.H. thanks the Fonds der Chemischen Industrie for continuous support.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bbrc.2007.10.081.
Appendix A. Supplementary data
Supplementary Figure 1.
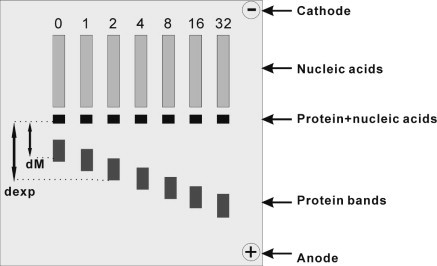
Schematic view of zone-interference gel electrophoresis (ZIGE). Adapted from Abrahams et al. (Nucleic Acid. Res. 16 (1988) 10099–10108).
Supplementary Figure 2.
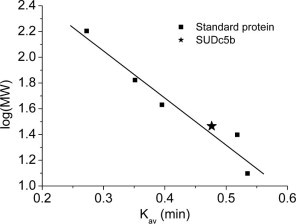
Calibration curve of analytical gel filtration for SUDc5b. Standard proteins and SUDc5b are marked as squares and asterisk, respectively ; Ve, elution volume; V0, void volume; Vt, total volume; MW, calculated molecular mass.
Supplementary Figure 5.
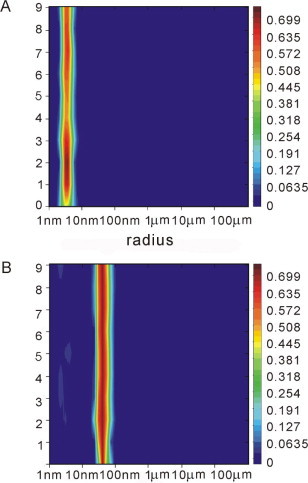
Dynamic light-scattering of SUDc5b (A) and SUDc5b incubated with (dG)8 (B). The abscissa shows the particle radius, the ordinate the number of measurements.
Supplementary Figure 4.
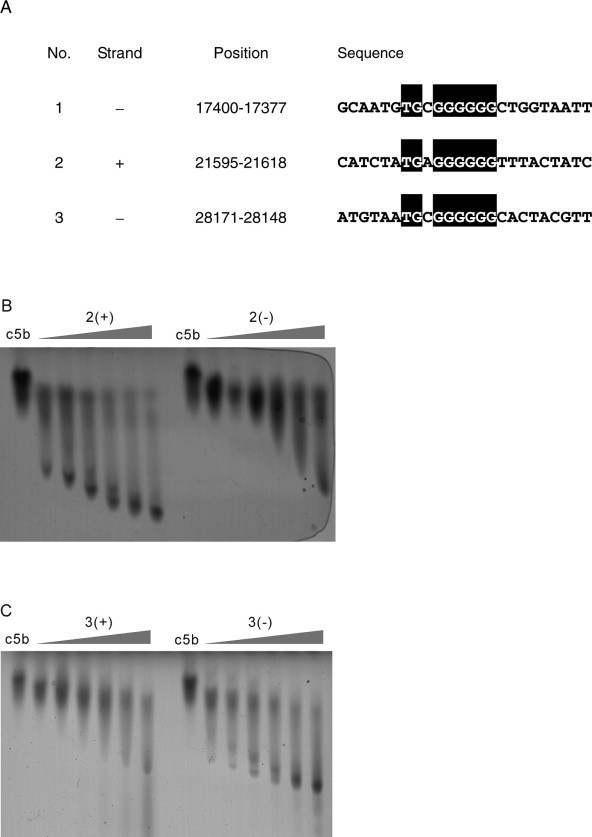
The interactions between SUDc5b and DNA fragments derived from the SARS-CoV genome. (A) G6-motives in the genome of the SARS coronavirus TOR2 (Accession No. AY274119). The numbering refers to the positions on the (+)-strand. The sequences from the (−)-strand are listed as the reverse complement. (B) ZIGE of SUDc5b in presence of increasing concentrations (1, 2, 4, 8, 16, and 32 μM) of 2(+) and 2(−). The sequences of 2(+) and 2(−) are 5′-CAT CTA TGA GGG GGG TTT ACT ATC-3′ and 5′-GAT AGT AAA CCC CCC TCA TAG ATG-3′, respectively. (C) ZIGE of SUDc5b in presence of increasing concentrations (1, 2, 4, 8, 16, and 32 μM) of 3(+) and 3(−). The sequence of 3(+) and 3(−) are 5′-AAC GTA GTG CCC CCC GCA TTA CAT-3′ and 5′-ATG TAA TGC GGG GGG CAC TAC GTT-3′, respectively.
References
- 1.Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Cheung M.T., Cheng V.C. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1978. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weißbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 5.Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K., Velasquez J., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1′′-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., Ziebuhr J., Ahola T., Canard B. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snijder E.J., Bredenbeek P.J., Dobbe1 J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;24:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;24:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.T., Baker S.C., Li K. Regulation of IRF-3 dependent innate immunity by the papain-like protease domain of the SARS coronavirus. J. Biol. Chem. 2007 doi: 10.1074/jbc.M704870200. M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanjanahaluethai A., Chen Z., Jukneliene D., Baker S.C. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology. 2007;361:391–401. doi: 10.1016/j.virol.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein–protein interactions of the SARS coronavirus ORFeome. PLoS ONE. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusov Y., Gauss-Müller V. In vitro RNA binding of the hepatitis A virus proteinase 3C (HAV 3Cpro) to secondary structure elements within the 5′ terminus of the HAV genome. RNA. 1997;3:291–302. [PMC free article] [PubMed] [Google Scholar]
- 18.Abrahams J.P., Kraal B., Bosch L. Zone-interference gel electrophoresis: a new method for studying weak protein–nucleic acid complexes under native equilibrium conditions. Nucleic Acid Res. 1988;16:10099–10108. doi: 10.1093/nar/16.21.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis C.D., Clark S.P., Felsenfeld G., Gould H. An erythrocyte-specific protein that binds to the poly(dG) region of the chicken beta-globin gene promoter. Genes Dev. 1988;2:863–873. doi: 10.1101/gad.2.7.863. [DOI] [PubMed] [Google Scholar]
- 20.Xiang M., Lu S.-Y., Musso M., Karsenty G., Klein W.H. A G-string positive c/s-regulatory element in the LpS1 promoter binds two distinct nuclear factors distributed non-uniformly in Lytechinus pictus embryos. Development. 1991;113:1345–1355. doi: 10.1242/dev.113.4.1345. [DOI] [PubMed] [Google Scholar]
- 21.Han J.-W., Flemington C., Houghton A.B., Gu Z., Zambetti G.P., Lutz R.J., Zhu L., Chittenden T. Expression of bbc3, a-pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc. Natl. Acad. Sci. USA. 2001;98:11318–11323. doi: 10.1073/pnas.201208798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acid Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollams E.M., Giles K.M., Thomson A.M., Leedman P.J. mRNA stability and the control of gene expression: implications for human disease. Neurochem. Res. 2002;27:957–980. doi: 10.1023/a:1020992418511. [DOI] [PubMed] [Google Scholar]
- 24.Giles K.M., Daly J.M., Beveridge D.J., Thomson A.M., Voon D.C., Furneaux H.M., Jazayeri J.A., Leedman P.J. The 3′-untranslated region of p21WAF1 mRNA is a composite cis-acting sequence bound by RNA-binding proteins from breast cancer cells, including HuR and poly(C)-binding protein. J. Biol. Chem. 2003;278:2937–2946. doi: 10.1074/jbc.M208439200. [DOI] [PubMed] [Google Scholar]
- 25.Kawai T., Lal A., Yang X., Galban S., Mazan-Mamczarz K., Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol. Cell. Biol. 2006;26:3295–3307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X-L., Andersen J.B., Ezelle H.J., Wilson G.M., Hassel B.A. Post-transcriptional regulation of RNase-L expression is mediated by the 3′-untranslated region of its mRNA. J. Biol. Chem. 2007;282:7950–7960. doi: 10.1074/jbc.M607939200. [DOI] [PubMed] [Google Scholar]
- 27.Wie J.S., Whiteford C.C., Cenacchi N., Son C.G., Khan J. BBC3 mediates fenretinide-induced cell death in neuroblastoma. Oncogene. 2005;24:7976–7983. doi: 10.1038/sj.onc.1208947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizutani T. Signal transduction in SARS-CoV-infected cells. Ann. NY Acad. Sci. 2007;1102:86–95. doi: 10.1196/annals.1408.006. [DOI] [PMC free article] [PubMed] [Google Scholar]


