Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) 3a protein is one of the opening reading frames in the viral genome with no homologue in other known coronaviruses. Expression of the 3a protein has been demonstrated during both in vitro and in vivo infection. Here we present biochemical data to show that 3a is a novel coronavirus structural protein. 3a was detected in virions purified from SARS-CoV infected Vero E6 cells although two truncated products were present predominantly instead of the full-length protein. In Vero E6 cells transiently transfected with a cDNA construct for expressing 3a, a similar cleavage was observed. Furthermore, co-expression of 3a, membrane and envelope proteins using the baculovirus system showed that both full-length and truncated 3a can be assembled into virus-like particles. This is the first report that demonstrated the incorporation of 3a into virion and showed that the SARS-CoV encodes a novel coronavirus structural protein.
Keywords: Severe acute respiratory syndrome, SARS coronavirus, 3a viral protein, Structural protein, Baculovirus, Virus-like particles
The recent severe acute respiratory syndrome (SARS) epidemic, which affected over 30 countries, resulted in more than 8000 cases of infection and more than 800 fatalities (World Health Organization, http://www.who.int/csr/sars/country/en/). A novel coronavirus was identified as the etiological agent of SARS [1]. Like all coronaviruses, the SARS-CoV genome encodes for four major structural proteins, nucleocapsid (N), spike (S), membrane (M), and envelope (E). The RNA is packaged by the N protein into a helical nucleocapsid [2]. The S protein, which forms morphologically characteristic projections on the virion surface, mediates binding to host receptors and membrane fusion [3]. The M protein is a triple-spanning integral membrane protein with a short ectodomain and a large carboxyl-terminus endodomain [4]. More recently, the E protein was showed to play a major role in coronavirus assembly [5], [6], [7], [8]. Another structural protein, the hemagglutinin esterase glycoprotein (HE), is found in only a subset of coronaviruses but its role in the virus life cycle has not been well established [9]. The SARS-CoV genome does not appear to encode for a HE protein [10].
In addition, the SARS-CoV genome also contains another nine open reading frames (ORFs) with no homologue in other known coronaviruses [10], [11]. 3a (also known as X1 in [10], as ORF3 in [11], and as U274 in [12], [13]) is the largest of these unique ORFs and consists of 274 amino acids. Three groups have independently reported the expression of 3a in SARS-CoV infected cells [13], [14], [15] and it was also detected in a SARS-CoV infected patient’s lung specimen [14]. Antibodies against 3a were also found in convalescent patients [12], [14], [16]. 3a was localized in the perinuclear region and was also transported to the cell surface, where it could undergo internalization [13], [14]. It is tempting to postulate that 3a is a structural protein as only the coronavirus structural proteins, S, HE, and E, have been shown to be transported to the plasma membrane/cell surface [17], [18], [19], [20], [21].
3a can also interact with M and E, which are two key players in the viral assembly of coronaviruses, as well as with the S protein [13], [15]; hence, it may be important for viral assembly and/or release of virus from infected cells. Indeed, Zeng et al. [15] detected disulfide-linked complexes of S and 3a in the culture supernatant of SARS-CoV infected cells, indicating that 3a could be secreted together with S, possibly through the formation of virions. However, it is necessary to confirm this finding with highly purified SARS-CoV virions as viral proteins could also be released into the culture supernatant through cell lysis.
Here we provide biochemical evidence showing that 3a is a novel coronavirus structural protein. First, 3a is not only secreted into the culture supernatant of SARS-CoV infected cells, but it can also be detected in purified virion. Second, co-expression of 3a with M and E in the baculovirus system resulted in the incorporation of 3a into viral-like particles. Similar to other coronaviruses [5], [6], [7], the co-expression of SARS-CoV M and E in the baculovirus system was sufficient for the formation of viral-like particles [22], [23]. Our data showed that 3a is a novel coronavirus structural protein that is not essential for virus assembly but is incorporated into the virion when present.
Materials and methods
Construction of plasmids. The cDNA construct for expressing SARS-CoV 3a protein was obtained from SARS-CoV 2003VA2774, an isolate from a SARS patient in Singapore, as previously described [13]. pXJ3′-3a for transient expression in mammalian cells was previously described [13], while pBacPAK8-3a used for the production of recombinant baculovirus was constructed for this study using the method previously described [22].
Transient transfection of Vero E6 culture and infection with SARS-CoV. Vero E6 (African green monkey kidney epithelial) cells were purchased from the American Type Culture Collection and cultured as previously described [13]. SARS-CoV isolate from a SARS patient in China was used to infect Vero E6 cells as previously described [13], [21]. For metabolic labeling, monolayer of cells was infected at multiplicity of infection of 0.1 for 2 h, then starved with methionine (Met) and cysteine (Cys) free medium (DMEM, NEN) for 0.5 h and were then labeled for 12 h by replacing with the medium supplemented with [35S]Met and [35S]Cys (100 Ci/ml, NEN). At approximately 14 h post-infection, the cells showed approximately 50% cytopathic effects (CPE) and the culture supernatant was collected. Lysis buffer was added to the culture supernatant to give a final concentration of 50 mM Tris–HCl, pH 8, 150 mM NaCl, 0.5% NP40 (BDH Laboratory Supplies), 0.5% sodium deoxycholate, and 0.005% SDS (1× lysis buffer). The remaining cells were lysed in 1× lysis buffer. Then both cell lysates and culture supernatant were subjected to immunoprecipitation with rabbit polyclonal anti-3a antibody and protein A–Sepharose beads (Roche Molecular Biochemicals) as previously described [13]. The immuno-complexes were separated on 15% SDS–PAGE and detected by autoradiography as previously described [24], [25]. A mock infection was performed in parallel as negative control.
For detection of 3a by Western blot analysis, a similar protocol was used except that no metabolic labeling was performed. The cell lysates were subjected to Western blot as previously described [13]. For transient expression of 3a, Vero E6 cells were transfected with pXJ3′-3a using Lipofectamine reagent (Invitrogen) according to the manufacturer’s protocol. Primary antibodies for Western blot analysis (anti-3a rabbit polyclonal antibody [13] or anti-tubulin monoclonal antibody (Sigma)) were used at 1:3000 dilutions.
Quantification of autoradiographs was performed using a model Bio-Rad/GS-700 Imaging Densitometer with Bio-Rad Multi-Analyst version 1.02/Mac software.
Purification of virions from culture supernatant of SARS-CoV infected Vero E6 cells. In order to obtain purified virion from SARS-CoV infected cells, Vero E6 were infected as described above except that in order to obtain the highest virus titer, the cells were left for ∼48 h post-infection when the CPE observed was ∼90%. Then, β-propiolactone was added to infected cell culture to a final concentration of 0.05% to inactivate infectivity. The inactivation was examined by titration of treated samples in Vero E6 cells. The viruses were harvested by freezing/thawing three times and cell debris was removed by centrifugation at 5000 rpm for 10 min. Ultrafiltration was performed to concentrate viruses (300,000 NMWL, Millipore). The concentrated sample was applied to Sepharose 4B fast flow column (Pharmacia) following manufacturer’s instruction. The eluted fractions were examined by transmission electron microscopy and the fraction containing virus particles was used for Western blot analysis.
Insect cell culture and production of recombinant baculoviruses. The Spodoptera frugiperda IPLB-Sf21 (Sf21) insect cell line was cultured as a monolayer in TNM-FH insect medium, containing 8% heat-inactivated fetal bovine serum as described previously [26]. It was used for the propagation and infection of recombinant baculoviruses; titers of viruses were determined by a newly developed quantitative real-time PCR-based method [27].
Recombinant baculoviruses carrying the M and E genes were produced in a previous study [22]. Here recombinant baculoviruses carrying the 3a gene were generated using Autographa californica multiple nucleopolyhedrovirus (AcMNPV) viral genome in a similar manner. Briefly, pBacPAK8-3a was co-transfected with linearized viral DNA (BaculoGold, BD Biosciences) by using Lipofectin (Invitrogen) into insect cells and successful recombinants were isolated by the indication of red fluorescence. The recombinant baculoviruses encoding the E, M, and 3a proteins, assigned as vABhRpE, vABhRpM, and vABhRp3a respectively, were obtained by two- or three-round serial dilutions, and all viral stocks were prepared and manipulated according to the standard protocol described by O’Reilly et al. [28].
Isolation of coronavirus-like particles (VLPs). Sf21 insect cells were co-infected with vABhRpE, vABhRpM, and vABhRp3a at a multiplicity of infection of 1:5:1. At 4 days post-infection, cells were collected by a cell scraper (Costar) and then re-suspended in Tris-buffered saline (TBS), containing a cocktail of protease inhibitors (1:1000 dilution, SET III, Calbiochem), and lysed by sonication. The post-nuclear supernatant was obtained by centrifugation at 1000 rpm for 10 min and was then placed on a 30% (w/w) sucrose cushion for centrifugation at 34,000 rpm for 20 h. Pellets were washed twice with TBSs resuspended in the same buffer, and then subjected to a 20–60% (w/w) sucrose gradient at 34,000 rpm for 60 h. Twenty-eight fractions, monitored at a wavelength of 280 nm, were extracted from the centrifuged sucrose gradient using a density gradient fractionation system (ISCO). Each fraction of sucrose gradient was separated on 12% SDS–PAGE and then transferred onto a PVDF membrane (Immobilin-P, Millipore). The membrane was blocked with 5% non-fat milk for 1 h and then probed (at a dilution of 1:5000) by antiserum at 4 °C overnight. The antibody used to detect the 3a protein has been described previously [13], [29] and was developed by immunizing rabbits with bacterially expressed recombinant proteins. And the antibody for detecting the M protein was purchased from Abgent. The antibody used to detect the E protein has been described previously [22] and was raised by immunizing rabbits with a synthetic peptide. After three washes with 0.1% Tween 20 containing phosphate-buffered saline (TPBS), a horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG was added at a dilution of 1:2500 for 1 h at room temperature. The blot was then washed with TPBS four times, followed by visualization using chemiluminescent reagent (Western Lightning, Perkin-Elmer) and developed on an X-ray film (BioMax, Kodak).
Results and discussion
SARS-CoV 3a was secreted into the culture supernatant of infected Vero E6 cells and was incorporated into virions
In order to determine if the SARS-CoV 3a protein can be secreted, cell lysates and culture supernatants from metabolically labeled SARS-CoV infected Vero E6 were subjected to immunoprecipitation with a polyclonal anti-3a antibody. As shown in Fig. 1 A, a specific protein band of ∼35 kDa was detected in cell lysates obtained from SARS-CoV infected cells but not in cell lysates from mock infected cells (lanes 1 and 2). Therefore, 3a was expressed in the SARS-CoV infected cells, which is consistent with previous studies [30]. The 3a protein was also specifically immunoprecipitated from the culture supernatant of SARS-CoV infected cells (Fig. 1A, lanes 3 and 4), indicating that 3a protein was secreted into the culture supernatant. Consistently, Zeng et al. [15] recently reported the detection of disulfide-linked complexes of S and 3a in the culture supernatant of SARS-CoV infected cells. Four other proteins, with molecular weights less than 35 kDa, in the cell lysates also co-immunoprecipitated with 3a (Fig. 1A, lane 2). The sizes of these proteins are ∼24, ∼20, ∼15, and ∼8 kDa and they could be host or viral proteins that bound to 3a or they could be cleaved/degraded forms of 3a. The smallest of these co-immunoprecipitated proteins (∼8 kDa) and another protein of ∼10 kDa that co-immunoprecipitated with 3a from the culture supernatant have similar molecular weight to that predicted for the E protein (Fig. 1A, lanes 2 and 4). As the interaction between 3a and E has been previously demonstrated [13], it is likely that the E protein (∼8 and ∼10 kDa in the cell lysate and culture supernatant, respectively) expressed in the infected cells has co-immunoprecipitated with 3a. Interestingly, the relative amount of E to 3a in the immuno-complexes from culture supernatant was significantly higher when compared to the cell lysate, suggesting that 3a may need to be complexed with E before secretion out of the cells, whilst there was a pool of free 3a intracellularly. The molar ratios of E:3a, as computed by quantification of the autoradiograph shown in Fig. 1A, were 3:2 in the culture supernatant and 7:8 in the cell lysate. Based on this assumption, we estimated that the molar ratio of E to 3a in the virion is 3:2. It should be noted that this is only an approximation and future analysis of highly purified virus samples by immunoprecipitation with specific monoclonal antibodies, as recently described for transmissible gastroenteritis coronavirus by Escors et al. [31], will be required to determine an accurate stoichiometric ratio of 3a in the virion relative to E and the other structural proteins.
Fig. 1.
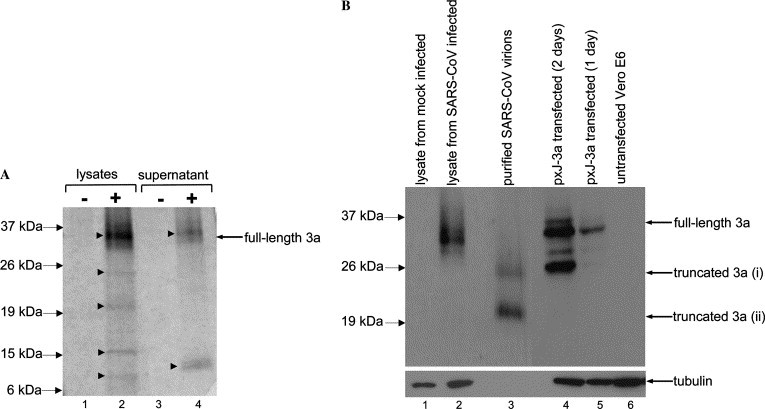
Expression of 3a in SARS-CoV infected Vero E6 cells, purified virions and transfected cells. (A) 35S metabolically labeled cell lysates or culture supernatants from mocked infected (lanes 1 and 3) or SARS-CoV infected cells (lanes 2 and 4) were subjected to immunoprecipitation with anti-3a polyclonal antibody. The 3a protein(s) or proteins that interacted with 3a were detected in SARS-CoV infected cells (marked by arrowheads, lanes 2 and 4), but not in mocked infect cells. (B) Western blot analysis, using specific anti-3a antibody (top panel), to detect for the expression of 3a in cells lysates obtained from mock infected cells (lane 1), SARS-CoV infected cells (lane 2), and in virions purified from SARS-CoV infected cells (lane 3). Cells transiently transfected with a DNA construct for expressing 3a were harvested at 1 or 2 days post-transfection and subjected to Western blot analysis (lanes 4 and 5). Untransfected Vero E6 was included as negative control (lane 6). The amounts of total cell lysates loaded for each samples were verified by measuring the level of endogenous tubulin (bottom panel). Note that no tubulin was present in the purified virion (lane 3).
Next, the presence of 3a in purified virion was determined in order to confirm if 3a was secreted as part of a virion (Fig. 1B). Due to the highly contagious nature of the SARS-CoV, it was not feasible to subject SARS-CoV to the standard sucrose gradient purification. Instead, the virus was subjected to size exclusion chromatography and the presence of virion in the eluted fractions was confirmed by electron microscopy studies (data not shown). As this was performed as part of a vaccine development program at the Wuhan Institute of Biological Products (China), infected cells were collected at late infection (90% CPE) in order to obtain the highest titer of virus. Western blot analysis showed that two truncated forms of 3a, ∼24 and ∼20 kDa, were detected in purified virion using specific anti-3a antibody (Fig. 1B, lane 3). These two truncated forms of 3a may correspond to the two protein bands of the similar sizes observed in Fig. 1A (lane 2). Curiously, the full-length 3a was not easily detectable although it was clear that the full-length 3a was expressed in and secreted from the infected cells at earlier phase of infection (50% CPE) (Fig. 1B, lane 2 and results above). No cellular tubulin, which is highly abundant in the cell lysates, was associated with the purified virion (Fig. 1B, lanes 1–3), indicating that the purification process successfully eliminated proteins that were not associated with the virion.
In order to understand the time-course for the cleavage of 3a, Vero E6 cells were transiently transfected with a cDNA construct to express 3a (Fig. 1B, lanes 4 and 5). The results showed that 3a can indeed be cleaved into smaller products by 2 days post-transfection. Collectively, the probable explanation for our observation of only truncated forms of 3a in the virion is that the virion was purified from infected cells at a much later phase of infection and that 3a is sensitive to cleavage over time. Unfortunately, it was not possible to repeat the purification of virion from an earlier phase of infection as the incidents of laboratory acquired SARS-CoV infections [32] have led to a restriction on unnecessary protocols, like purification of virions that requires high virus titer and specialized instrumentations, using live SARS-CoV.
SARS-CoV 3a was incorporated into virus-like particles in insect cells co-infected with recombinant baculoviruses expressing the E, M, and 3a
To express SARS-CoV 3a protein in the baculovirus-insect system, recombinant virus vABhRp3a was generated as previously described [22]. Expression of 3a in Sf21 insect cells after infection with vABhRp3a was verified by Western blot analysis (Fig. 2 ). Interestingly, both the full-length 3a and a truncated 3a product of ∼24 kDa were specifically observed in the infected insect cells (lane 1) but not in the control cells (lanes 2 and 3). The cells were harvested at 4 days post-infection and therefore the amount of the truncated 3a was high, similar to what was observed in Vero E6 cells 2 days post-transfection with pXJ-3a construct (Fig. 1B). Hence, similar processing of 3a was observed in both mammalian and insect cells.
Fig. 2.
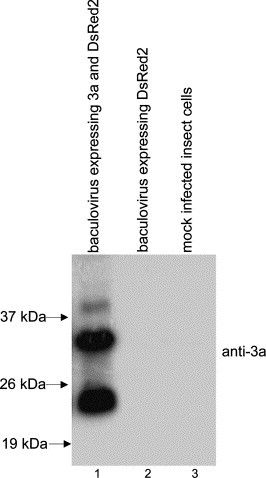
Expression of 3a in insect cells. Sf21 insect cells were infected with recombinant baculovirus carrying both 3a and DsRed2 (reporter gene) genes (lane 1). Cells were harvested at 4 days post-infection, lysed, and the cell lysate was subjected to Western blot analysis using anti-3a antibody. For negative controls, the cell lysates from Sf21 cells infected with recombinant baculovirus carrying only DsRed2 gene (lane 2) or mock infected Sf21 cells (lane 3) were used.
Previously, the formation of VLPs in insect cells expressing the SARS-CoV structural proteins M and E was demonstrated [22], [23]. The S protein could also be incorporated into VLPs when it is expressed together with M and E [22], [23]. In order to determine if SARS-CoV 3a protein can be incorporated into VLPs, insect cells co-expressing 3a, M, and E were subjected to sucrose gradient centrifugation, followed by Western blot analysis. As shown in Fig. 3 A, the 3a protein was found mainly in the fractions 17 and 18. The same fractions also contained the majority of the M and E proteins (Figs. 3B and C). Previously, electron microscopy showed that VLPs containing S were also found in fraction 17, which corresponded to 43% (w/w) sucrose and a density of 1.2 mg/ml [22]. Therefore, our data showed 3a, like the S protein, is not essential for the formation of VLPs but it is incorporated when present. In addition, both the full-length 3a and the truncated 3a were efficiently incorporated into VLPs, which is consistent with the observation of truncated 3a products in virions purified from SARS-CoV infected cells (Fig. 1B). In the VLPs from insect cells (Fig. 3A), the 3 forms of 3a observed were also similar in size to those observed in the purified virion, i.e., ∼35 kDa (and ∼32 kDa, which was not clearly resolved from the 35 kDa band) for full-length 3a, ∼24 and ∼20 kDa for the truncated products, indicating that the post-translational modifications of 3a in insect and mammalian cells were similar. However, the relative amounts of the different forms of 3a in virus or virus-like particles obtained from these 2 systems were quite different. In insect cells, the percentages of the 35, 24, and 20 kDa, products are 23%, 35%, and 42% respectively (Fig. 3A, fraction 17). And in mammalian cells, the corresponding percentages are 7%, 32%, and 61% respectively (Fig. 1B, lane 3). This indicates that the kinetics of processing as well as the incorporation into virions in insect and mammalian cells may not be identical.
Fig. 3.
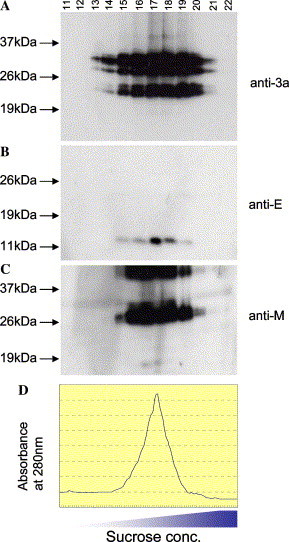
Incorporation of 3a into virus-like particles (VLPs). The VLPs were isolated from Sf21 insect cells co-expressing 3a, E, and M proteins. After sucrose gradient centrifugation, 28 fractions were collected and equal amounts of gradient fractions 11–22 were subjected to Western blot analysis using (A) anti-3a, (B) anti-E, and (C) anti-M antibodies. (D) Chromatograph showing the absorbance of each fraction at 280 nm.
In conclusion, our data showed that full-length 3a and its truncated products are incorporated into virion in both mammalian as well as insect cells, and provided the first direct evidence to prove that 3a is a novel coronavirus structural protein. 3a is predicted to have 3 transmembrane domains [10], [11] and when it is expressed on the cell surface, its N-terminus is facing the extracellular matrix and its C-terminus is facing the cytoplasm [13] As such, when 3a is incorporated into the virion, it would be expected to have a short N-terminus protruding out of the viral membrane while the C-terminus of the protein will be on the inside of the virion. Interestingly, when Liu et al. [33] used phage-display technology to characterize B cell epitopes recognized by antibodies from SARS patients, they found one consensus motif VKIXN which corresponded uniquely to 18–22 amino acids of the N-terminus ectodomain of 3a. Detailed studies, like electron microscopy analysis, will be necessary to determine the precise topology of full-length 3a (and its truncated products) in virus particles. As 3a can interact with S, M, and E proteins [13], [15], it is also critical to determine how these viral–viral interactions contribute to the incorporation of 3a and/or S into the virion. Interestingly, sequence comparison of isolates from different clusters of infection showed that both S and 3a [15], [34], [35] showed a positive selection during virus evolution. Collectively, these findings suggest that 3a could play important roles during viral infection, replication, and/or pathogenesis. Future studies will yield more information on the structural and functional significance of the incorporation of 3a into virions of SARS-CoV and such knowledge may have important implications for anti-viral or vaccine developments.
Acknowledgments
This work was supported by grants from the Agency for Science, Technology and Research (A*STAR), Singapore, and from the National Science Council (NSC93-2751-B-001-005-Y) and Academia Sinica (IMB 3/15 and AS93-AB-IMB-01), Taiwan.
References
- 1.Drosten C., Preiser W., Gunther S., Schmitz H., Doerr H.W. Severe acute respiratory syndrome: identification of the etiological agent. Trends Mol. Med. 2003;9:325–327. doi: 10.1016/S1471-4914(03)00133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laude H., Masters P.S. The coronavirus nucleocapsid protein. In: Siddell S.G., editor. The Coronaviridae. Plenum Press; New York: 1995. pp. 141–163. [Google Scholar]
- 3.Cavanagh D. The coronavirus surface glycoprotein protein. In: Siddell S.G., editor. The Coronaviridae. Plenum Press; New York: 1995. pp. 73–113. [Google Scholar]
- 4.Rottier P.J.M. The coronavirus membrane glycoprotein. In: Siddell S.G., editor. The Coronaviridae. Plenum Press; New York: 1995. pp. 115–139. [Google Scholar]
- 5.Bos E.C., Luytjes W., van der Meulen H.V., Koerten H.K., Spaan W.J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baudoux P., Carrat C., Besnardeau L., Charley B., Laude H. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 1998;72:8636–8643. doi: 10.1128/jvi.72.11.8636-8643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corse E., Machamer C.E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 2000;74:4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brian D.A., Hogue B.G., Kienzle T.E. The coronavirus hemagglutinin esterase glycoprotein protein. In: Siddell S.G., editor. The Coronaviridae. Plenum Press; New York: 1995. pp. 165–179. [Google Scholar]
- 10.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 11.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 12.Tan Y.-J., Goh P.–Y., Fielding B.C., Shen S., Chou C.–F., Fu J.–L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E., Lim S.G., Hong W. Profile of antibody responses against SARS-Coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11:362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan Y.-J., Teng E., Shen S., Tan T.H.P., Goh P.–Y., Fielding B.C., Ooi E.-E., Tan H.-C., Lim S.G., Hong W. A novel SARS coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu C.-J., Chen Y.-C., Hsiao C.-H., Kuo T.-C., Chang S.C., Lu C.-Y., Wei W.-C., Lee C.-H., Huang L.-M., Chang M.-F., Ho H.-N., Lee F.-J.S. Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett. 2004;565:111–116. doi: 10.1016/j.febslet.2004.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng R., Yang R.F., Shi M.D., Jiang M.R., Xie Y.H., Ruan H.Q., Jiang X.S., Shi L., Zhou H., Zhang L., Wu X.D., Lin Y., Ji Y.Y., Xiong L., Jin Y., Dai E.H., Wang X.Y., Si B.Y., Wang J., Wang H.X., Wang C.E., Gan Y.H., Li Y.C., Cao J.T., Zuo J.P., Shan S.F., Xie E., Chen S.H., Jiang Z.Q., Zhang X., Wang Y., Pei G., Sun B., Wu J.R. Characterization of the3a protein of SARS-associated coronavirus in infected Vero E6 cells and SARS patients. J. Mol. Biol. 2004;341:271–279. doi: 10.1016/j.jmb.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo J.P., Petric M., Campbell W., McGeer P.L. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology. 2004;324:251–256. doi: 10.1016/j.virol.2004.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kienzle T.E., Abraham S., Hogue B.G., Brain D.A. Structure and orientation of expressed bovine coronavirus hemagglutinin–esteraprotein protein. J. Virol. 1990;64:1834–1838. doi: 10.1128/jvi.64.4.1834-1838.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith A.R., Boursnell M.E., Binns M.M., Brown T.D., Inglis S.C. Identification of a new membrane-associated polypeptide specified by the coronavirus infectious bronchitis virus. J. Gen. Virol. 1990;71:3–11. doi: 10.1099/0022-1317-71-1-3. [DOI] [PubMed] [Google Scholar]
- 19.Vennema H., Heijnen L., Zijderveld A., Horzinek M.C., Spaan W.J. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J. Virol. 1990;64:339–346. doi: 10.1128/jvi.64.1.339-346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou C.-F., Shen S., Tan Y.-J., Fielding B.C., Tan T.H.P., Fu J., Xu Q., Lim S.G., Hong W. A novel cell-based binding assay system reconstituting interaction between SARS-CoV S protein and its cellular receptor. J. Virol. Methods. 2004;123:41–48. doi: 10.1016/j.jviromet.2004.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keng C.-T., Zhang A., Shen S., Lip K.M., Fielding B.C., Tan T.H.P., Chou C.-F., Loh C.B., Wang S., Fu J., Yang X., Lim S.G., Hong W., Tan Y.-J. Amino acids 1055 to 1192 in the S2 region of SARS coronavirus S protein induces neutralizing antibodies: implications for the development of vaccine and anti-viral agent. J. Virol. 2005;79:3289–3296. doi: 10.1128/JVI.79.6.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho Y., Lin P.H., Liu C.Y., Lee S.P., Chao Y.C. Assembly of human severe acute respiratory syndrome coronavirus-like particles. Biochem. Biophys. Res. Commun. 2004;318:833–838. doi: 10.1016/j.bbrc.2004.04.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.E Mortola, Roy P. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 2004;576:174–178. doi: 10.1016/j.febslet.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen S., McKee T.M., Wang Z.D., Desselberger U., Liu D.X. Sequence analysis and in vitro expression of genes 6 and 11 of an ovine group B rotavirus isolate, KB63: evidence for a non-defective, C-terminally truncated NSP1 and a phosphorylated NSP5. J. Gen. Virol. 1999;80:2077–2085. doi: 10.1099/0022-1317-80-8-2077. [DOI] [PubMed] [Google Scholar]
- 25.Fielding B.C., Tan Y.–J., Shen S., Tan T.H.P., Ooi E.-E., Lim S.G., Hong W., Goh P.–Y. Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome (SARS) coronavirus. J. Virol. 2004;78:7311–7318. doi: 10.1128/JVI.78.14.7311-7318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J.C., Chen H.H., Chao Y.C. Persistent baculovirus infection results from deletion of the apoptotic suppressor gene p35. J. Virol. 1998;72:9157–9165. doi: 10.1128/jvi.72.11.9157-9165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo H.R., Chao Y.C. Rapid titer determination of baculovirus by quantitative real-time polymerase chain reaction. Biotechnol. Prog. 2004;20:354–360. doi: 10.1021/bp034132i. [DOI] [PubMed] [Google Scholar]
- 28.O’Reilly D.R., Miller L.K., Luckow V.A. Oxford University Press; New York: 1994. Baculovirus Expression Vectors: A Laboratory Manual. [Google Scholar]
- 29.Guan M., Chen H.Y., Tan P.H., Shen S., Goh P.Y., Tan Y.J., Pang P.H., Lu Y., Fong P.Y., Chin D. Use of viral lysate antigen combined with recombinant protein in Western immunoblot assay as confirmatory test for serodiagnosis of severe acute respiratory syndrome. Clin. Diagn. Lab. Immunol. 2004;11:1148–1153. doi: 10.1128/CDLI.11.6.1148-1153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan Y.-J., Lim S.G., Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antiviral Res. 2005;65:69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Escors D., Capiscol C., Enjuanes L. Immunopurification applied to the study of virus protein composition and encapsidation. J. Virol. Methods. 2004;119:57–64. doi: 10.1016/j.jviromet.2004.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim P.L., Kurup A., Gopalakrishna G., Chan K.P., Wong C.W., Ng L.C., Se-Thoe S.Y., Oon L., Bai X., Stanton L.W., Ruan Y., Miller L.D., Vega V.B., James L., Ooi P.L., Kai C.S., Olsen S.J., Ang B., Leo Y.S. Laboratory-acquired severe acute respiratory syndrome. N. Engl. J. Med. 2004;350:1740–1745. doi: 10.1056/NEJMoa032565. [DOI] [PubMed] [Google Scholar]
- 33.Liu I.J., Hsueh P.R., Lin C.T., Chiu C.Y., Kao C.L., Liao M.Y., Wu H.C. Disease-specific B cell epitopes for serum antibodies from patients with severe acute respiratory syndrome (SARS) and serologic detection of SARS antibodies by epitope-based peptide antigens. J. Infect. Dis. 2004;190:797–809. doi: 10.1086/422753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guan Y., Peiris J.S., Zheng B., Poon L.L., Chan K.H., Zeng F.Y., Chan C.W., Chan M.N., Chen J.D., Chow K.Y., Hon C.C., Hui K.H., Li J., Li V.Y., Wang Y., Leung S.W., Yuen K.Y., Leung F.C. Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet. 2004;363:99–104. doi: 10.1016/S0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeh S.H., Wang H.Y., Tsai C.Y., Kao C.L., Yang J.Y., Liu H.W., Su I.J., Tsai S.F., Chen D.S., Chen P.J. National Taiwan University SARS Research Team., Characterization of severe acute respiratory syndrome coronavirus genomes in Taiwan: molecular epidemiology and genome evolution. Proc. Natl. Acad. Sci. USA. 2004;101:2542–2547. doi: 10.1073/pnas.0307904100. [DOI] [PMC free article] [PubMed] [Google Scholar]


