Abstract
HCoV-229E spike (S) protein mediates virion attachment to cells and subsequent fusion of the viral and cellular membranes. This protein is composed of an N-terminal receptor-binding domain (S1) and a C-terminal trans-membrane fusion domain (S2). S2 contains a highly conserved heptad repeat 1 and 2 (HR1 and HR2). In this study, the HRs sequences were designed and connected with a flexible linker. The recombinant fusion core protein was crystallized and its structure was solved at a resolution of 2.45 Å. Then we characterized the binding of HR1s and HR2s via both sequence alignment and structural analysis. The overall structures, especially the residues in some positions of HR2 are highly conserved. Fourteen hydrophobic and three polar residues from each HR1 peptide are packed in layers at the coiled-coil interface. These core amino acids can be grouped into seven heptad repeats. Analysis of hydrophobic and hydrophilic interactions between HR2 helix and HR1 helices, shows that the HR1 and HR2 polypeptides are highly complementary in both shape and chemical properties. Furthermore, the available knowledge concerning HCoV-229E fusion core may make it possible to design small molecule or polypeptide drugs targeting membrane fusion, a crucial step of HCoV-229E infection.
Keywords: Human coronavirus 229E, Spike, Fusion core
1. Introduction
The genus Coronavirus (CoVs) belong to the order Nidovirales, family Coronaviridae and subfamily Coronavirinae. Recently, the Coronavirus Study Group of the International Committee for Taxonomy of Viruses has proposed three genera: including the mammalian genera Alpha- and Beta- and the bird-associated genera Gamma- and Detacoronavirus recently [[1], [2], [3]], to replace the traditional CoV groups 1, 2, and 3 [4,5]. CoVs are a diverse group of enveloped, plus-stranded RNA viruses that infect humans and many animal species, in which they can cause respiratory, enteric, hepatic, central nervous system and neurological diseases of varying severity [3]. In 2003, a novel aggressive human coronavirus termed Severe Acute Respiratory Syndrome coronavirus (SARS-CoV), which is most related to genus Betacoronavirus, caused a large epidemic with a case fatality rate of ∼10%, creating a global panic and economic damage [6]. In 2012, the newly emerging Middle East respiratory syndrome coronavirus (MERS-CoV), a highly pathogenic virus with an approximately 50% fatality rate, can cause a serious respiratory manifestations, including fever, cough, shortness of breath, and acute respiratory distress syndrome (ARDS) [7,8]. To date, it has been shown that four other human coronaviruses HCoV-229E, HCoV-OC43, HCoV-NL63 and HKU1 [[9], [10], [11]], cause a significant portion of upper and lower respiratory tract infections in humans, including common colds, bronchiolitis and neurological disease [12]. To date, no medication is available to effectively prevent or cure HCoV-229E-related diseases. Thus, it is important to design and develop new vaccines or drugs that are specific for the HCoV-229E-related diseases.
CoVs are enveloped virus, suggesting that the fusion between the cell membrane and the viral envelope is essential for the initiation of viral invasion. Following binding with host receptor, CoVs release viral genetic materials into the cytoplasm through direct membrane fusion [13,14]. Membrane fusion is mediated by spike protein S2 subunit. This subunit is expected to present rearrangement of its characteristic elements called heptad repeats (HRs) to form a stable 6-helix bundle fusion core [14]. Although great advances in the knowledge of CoVs protein and host factors have been achieved, no licensed vaccines or specific drugs are available to prevent CoVs infection [15]. In view of prevalence of epidemics due to CoVs, it is essential to develop novel broad-spectrum antivirus drugs. Since fusion with the host membrane is a vital early step in the life cycle of the virus, fusion inhibitors could be designed as antiviral drugs. Such as Human immunodeficiency virus I inhibitor T20, a peptide mimic of HR2 and acting through competitively binding to HR1, is the first fusion inhibitor targeting membrane fusion [16,17].
To date, some studies discovered the highly conserved conformation of fusion core, based on four solved crystal structures namely that from NL-63, MHV, SARS and MERS [[18], [19], [20], [21]]. Little if any experimental evidence has been presented to propose that the HCoV-229E fusion core shares the same features. In order to unravel the structure basis of HCoV-229E membrane fusion, we solved the structure of fusion core of HCoV-229E and characterized the binding of HR1s and HR2s via both sequence alignment and structural analysis. The overall structures shows that the HR1 and HR2 polypeptides are highly complementary in both shape and chemical properties. Although HCoV-229E fusion core presenting the highly conserved sequences copmare with MHV, SARS, MERS, and NL-63, the HR2 have some differences in stereo configuration. Furthermore, the available knowledge concerning HCoV-229E fusion core may provide inportant details for the design of small molecule or polypeptide drugs that target HCoV-229E infection.
2. Materials and methods
2.1. Cloning, expression and purification
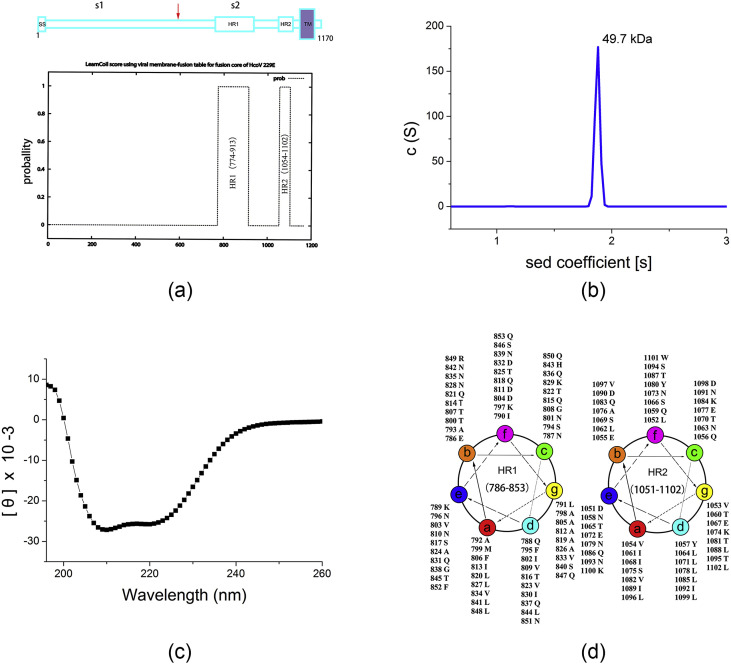
The fusion core construct (FC1), including different truncated heptad-repeat regions (HR1 and HR2) of HcoV-229E spike protein (GenBank™ accession number BAL45641.1), were predicted by LearnCoil-VMF [22] (Fig. 1 ) and further modified according to comparison with the available data on CoV pathogenesis in their corresponding conserved regions. In the FC1 construct, HR1 and HR2 regions cover the amino acids 786–853 and 1051–1102, respectively. Subsequently, truncated HR1 and HR2 were connecting with a flexible linker (LVPRGSGGSGGSGGLEVLFQGP). The coding sequence, optimized codons for Escherichia coli, was synthesized by BeijingAuGCT Biotechnology Co., Ltd., amplified by PCR and inserted between the BamH I and XhoI restriction sites of pET28a vector (Novagen). The protein production information is showed in Table 1 .
Fig. 1.
a) Cartoon characterization of HCoV-229E fusion protein and prediction of the two heptad repeat regions. A schematic diagram of S protein (amino acids 1–1170) is shown in the upper panel. The cleavage site of S1 and S2 is indicated with a red arrow. SS, signal sequence; S1 and S2, two cleaved fragments of the fusion protein; HR, heptad repeat region; and TM, trans-membrane region. In the lower panel, the likelihood of HR1 and HR2 predicted by LearnCoil-VMF program is characterized. b) Analytic ultracentrifugation (AUC) data for FC1. c) CD spectrum for FC1. d) Helical-wheel representation of the HR regions of HCoV-229E fusion protein (FC1). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Protein production information.
| Source organism | HcoV-229E |
|---|---|
| DNA source | synthesized by BeijingAuGCT Biotechnology Co.,Ltd. |
| Forward primer | CGGGATCCGAAAACCAGAAAATTCT |
| Reverse primer | CCGCTCGAGCAGCCATTTCAGATC |
| Cloning vector | pET28a |
| Expression vector | pET28a |
| Expression host | Escherichia coli, |
| Complete amino acid sequence of the construct produced | ENQKILAASFNKAMTNIVDAFTGVNDAITQTSQALQTVA TALNKIQDVVNQQGNSLNHLTSQLRQNFQLVPRGSGGSGGSGG LEVLFQGPDLVVEQYNQTILNLTSEIST LENKSAELNYTVQKLQ TLIDNINSTLVDLKWLHHHHHH* |
The construct expressed protein with an C-terminal hexa histidine tag. Corresponding proteins were over expressed in E. coli BL21 (DE3) (TransGen Biotech). Cells were grown aerobically at 37 °C in Luria-Bertani medium supplemented with 100 μg/ml kanamycin. When the cell density (OD600) reached 0.6, the culture was first cooled for 2 h at 4 °C and then induced at 16 °C for 18 h with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG).
Cells were harvested by centrifugation at 8000g for 15 min. The cell pellet was re-suspended in McAc 0 buffer (25 mM Tris-HCl, 500 mM NaCl, pH 8.0) and lysed by sonication. Unbroken cells and debris were removed by spinning the lysate at 12000 g for 40 min at 4 °C. Supernatant containing soluble target protein was then loaded onto a Ni-NTA column (GE Healthcare) previously equilibrated with McAc 0 buffer. After thorough or washing with buffer, protein bound to the column was eluted with McAc 500 buffer supplemented with 500 mM imidazole, The protein solution was concentrated to 1 ml and applied to a Resource™ Q anion-exchange chromatography column (GE Healthcare) equilibrated with buffer A. Proteins were then eluted with a 0–1 M gradient of NaCl in buffer A. The protein eluted at approximately 300 mM NaCl. The fractions were pooled, concentrated to 500 μl, and applied to a Superdex® 200 gel filtration column (GE Healthcare) equilibrated with buffer A containing 150 mM NaCl. The purity of the protein was estimated to be greater than 95% by SDS-PAGE analysis. Then the target protein were concentrated to 15–20 mg/ml and stored at −20 °C until further use.
2.2. Crystallization
Crystals of HCoV 229E fusion core were grown at 293 K using the sitting drop vapour diffusion technique. A 1 μl protein solution (10 mg/ml, 25 mM Tris, 150 mM NaCl, pH 8.0) was mixed with 1 μl reservoir solution and equilibrated over 100 μl of reservoir solution. Crystals were obtained in condition containing 1.5 M ammonium sulfate, 12% (v/v) glycerol, 100 mM Tris/HCl, pH 8.5; The crystal was mounted in a nylon loop before being flash-frozen in liquid nitrogen. Crystallization details are list in Table 2 . X-ray diffraction data were collected at beamline 5 A of the Photon Factory (Japan). All data were processed with the HKL2000 Suite of programs [23].
Table 2.
Crystallization conditions.
| Method | sitting drop vapour diffusion |
|---|---|
| Plate type | 48 well |
| Temperature (K) | 293 |
| Protein concentration | 10 mg/ml |
| Buffer composition of protein solution | 25 mM Tris, 150 mM NaCl, pH 8.0 |
| Composition of reservoir solution | 1.5 M Ammonium sulfate, 12% (v/v) Glycerol, 100 mM Tris/HCl, pH 8.5 |
| Volume and ratio of drop | 1:1 (2 μl) |
| Volume of reservoir | 100 ul |
2.3. Data processing, structure solution and refinement
The crystal structure of HCoV-229E fusion core was determinated by molecular replacement with the program Phaser [24] using the structure of the HCoV-NL63 N57C42 (PDB code 2IEQ) trimer as the search template. The initial model was first refined with the Phenix AutoBuild wizard [25]. The model was then improved further with iterative cycles of manual rebuilding in coot [26] and refinement by Phenix. refine [27]. All structures were judged to have good stereochemistry according to the Ramachandran plot calculated by MolProbity [28]. A summary of the data collection, phasing and structure refinement statistics is listed in Table 3 and Table 4 .
Table 3.
Values for the outer shell are given in parentheses.
| Diffraction source | Photon Factory (Japan) |
|---|---|
| Wavelength (Å) | 1.0000 |
| Crystal-detector distance (mm) | 350 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 180 |
| Space group | P1 |
| a, b, c (Å) | 30.1, 77.3, 77.4 |
| α, β, γ (°) | 76.9, 89.8, 90.2 |
| Mosaicity (°) | 0.3 |
| Resolution range (Å) | 50.00–2.44 (2.49–2.44) |
| Total No. of reflections | 46795 |
| No. of unique reflections | 24629 |
| Completeness (%) | 97.9 (94.4) |
| Redundancy | 1.9 (1.9) |
| 〈 I/σ(I)〉 | 17.2 (2.9) |
| Rr.i.m. | 3.9 (23.5) |
| Overall B factor from Wilson plot (Å2) | 48.8 |
Table 4.
Values for the outer shell are given in parentheses.
| Resolution range (Å) | 31.00–2.44 (2.50–2.44) |
|---|---|
| Completeness (%) | 97.9 (94.4) |
| No. of reflections, working set | 24511 (1504) |
| No. of reflections, test set | 2018 (134) |
| Final Crist | 21.9 |
| Final Free | 26.1 |
| No. of non-H atoms | |
| Protein | 4039 |
| Water | 11 |
| Total | 4050 |
| RMSD values | |
| Bonds (Å) | 0.009 |
| Angles (°) | 1.045 |
| Average B factors (A) | |
| Protein | 64.0 |
| Water | 69.1 |
| Ramachandran plot | |
| Most favoured (%) | 99.22 |
| Allowed (%) | 0.78 |
2.4. Analytic ultracentrifugation (AUC)
Sedimentation velocity (SV) experiments were performed with a Beckman/Coulter Optima XL-1 analytical ultracentrifuge or AUC equipped with two-channel centerpieces and sapphire windows (Beckman Coulter). The experiment was carried out at 4 °C using purified protein in buffer containing 25 mM Tris, 150 mM NaCl, pH 8. Protein sample (0.3 mg/ml) was centrifuged at 4 °C at 42000 rpm and migration of protein was monitored based on absorbance at 280 nm. The SV data were analyzed with the SEDFIT program [29].
2.5. Circular dichroism spectroscopy
Circular dichroism (CD) spectra were performed on a Biologic M450 spectropolarimer (Biologic Science Inc.) equipped with a thermoelectric temperature controller. Spectra of HCoV-229E fusion core (0.1 mg/ml) were measured by taking data points every 1.0 nm from 195 nm to 260 nm at 20 °C in 25 mM Tris, 150 mM NaCl, pH 8.0.
3. Results and discussion
3.1. Generation and characterization of HCoV-229E fusion core
From the NCBI database, we obtained the primary sequence of HCoV-229E spike protein (Gen Bank: BAL45641.1). In view of the prediction of the LearnCoil-VMF program and the multiple alignments of the putative heptad repeat regions of HCoV-229E with those of several other coronaviruses, the final construct of the HCoV-229E fusion core was designed. The fusion core consists of HR1, a truncated type of the LearnCoil-VMF predicted HR1 (residues 786–853), connected with HR2, an extended version of the LearnCoil-VMF predicted HR2 (1051–1102) (Fig. 1a) and a flexible 22-residue linker (LVPRGSGGSGGSGGLEVLFQGP). This flexible linker has been shown to work successfully with the fusion core of SARS-CoV and MERS-CoV that allows for easy expression and purification of the fusion core complex [19,30]. The recombinant protein, designated fusion core (FC1), was expressed in E. coli and purified by affinity chromatography and size exclusion chromatography. FC1 was eluted at approximately 15.8 ml on a calibrated Superdex 200 10/300 column, corresponding to a molecular mass of ∼45 kDa. AUC was also used to investigate the molecular mass Fig. 1b which gave a value of 49.7 kDa. As the single HR1-linker-HR2 chain is ∼15.4 kDa, these data indicate a trimer has formed in solution, as expected. We further tested the protein by CD spectroscopy. Double minima at 208 and 222 nm were observed, demonstrating that the FC1 has a high alpha-helical content (Fig. 1c). The fusion core designed here shared the feature of the helix wheels (Fig. 1d), a typical characteristic of class I fusion protein [19].
3.2. Overall structure of the HCoV-229E fusion core
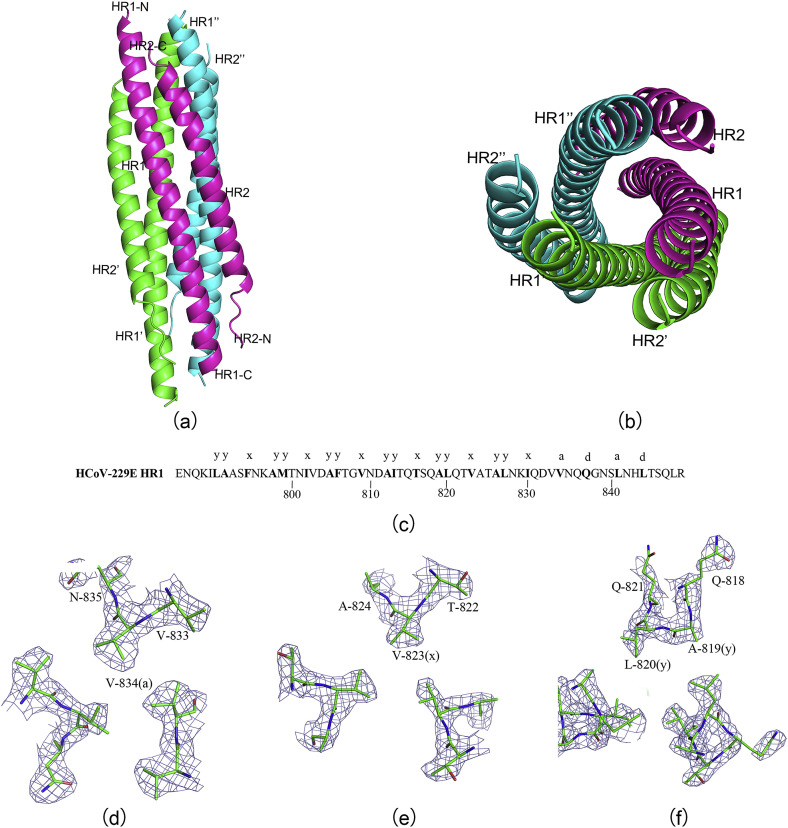
In the three-dimensional structure of HCoV-229E fusion core, clear electron densities could be traced for residues Q788-G838 in HR1 and T1060-V1097 in HR2. Residues L791-V836 of HR1 fold into a 13-turn α-helix stretching the entire length of the coiled coil, whereas residues Q788-I790 at the N terminus and residues Q837-G838 at the C terminus form two extended conformations, respectively. While in HR2, residues S1066-S1094 form an eight-turn amphipathic α-helix, whereas residues T1060-T1065 at the N terminus and residues T1095-V1097 at the C terminus form two extended loops, respectively. The HR1 helices form an interior, parallel trimeric coiled-coil. Then three HR2 helices pack in an oblique, left-handed and antiparallel direction into hydrophobic grooves on the surface of this trimeric coiled-coil (Fig. 2 a). This leads to a 6-helix bundle of approximately 13 Å in semidiameter and approximately ∼80 Å in height (Fig. 2b).
Fig. 2.
a) Overall structure of HCoV-229E fusion core. Side view of the fusion core 6-helix bundle of approximately 13 Å in semidiameter, b) top view of the fusion core 6-helix bundle approximately ∼80 Å in height. c) Sequences of the fusion core HR1 peptide with the observed heptad-repeat positions. Amino acids at the a, d, x, and y positions are in boldface. The residues are numbered according to their position in the HCoV-229E S sequences. d) Cross-section of the trimer in the Val832 (a) layer. (d–f) The 2Fo - Fc electron density map (contoured at 1.5 σ) with the refined molecular model overlaid. e) The “x-like” packing of Val823 pointing toward the 3-fold symmetry axis. g) The “y-like” packing of Ala819 and Leu820 with the alternating small and large side-chains facing inward to form a hydrophobic core.
Similar to the core structure of S2 from HCoV-NL63 [18], fourteen hydrophobic and three polar residues from each HR1 peptide are packed in layers at the coiled-coil interface. These amino acids had been grouped into seven heptad repeats (Fig. 2c). In the last two of these repeats, the side-chains of the “a” and “d” residues in one α-helix point directly into the hole formed between the side-chains of four residues in an adjacent HR1 helix that exhibits canonical knobs-into-holes packing (Fig. 2d). In contrast, the first six heptad repeats of HR1 lack any regular 3–4 hydrophobic periodicity. Instead, cross-sectional layers containing an “x-like” symmetric pattern (Fig. 2e) [31] alternate with layers containing a da-like two-residue pattern (“y-like”packing; see Fig. 2f) [32]. Side chains at the x position project toward the center of the hydrophobic core (Fig. 2e), and similarly alternating side chains at the y positions pack in a hexagonal arrangement (Fig. 2f) [18]. All these core side chains (excluding alanine) adopt their well-populated rotamer conformations in R-helices.
3.3. Interactions between the HCoV-229E fusion core helices
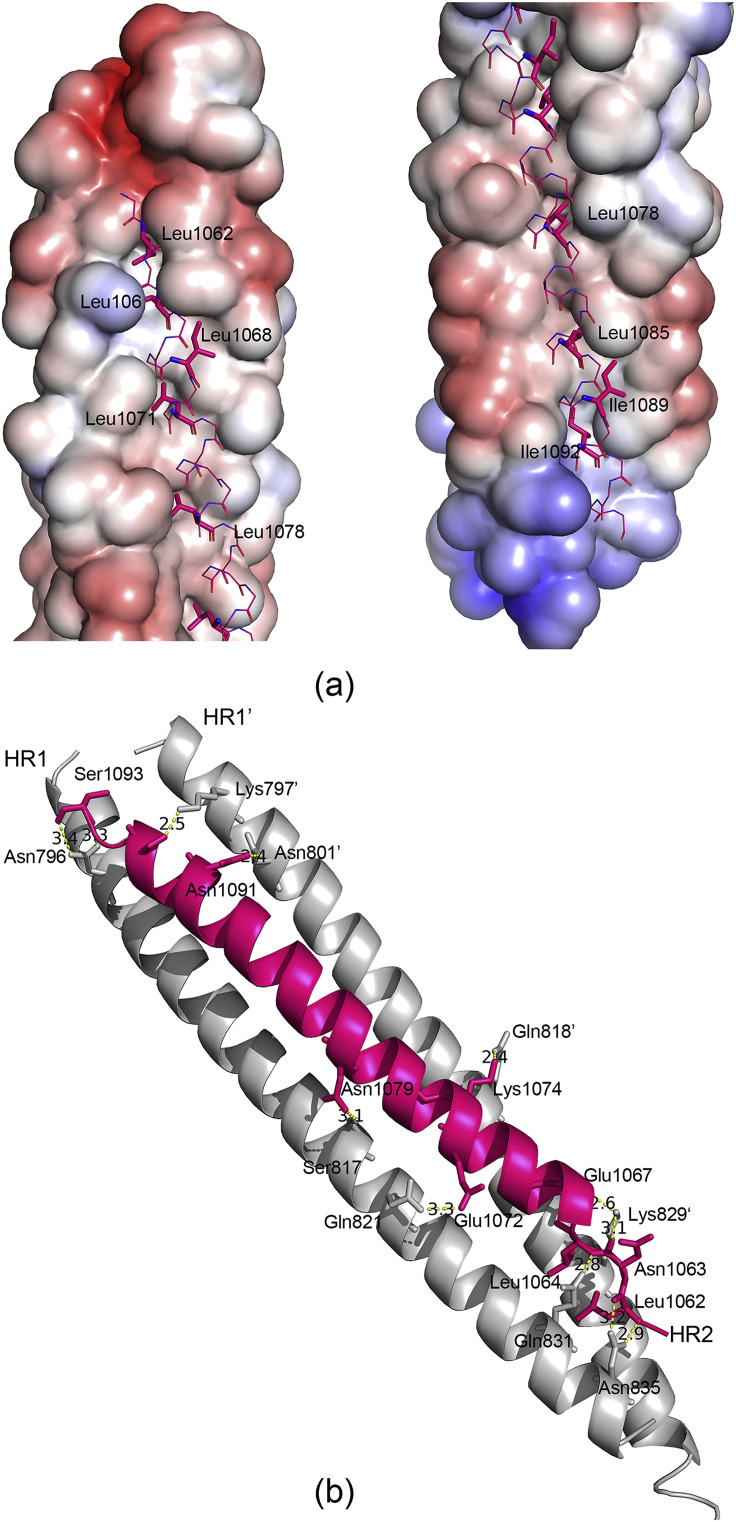
We further characterized the atomic details mediating the formation of the hairpin trimer. Consistent with the HR features, the hydrophobic residues at positions “a” and “d” in HR1 are aligned on one side of the helix, forming an interface of strong hydrophobicity. Three HR1 helices thereby pack against each other and stack the hydrophobic helical laterals in the center of the coiled-coil core. These three hydrophobic grooves become the binding sites of three HR2 helices. Although a notable difference in length is observed between the HR1 and HR2 helices (∼13 turns versus ∼8 turns), both the N- and C-terminal tails of the HR2 region pack in an orderly manner against the hydrophobic grooves of a central three-helical coiled-coil, mainly through hydrophobic interactions involving L1062, L1064, L1068, L1071 and L1078 at the N-terminal portion and L1085, I1089 and I1092 at the C-terminal portion of the HR2 helix (Fig. 3 a).
Fig. 3.
a) Electrostatic potential surface of three central HR1 helices, as calculated using Pymol, shows the hydrophobic grooves formed between each of the two adjacent HR1 helices. one of three HR2 segments in the grooves are shown in lines representation. The residues that insert into hydrophobic grooves are shown in stick representation. b) The hydrophilic interactions between HR1 and HR2 helices. The residues involved in forming hydrogen bonds are shown in stick representation and are labelled, as well as hydrogen bonds in yellow dashed lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The hydrophilic interactions between one HR2 helix and its neighbouring HR1 helices consist of 12 hydrogen bonds, mostly distributed in the regions around the N- and C-terminal ends of the HR2 helices (Fig. 3b). Around the N-terminal portion of the HR2 helix, its L1062, L1064, E1067 and K1074 form hydrogen bonds with Q821, Q831 and N835 of its fused HR1 helix and Q818, K829 of the adjacent HR1 helix. Around the C-terminal portion of the HR2 helix, its N1091, S1093 and V1097 form hydrogen bonds with N796 of its fused HR1 helix and K797 and N801 of the adjacent HR1 helix. The concentrated arrangement of hydrogen bonds constitute two anchoring points at both ends of one short HR2 helix; further stabilizing its binding with the central hydrophobic grooves.
3.4. Sequence alignment and structure comparison
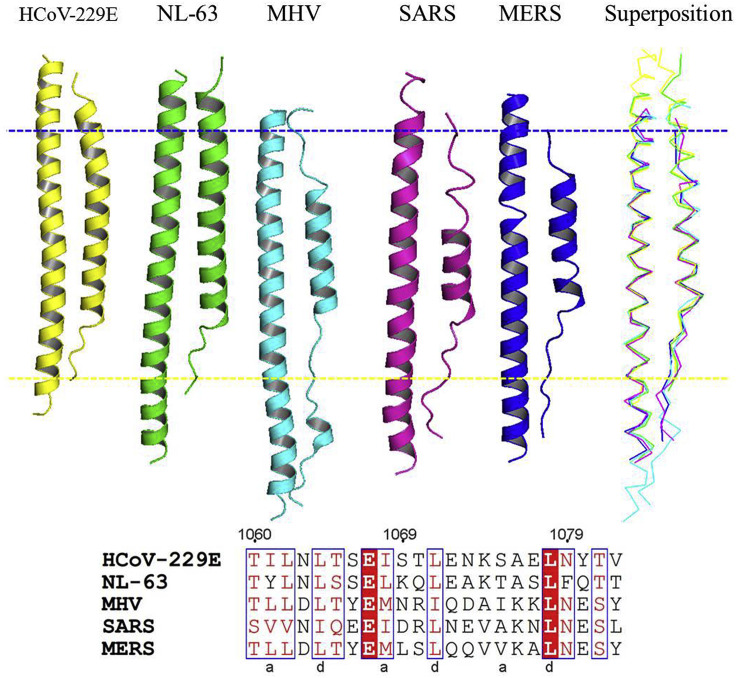
Since fusion core proteins presented comparatively high sequence similarity within each CoVs genera, representatives of each genera were chosen for comparison. In genera Betacoronavirus, structures from MHV, SAES and MERS are available; in genera Alphacoronavirus, only NL-63 and HCoV-229E; while in genera Gamma- and Delta-coronavirus, no structures are presently available. Superposition of the 2-helix bundle crystal structures of HCoV-229E with NL-63, MHV, SARS and MERS showed ∼0.60 Å, ∼0.94 Å, ∼0.70 Å and ∼9.37 Å root mean square deviation (rmsd) for ∼80 Cα (Fig. 4 ), respectively. The common regions include the following peptide fragments: residues 1060–1082 in HCoV-229E, residues 1244–1266 in NL-63, residues 1227–1249 in MHV, residues 1156–1178 in SARS and residues 1258–1280 in MERS. The residues in the a position (I1061, T1068, S1075, V1082) and d position (L1064, L1071, L1078), showing relatively conservation, pack against hydrophobic grooves formed on the surface of the HR1 core. All of the helical elements and a majority of the extended HR2 loops could be well aligned. However, some terminal residues exhibit conformational variance, and the helices in HR2 diffe in length. As described above, all of the residues at a/d position of HR2 are highly conserved (either Leu and Ile). Their presence is essential to form an interface of strong hydrophobicity. In addition, e/g positions of HR1 are also conserved. Thus, the overall structures and the hydrophobic character of the interaction between HR1 and HR2 is conserved in both fusion cores, suggesting the possibility for broad-spectrum inhibitor design targeting fusion core proteins of all CoVs.
Fig. 4.
In the upper panel, representatives of each genera were chosen for comparison. Superposition of the 2-helix bundle crystal structures of HCoV-229E (yellow) with NL-63 (green), MHV (blue), SARS (violet) and MERS (dark blue) showed ∼0.61 Å, ∼0.94 Å, ∼0.70 Å and ∼9.36 Å root mean square deviation for approximate 80 Cα. In the lower panel, it shows that all of the residues at d position of HR2 are highly conserved. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Acknowledgements
We thank the staff at the Shanghai Synchrotron Radiation Facility (China) and Photon Factory (Japan) for their assistance in data collection. This work was supported by grants from National Key R&D Program of China (grant No. 2017YFC0840300), MOST 973 Project (Grant No. 2014CB542800, No. 2014CBA02003), CAS SPR program B (Grant No. XDB08020200) and National Science Foundation of China (Grant Nos. 81330036 and 81520108019).
Footnotes
Transparency document related to this article can be found online at https://doi.org/10.1016/j.bbrc.2018.02.136.
Transparency document
References
- 1.Adams M.J., Lefkowitz E.J., King A.M., Carstens E.B. 2014. Ratification Vote on Taxonomic Proposals to the International Committee on Taxonomy of Viruses. Arch Virol (2014) [DOI] [PubMed] [Google Scholar]
- 2.Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 7.Bermingham A., Chand M.A., Brown C.S., Aarons E., Tong C., Langrish C., Hoschler K., Brown K., Galiano M., Myers R., Pebody R.G., Green H.K., Boddington N.L., Gopal R., Price N., Newsholme W., Drosten C., Fouchier R.A., Zambon M. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17:20290. [PubMed] [Google Scholar]
- 8.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 9.Chiu S.S., Chan K.H., Chu K.W., Kwan S.W., Guan Y., Poon L.L., Peiris J.S. Human coronavirus NL63 infection and other coronavirus infections in children 306 hospitalized with acute respiratory disease in Hong Kong. Clin. Infect. Dis.: official publ. Infect. Dis. Soc. Am. 2005;40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vabret Astrid, Mourez Thomas, Dina Julia, van der Hoek Lia, Gouarin Stéphanie, Petitjean Joëlle, Brouard Jacques, Freymuth F. Human coronavirus NL63, France. Emerg. Infect. Dis. 2005;11:1225–1229. doi: 10.3201/eid1108.050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sloots T.P., McErlean P., Speicher D.J., Arden K.E., Nissen M.D., Mackay I.M. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J. Clin. Virol. 2006;35:99–102. doi: 10.1016/j.jcv.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Hoek L. Human coronaviruses: what do they cause? Antivir. Ther. 2007;12:651–658. [PubMed] [Google Scholar]
- 13.Badani H., Garry R.F., Wimley W.C. Peptide entry inhibitors of enveloped viruses: the importance of interfacial hydrophobicity. Biochim. Biophys. Acta. 2014;1838:2180–2197. doi: 10.1016/j.bbamem.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plemper R.K. Cell entry of enveloped viruses. Curr. Opin. Virol. 2011;1:92–100. doi: 10.1016/j.coviro.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes K.V. Enteric infections with coronaviruses and toroviruses. Novartis Found. Symp. 2001;238:258–269. doi: 10.1002/0470846534.ch16. discussion 269–275. [DOI] [PubMed] [Google Scholar]
- 16.Wild C.T., Shugars D.C., Greenwell T.K., McDanal C.B., Matthews T.J. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilby J.M., Hopkins S., Venetta T.M., DiMassimo B., Cloud G.A., Lee J.Y., Alldredge L., Hunter E., Lambert D., Bolognesi D., Matthews T., Johnson M.R., Nowak M.A., Shaw G.M., Saag M.S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 18.Zheng Q., Deng Y., Liu J., Hoek L.v.d., Berkhout B., Lu M. Core structure of S2 from the human coronavirus NL63 spike glycoprotein. Biochemistry. 2006;45:15205. doi: 10.1021/bi061686w. [DOI] [PubMed] [Google Scholar]
- 19.Gao J., Lu G., Qi J., Li Y., Wu Y., Deng Y., Geng H., Li H., Wang Q., Xiao H., Tan W., Yan J., Gao G.F. Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S Protein of Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:13134–13140. doi: 10.1128/JVI.02433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y., Liu Y., Lou Z., Qin L., Li X., Bai Z., Pang H., Tien P., Gao G.F., Rao Z. Structural basis for coronavirus-mediated membrane fusion. Crystal structure of mouse hepatitis virus spike protein fusion core. J. Biol. Chem. 2004;279:30514–30522. doi: 10.1074/jbc.M403760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Supekar V.M., Bruckmann C., Ingallinella P., Bianchi E., Pessi A., Carfi A. Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17958–17963. doi: 10.1073/pnas.0406128102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh M., Berger B., Kim P.S. LearnCoil-VMF: computational evidence for coiled-coil-like motifs in many viral membrane-fusion proteins. J. Mol. Biol. 1999;290:1031–1041. doi: 10.1006/jmbi.1999.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z., Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terwilliger T.C., Grosse-Kunstleve R.W., Afonine P.V., Moriarty N.W., Zwart P.H., Hung L.W., Read R.J., Adams P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., Zwart P.H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y., Lou Z., Liu Y., Pang H., Tien P., Gao G.F., Rao Z. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 2004;279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lupas A.N., Gruber M. The structure of alpha-helical coiled coils. AdV. Adv. Protein Chem. 2005;70:37–38. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 32.Deng Y., Liu J., Zheng Q., Yong W., Lu M. Structures and polymorphic interactions of two heptad-repeat regions of the SARS virus S2 protein. Structure. 2006;14:889–899. doi: 10.1016/j.str.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.