Abstract
Loss of genetic variation may render populations more vulnerable to pathogens due to inbreeding depression and depletion of variation in genes responsible for immunity against parasites. Here we review the evidence for the significance of variation in genes of the Major Histocompatibility Complex (MHC) for conservation efforts. MHC molecules present pathogen-derived antigens to the effector cells of the immune system and thus trigger the adaptive immune response. Some MHC genes are the most variable functional genes in the vertebrate genome. Their variation is clearly of adaptive significance and there is considerable evidence that its maintenance is mainly due to balancing selection imposed by pathogens. However, while the evidence for selection shaping MHC variation on the historical timescale is compelling, a correlation between levels of MHC variation and variation at neutral loci is often observed, indicating that on a shorter timescale drift also substantially affects MHC, leading to depletion of MHC diversity. The evidence that the loss of MHC variation negatively affects population survival is so far equivocal and difficult to separate from effects of general inbreeding. Some species with depleted MHC variation seem to be particularly susceptible to infection, but other species thrive and expand following severe bottlenecks that have drastically limited their MHC variation. However, while the latter demonstrate that MHC variation is not always critical for population survival, these species may in fact represent rare examples of survival despite of the loss of MHC variation. There is clearly a compelling need for data that would disclose the possible consequences of MHC diversity for population viability. In particular, we need more data on the impact of MHC allelic richness on the abundance of parasites or prevalence of disease in populations, while controlling for the role of general inbreeding. Before such evidence accumulates, captive breeding programs and other conservation measures aimed at inbreeding avoidance should be favoured over those protecting only MHC variation, especially since inbreeding avoidance programs would usually conserve both types of genetic diversity simultaneously.
Keywords: Major Histocompatibility Complex, Polymorphism, Conservation, Positive selection, Infectious diseases, Extinction
1. Introduction
Pathogens are considered as one of the major extinction factors (Smith et al., 2009, Wilcove et al., 1998). Arguably, depletion of genetic diversity within populations may make them more vulnerable to pathogen assault (Altizer et al., 2003, de Castro and Bolker, 2005, O’Brien and Evermann, 1988). First, inbreeding depression associated with population bottlenecks (Keller and Waller, 2002) may limit the ability of individuals to mount an effective immune response. Indeed, inbreeding has been demonstrated to increase susceptibility to infections (Acevedo-Whitehouse et al., 2003, Acevedo-Whitehouse et al., 2005, Coltman et al., 1999, Ilmonen et al., 2008, Reid et al., 2007, Ross-Gillespie et al., 2007, Spielman et al., 2004) (Fig. 1 ). Second, the loss of variation at genes responsible for resistance to parasites may render populations more susceptible to infection. This argument applies to highly polymorphic vertebrate Major Histocompatibility Complex (MHC) genes, coding for proteins presenting pathogen-derived antigens to T-cells, thus initiating the adaptive immune response (Janeway et al., 2004). Hughes (1991) suggested that retention of variation in these genes is an essential element of effective conservation programs, but this argument remains controversial (Hedrick, 2001). Apart from MHC, other polymorphic genes can influence the effectiveness of defences against pathogens (Acevedo-Whitehouse and Cunningham, 2006). Here, however we concentrate on MHC genes only, as they are the most polymorphic genes known in vertebrates, and their function and evolution is better understood than that of other genes involved in the immune response. Despite this, as shown in our review, there is few data linking MHC diversity to population viability and some of it is equivocal. We hope that by drawing attention to gaps in our knowledge, our review will stimulate research that will facilitate better design of conservation programs.
Fig. 1.
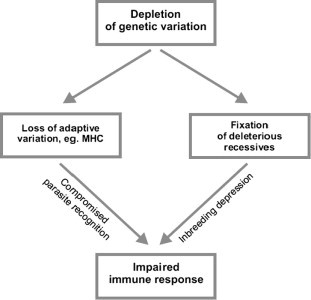
Depletion of genetic variation may affect resistance to pathogens via different paths; drift may fix deleterious mutations impairing organismal function including immune response, or cause the loss of alleles capable of recognition of assault by novel pathogens. Thus, association between infection and genetic diversity within populations need not be due to reduced MHC variation.
1.1. Adaptive significance of MHC variation
MHC codes for the most polymorphic genes in vertebrates (Garrigan and Hedrick, 2003), some of which (A, B within MHC I and DRB within MHC II) have hundreds of alleles described in human populations (http://www.ebi.ac.uk/imgt/hla/stats.html), and dozens of alleles are typically found in large vertebrate populations. Most variation in individual MHC genes is concentrated in regions coding for extracellular domains forming a groove that binds antigens from either intracellular (MHC class I) or extracellular (MHC II) pathogens (Hedrick and Kim, 1999). Polymorphism within these peptide binding regions is thought to be maintained by some form of balancing selection imposed by pathogens. Fast-evolving parasites may adapt to the most common host genotype and escape presentation of their antigens to the adaptive immune system of the host. Rare allelic variants of MHC genes to which parasites are unlikely to adapt would thus be favoured by negatively frequency-dependent selection (Borghans et al., 2004, Snell, 1968). Additionally, heterozygote advantage in resistance to parasites can contribute to polymorphism at MHC loci, as heterozygotes should be able to present a broader range of antigens (Doherty and Zinkernagel, 1975, Hughes and Nei, 1992, Takahata and Nei, 1990). Both these mechanisms would result in retention of allelic lineages over long evolutionary timescales, resulting in transspecific polymorphism (Klein, 1987), and in the establishment of novel alleles that arise via mutation or micro-recombination. Indeed, in most species investigated so far, positions in MHC sequences coding for residues involved in binding antigens consistently show an excess of non-synonymous over synonymous substitutions, which is the hallmark of positive selection for amino acid substitutions (Garrigan and Hedrick, 2003, Piertney and Oliver, 2006, Sommer, 2005). There is growing evidence for the association of MHC genotypes or individual alleles and susceptibility to infection (Bonneaud et al., 2006a, Deter et al., 2008, Schwensow et al., 2007; reviewed in Sommer, 2005) confirming that pressure from parasites is the primary source of selection on MHC. Indeed, in sticklebacks, parasite diversity was positively associated with MHC diversity across populations (Wegner et al., 2003). No correlation of parasite load with microsatellite diversity was detected, and no indication of interpopulation differences in N e was found. The authors thus argue that it is not the MHC diversity that affected population-level parasite load, but conversely, it is the prevalence of parasites that has promoted maintenance of high MHC variation. Similarly, Dionne et al. (2007) observed that MHC diversity changed with latitudinal temperature gradient in Atlantic salmon, but also with bacterial diversity in the environment. Their analysis suggested that bacterial diversity is predominant in shaping MHC variability. At the interspecific level, de Bellocq et al. (2008) found a positive correlation between parasite species richness and MHC diversity among rodents.
Balancing selection can also result from MHC-dependent choice of mates (Apanius et al., 1997). Several species show preferences for MHC-dissimilar mates (Olsson et al., 2003, Radwan et al., 2008, Schwensow et al., 2008, Yamazaki et al., 1976), which may be an adaptation to avoid mating with relatives or a way of producing MHC-heterozygous progeny, immune to a wider range of pathogens (Milinski, 2006, Penn, 2002). However, as preference for MHC-dissimilar mates does not seem to be the rule (e.g. Bonneaud et al., 2006b, Lampert et al., 2006, Miller et al., 2001, Paterson et al., 1998), it seems likely that pressure from parasites is the primary source of selection on MHC.
1.2. MHC in conservation
O’Brien and Evermann (1988) suggested that the high susceptibility of cheetah and other bottlenecked species to diseases may result from the loss of variation at genes responsible for defence against diseases, such as MHC genes. As discussed above, there is growing evidence for an association of MHC types with susceptibility to infection, which seems to support the hypothesis of O’Brien and Evermann. In large populations, genotypes resistant to many pathogens should be present due to ample MHC variation. However, in bottlenecked populations rare MHC alleles may be lost due to strong random drift, thus compromising the ability of a population to respond to fast-evolving parasites: none of the few alleles remaining may be able to present antigens of a novel parasite type. Furthermore, parasites are expected to adapt to the most common host genotypes. For example, the human HLA-A11 allele confers resistance to Epstein–Barr disease, but only in populations in which it is rare, implying that an increase in frequency of the resistant allele facilitates adaptation in the parasites (de Campos-Lima et al., 1993). Consequently, parasites may evolve ways to evade host immune systems faster in populations with limited MHC diversity. Finally, the recovery of MHC variation, which to some extent arises via micro-recombination between existing alleles within MHC loci (Richman et al., 2003), can be hindered in bottlenecked populations due to their lower effective recombination rate. However persuasive the arguments listed above, evidence for the effect of MHC diversity on population viability is needed before reliable recommendations for conservation programmes are made. The evidence for associations between MHC types and individual susceptibility to parasites, discussed in the previous section, gives us only a limited insight into effects of MHC diversity on population viability. Existence of such associations implies that some alleles may indeed not be able to respond to some parasite genotypes, but if few remaining alleles in a bottlenecked host population are highly divergent functionally, they may be able to bind antigens from a wide range of parasites (Hedrick, 2003), and population-level susceptibility to infection may not be substantially affected. Furthermore, host genotype – parasite load associations do not allow us to predict if, as discussed above, the adaptation of parasites, allowing them to overcome host defences, is facilitated in populations with limited MHC diversity. Likewise, widespread evidence for historical positive selection acting on MHC (Bernatchez and Landry, 2003; Garrigan and Hedrick, 2003; Piertney and Oliver, 2006) does not imply that there is a link between selection on MHC polymorphism and population dynamics. MHC variants controlling infection may give their bearers a large competitive advantage, even if the parasites do not have a large effect on survival in uncompetitive situations. This would cause soft selection on MHC genes without much effect on population dynamics, which in turn may be determined mostly by the carrying capacity of the environment (Babik et al., 2005). Thus, conservation biologists need to focus on consequences of population-level MHC diversity. After reviewing relevant evidence over a decade ago, Edwards and Potts (1996) concluded that “None of the studies [on species exhibiting low MHC polymorphism] identified the causes of low variability experimentally, and none has established causal relationship between low MHC variability and disease susceptibility”. Here, we review studies that have accumulated since the article by Edwards and Potts (1996) was published. We concentrated on three questions: (1) whether drift can render MHC loci effectively neutral and thus reduce their diversity in populations; (2) whether reduced variation at MHC increases population-level parasite load or prevalence of disease; (3) whether reduced variation at MHC increases probability of population extinction.
2. Methods
This review followed systematic review methodology according to a protocol available at www.environmentalevidence.org/SR65.html. Published studies were identified through searching the ISI Web of Knowledge (1996–2008) and Scopus database and by examining lists of references cited in these studies. We searched Scopus database since the beginning of records, as this database was not available at the time of the Edwards and Potts (1996). The search strategies used combination “MHC AND drift” for the first question, “MHC AND (diversity OR variation) AND (infection OR disease OR parasite* OR pathogen*)” for the second question; “MHC AND (extinction OR population survival OR population viability)” for the third question. The relevance of a study was first assessed by reading the title and abstract, and then by reading full texts of the papers considered relevant. To be included, studies concerning MHC and drift must have reported one or more tests of neutrality of MHC genes based on several populations, of which at least some have undergone a decrease in population size. Thus, some studies (e.g. Loiseau et al., 2009) were not included as no decrease in the size of any of the populations studied were reported. Sequences of MHC genes must have shown the signatures of balancing selection (to ensure that the locus examined has been subject to historical selection). Apart from MHC variation, data on neutral variation must have been reported to control for demographic processes that might produce signal mimicking selection (Nielsen, 2005). We identified 14 such studies of 12 species. A variety of methodologies were used to test for selection (Table 1 ). Seven studies reported correlations between neutral markers and MHC allelic richness. We have summarised the results of these studies by calculating the mean correlation coefficient weighted by sample size (i.e. the number of populations). We tested for significance of this mean r by converting r values from particular studies into standard normal deviates (Z s) using Fisher’s r to Z transformation and calculating combined, weighted Z c according to Eq. (23) in Wolf (1993). A set of seven studies tested whether structure in MHC is more/less pronounced than for neutral variation using F ST outlier (Beaumont and Nichols, 1996) or similar methods. Unfortunately most authors did not report exact P-values, and no standardised effect sizes are available for this simulation-based technique, which precluded formal meta-analysis. Instead, we employed simple tabulation of this and other, less commonly employed tests.
Table 1.
MHC and neutral variation in natural populations. Npop – number of studied populations; Nind – number of studied individuals. The evidence for historical selection on MHC comes from a dN/dS ratio >1 in the Antigen Binding Sites (ABS); Recent selection is inferred from deviation of genotype frequencies from expected Hardy–Weinberg proportions (H–W), lack of correlation of allelic richness with that for neutral alleles (correlation), lack of significant isolation by distance (IBD), higher or lower population differentiation than for neutral alleles (FST outlier, population differentiation FST), departures of allele frequency spectra from those expected under neutrality (Ewens–Watterson (E–W) and Slatkin’s P and bottleneck tests for allele frequency data, Tajima’s D test for nucleotide sequence data).
| Species (Taxon) | Npop | Nind | Evidence for departure from neutrality |
Populations examined | |
|---|---|---|---|---|---|
| Historical | Recent (type of evidence) | ||||
| Teleosts | |||||
| Atlantic salmon (Salmo salar)a | 7 | 666 | YES (dN/dS) | YES (E–W) NO (H–W, correlation) | Comparison of land-locked and river populations |
| Brown trout (Salmo trutta)b | 9 | 180 | YES (dN/dS) | NO (Tajima D, E–W, population differentiation (FST), correlation) | Small isolated populations |
| Brown trout (Salmo trutta)c | 7 | 492 | Not estimated | YES (FST outlier significant (diversifying selection) for MHC linked microsatellite locus in large populations, in small populations effect masked by immigration) | Comparison of large populations and those that have declined in size |
| California coastal steelhead (Oncorhynchus mykiss)d | 24 | 444 | YES (dN/dS) | NO (correlation, population differentiation FST), YES (FST outlier (diversifying selection) in one of three regions) | Populations that have experienced recent declines in size |
| Gila trout (Oncorhynchus gilae gilae)e | 10 | 142 | YES (dN/dS) | NO (FST outlier) | Populations that have declined in size |
| Sockeye salmon (Oncorhynchus nerka)f | 31 | 5400 | YES (dN/dS) | YES (E–W (16% of pops), Stalkin’s P (9% of pops), population differentiation FST) | Thirty one river populations compared with one lake population |
| Amphibians | |||||
| Alpine newt (Mesotriton alpestris)g | 7 | 149 | YES (dN/dS) | NO (correlation, FST outlier) | Groups of allopatric populations of postglacial origin |
| Crested newt (Triturus cristatus)h | 7 | 100 | YES (dN/dS) | NO (correlation) | Comparison between refugial populations and populations from the postglacial expansion area |
| Birds | |||||
| Great snipe (Gallinago media)i | 10 | 175 | YES (dN/dS) | YES (Tajima’s D, high structure between regions, not explained by neutral marker diversity, IBD) | Scandinavian mountain vs. East European population |
| Lesser kestrel (Falco naumanni)j | 7 | 121 | YES (dN/dS) | YES (Tajima’s D) NO (correlation, IBD pattern) | Free ranging but fragmented wild populations |
| South island robin (Petroica australis australis)k | 3 | 26 | YES (dN/dS) | NO (correlation) | Small, bottlenecked population |
| Mammals | |||||
| Spotted suslik (Spermophilus suslicus)l | 10 | 195 | YES (dN/dS) | NO (correlation, FST outlier) | Small, bottlenecked populations |
| Water vole (Arvicola terrestris)m | 7 | 591 | YES (dN/dS) | NO (global FST outlier) YES (bottleneck, higher diversity in MHC at the phase of low population density, stronger selection at DQA1 locus, at high density phase effect masked by migration) | Demographically fluctuating populations, comparison of low and high density phase |
| Water vole (Arvicola terrestris)n | 3 | 1303 | YES (dN/dS) | YES (H–W: excess of heterozygotes in MHC but not in microsatellites; GST among metapopulations higher for MHC than for microsatellites), NO (between-year correlation of MHC and microsatellite differentiation at the metapopulation level) | Metapopulations sampled over multiple years |
References.
Studies included to address the second question (MHC diversity and population-level infection) must have compared parasite/pathogen loads or disease prevalence in populations differing in the level of MHC variation, the number of populations examined should have allowed statistical analysis, and genome-wide inbreeding should have been taken into account. Crucially, the differences in MHC variation between populations must have been ascribed to drift, rather than to differences in pressure form parasites. Thus, studies on populations which have not undergone bottlenecks were not included, as it is likely that pressure from parasites selects for increased MHC diversity, resulting in positive correlation with parasite load (compare examples of sticklebacks and Atlantic salmon discussed above), rather than negative correlation predicted if drift leads to the loss of alleles conferring resistance. As we identified only one study that met these criteria, we also discuss results of other relevant papers.
The studies bearing on the third question (MHC variation and population extinction) mostly reported short or long-term survival of populations despite low MHC variation. However, as we discuss below, this kind of data can be inherently biased because it is rarely possible to attribute past extinctions to reduced MHC diversity.
3. Results and discussion
Full review is available at www.environmentalevidence.org/SR65.html (review no 65).
3.1. MHC diversity and genetic drift
Alleles will be effectively neutral if s < 1/2 N e, where s is the selection coefficient and N e is the effective population size. Thus, MHC alleles in even moderate frequencies will be subject to loss due to drift if population size is small or if balancing selection acting on MHC is relatively weak. A number of species that have experienced extreme population bottlenecks do indeed show depletion of variation at MHC, e.g. cheetah (O’Brien et al., 1985), Eurasian beaver (Babik et al., 2005, Ellegren et al., 1993), European bison (Radwan et al., 2007), giant panda (Zhu et al., 2007), Seychelles warbler (Hansson and Richardson, 2005), mountain goat (Mainguy et al., 2007) and Galapagos penguin (Bollmer et al., 2007). In most of these cases, the DNA sequences of the MHC genes show signatures of historical positive selection, so limited MHC variation can be attributed to drift, rather than to the lack of selection on MHC (Babik et al., 2005, Mikko and Andersson, 1995, Radwan et al., 2007).
However, strong balancing selection may resist drift even in drastically bottlenecked populations. Aguilar et al. (2004) have found that two MHC alleles were present in an island fox population totally devoid of variation in microsatellites and multilocus fingerprints and argued that MHC variation was preserved due to balancing selection. Hedrick (2004), however, pointed out that alternative scenarios, such as a founder effect from a nearby larger population, can also explain the observed pattern. Recently, van Oosterhout et al. (2006) found a higher parasite load in the smaller of two isolated populations of the guppy, Poecilla reticulata. Interestingly, MHC variation was similar in the two populations, in spite of the difference in microsatellite diversity. Van Oosterhout et al. (2006) thus concluded that it was the parasite load that imposed stronger balancing selection on MHC and maintained variation in the small population. Another interesting example is the great crested newt (discussed in detail below), in which three geographically distant populations inhabiting postglacial expansion areas share the same two alleles (Babik et al., 2009). This suggests that the alleles could have been maintained by balancing selection for several thousands of years since the expansion.
The study of moderately bottlenecked fragmented populations within species offers more stringent tests of the role of drift in shaping MHC variation. Several recent studies compared levels of MHC diversity across populations with that of neutral variation, and most have found a significant correlation (Table 1). The mean weighted r calculated from these studies was 0.76 (Z c = 2.59, P = 0.01). This indicates that on the short timescale MHC variation is shaped predominately by demographic processes rather than by selection. Nevertheless, some tests of selection, such as these of Ewens-Watterson or Tajima, suggest the role of selection in shaping MHC allele frequencies (Table 1). In two of 12 species included in Table 1 evidence of balancing selection and not of drift shaping MHC variation was found. In five species the evidence was mixed (different tests yielded disparate conclusions), indicating that both drift and selection may have an impact, and five studies found evidence of drift only. In one of the two positive studies, the sockeye salmon, the evidence of positive selection was found only in a fraction of populations (Table 1).
Rather than selection, population demographic history may account for the significant outcomes of neutrality tests (Nielsen, 2005), although it can be to some extent corrected for by running simultaneous test on neutral markers (e.g. Bryja et al., 2007). A more rigorous test of the role of selection is offered by the F ST outlier analysis (Beaumont and Nichols, 1996). The rationale behind this analysis is that demographic processes affect neutral loci, distributed over the genome, in the same way, leading to a single expectation for the level of population differentiation (measured by F ST). Loci which fall outside of confidence intervals of the neutral F ST values are likely to be under selection – diversifying if they show significantly higher F ST and spatially uniform if their F ST is lower than neutral expectations (Beaumont, 2005). The spatially uniform selection is a likely result of balancing selection, preventing the loss of rare alleles and homogenising their frequencies across populations, whereas diversifying selection may result from directional selection, which could enhance the loss of genetic diversity rather than prevent it. Applications of this test to microsatellite and MHC variation revealed significant deviation from neutrality in two of six studies (Table 1). In both these studies, however, F ST outlier analysis indicated action of diversifying selection. Overall, MHC alleles seem to often be affected by drift even if reduction of population size is not extreme.
3.2. MHC diversity and susceptibility to infection
Some studies reported positive associations between parasite species richness and MHC variation (Wegner et al., 2003, Dionne et al., 2007), but as discussed above, the differences in MHC diversity were ascribed to differential pressure from parasites, rather than to genetic drift. As such, results of these studies are not relevant to the question of what are the consequences of the loss of MHC variation in bottlenecked populations. We have found only a single study of susceptibility to infection across populations that differed in MHC diversity due to recent bottlenecks. Giese and Hedrick (2003) found no evidence that MHC variation was associated with mortality caused by the bacterial pathogen Listonella anguillarum in the endangered gila topminnow. The same was true of other measures of genetic diversity, including variation at microsatellite loci. As results of a single study are hard to generalise, we summarise below other studies that provide some suggestive data, and we highlight the need for further research.
Perhaps the most convincing is the case of the Tasmanian devil, reported recently by Siddle et al. (2007), in which the spread of a transmissible tumour is apparently facilitated by very low variation at MHC I. The tumour, transferred between individuals by biting, may be considered as an allograft, and should thus be rejected by the host immune system following recognition of foreign MHC. The lack of rejection might result from the down-regulation of classical MHC molecule expression by the tumour, but Siddle et al. found no evidence of this; rather, because of low MHC I variation, the tumour is often perceived as “self” by the host immune system.
While MHC monomorphism prevents graft rejection, it may still allow presentation of a large number of antigens from pathogens due to the promiscuity of peptide binding by MHC molecules (Nikolich-Zugich et al., 2004). Thus, the susceptibility of populations with limited MHC variation to an unusual transmissible tumour may not be representative of threats posed by pathogens to population survival. Mainguy et al. (2007) found no evidence for increased susceptibility to disease in a bottlenecked population of mountain goats, in which only two MHC DRB alleles were retained. On the other hand, high mortality in a breeding colony of cheetah caused by coronavirus-associated feline infectious peritonitis has been attributed to reduced genetic variation at MHC, ascertained in this species (O’Brien et al., 1985). However, general inbreeding may have also brought about the same outcome (Fig. 1). Indeed, separation of the effect of general inbreeding from that of depletion of MHC variation may often be difficult. In several studies infections were shown to be correlated with decreased genetic diversity, although variation at MHC had not been directly assessed. The stability of populations of critically endangered Ethiopian wolf (Canis simensis) is affected by rabies virus epidemics, particularly likely to eradicate smaller populations (van de Bildt et al., 2002). As small populations are expected to show lower levels of genetic variation, this suggests a link between susceptibility to infection and genetic variation. Pearman and Garner (2005) compared susceptibility to infection with FV3 (frog virus 3) among populations of the Italian agile frog Rana latastei. They found that populations with lower microsatellite diversity suffered higher mortality imposed by this emerging pathogen. On a smaller scale, a bottlenecked population of a New Zealand robin Petroica australis had similar parasite load, but lower immune response compared to a more diverse source population (Miller and Lambert, 2004). Whiteman et al. (2006) found a significant association of minisatellite diversity and louse abundance across populations of the Galapagos hawk (Buteo galapagoensis). They also found that genetic diversity correlated with the level of natural antibodies (Nabs), which in turn was also correlated with louse abundance. As expression of Nabs is constitutive and not antigen-dependent, the increased susceptibility of genetically depauperate populations to louse infestation is likely due to factors other than depletion of MHC variation. Whether this is also true for other cases of inverse correlations between genetic diversity (including MHC diversity) and infection susceptibility remains to be investigated. Future studies should make an effort to separate effects of genome-wide inbreeding from the effect of reduced MHC variation. In natural populations this will often be difficult, as MHC and neutral variation are often correlated (Table 1), unless strong selection will maintain substantial MHC diversity despite low N e (see previous section). Experimental approaches (e.g. Giese and Hedrick, 2003) are potentially very informative, but limited to species that can be subjected to experimental breeding.
3.3. Does MHC variation affect population survival?
We have found a number of studies reporting a limited MHC variation in species that have undergone population bottlenecks, but yet the populations survived and even increased in number (reviewed below). We found no studies reporting populations or species that have gone extinct following reduction of MHC variations. However, Tasmanian devils discussed above seem to be declining since emergence of transmissible tumor epidemics and no compensatory population responses are observed, making extinction of the population a serious threat (Lachish et al., 2007). Furthermore, we want to stress that this data set is likely to be heavily biased. While it is possible to determine MHC variation in species that have survived despite bottlenecks, there is usually no way of telling if species which went extinct in the past did so because of genetic factors (Frankham, 2005). Thus, the examples of species whose MHC variation has been depleted during extreme bottlenecks, but which nevertheless were able to recover and increase in numbers, can be taken as evidence that not all such species are bound to go extinct, but not as a proof that MHC diversity does not affect probability of extinction.
Several studies concluded that MHC variation has little effect on population dynamics on the short timescale (Babik et al., 2005, Ellegren et al., 1993, Mainguy et al., 2007, Mikko and Andersson, 1995, Weber et al., 2004). An extreme example is provided by the Scandinavian population of Eurasian beaver, lacking variation at MHC altogether (Ellegren et al., 1993). Despite this, the population size increased from about 100 to more than 150,000 during the 20th century (Halley and Rosell, 2002). The speculation that beavers may have experienced little selection on MHC (Ellegren et al., 1993) has been rejected by data from other relict Eurasian beaver populations. Babik et al. (2005) have found that while six out of seven of these populations were monomorphic, each was fixed for a different MHC II DRB allele. This pattern of variation, and a strong signal of historical selection acting on DRB sequences, indicate that MHC variation was lost due to the extreme bottleneck the species experienced at the end of the 19th century (Babik et al., 2005). Most of these populations recovered after the species was granted legal protection (Halley and Rosell, 2002, Nolet and Rosell, 1998), suggesting that populations can thrive despite MHC monomorphism. However, a century that has passed since the bottleneck may be too brief a period for the conclusion that MHC variation does not affect population dynamics. A longer timescale is provided by species whose genetic variation was reduced during glaciations. In great crested newts, ample variation at MHC II has been retained in the southern glacial refugium, but northern populations, founded during postglacial expansion, harbour only two expressed alleles. Despite such limited variation, populations in the expansion area have been viable for several thousands of years (Babik et al., 2009). In moose MHC variation was probably lost during a bottleneck that occurred about 100,000 years ago and since then new mutations accumulated and additional alleles have apparently arisen via micro-recombination (Mikko and Andersson, 1995). Most of the variation in the newly emerged MHC alleles is non-synonymous, indicating that they were quickly driven to high frequencies via positive Darwinian selection. This is an informative case indicating that loss of MHC is not necessarily conductive to extinction, but this does not imply a lack of selection on MHC. This is to be expected if selection resulting from parasites arises from reduced competitiveness of infected individuals instead of from mortality (Babik et al., 2005). Alternatively, mortality may be of compensatory, rather than of additive character (or soft, rather than hard), i.e. parasites may cause the death of the proportion of infected individuals that exceeds the carrying capacity of their environment (Combes, 1996, Combes, 1997). This scenario would result in long-term selection, evident in sequences, but with little effect on population dynamics. Indeed, recent reviews concluded that despite infectious disease being considered one of five major extinction factors (Wilcove et al., 1998), there is little hard evidence to support this conviction (de Castro and Bolker, 2005, Smith et al., 2006). Extinction was attributed, at least partially, to infectious disease in only 3.5% of species reported extinct in the last 500 years by IUCN Red list, but in no case was it listed as the single cause (Smith et al., 2006). The authors emphasize, however, that there is a large degree of uncertainty concerning the inferred causes of extinction reported by the IUCN, so the role of parasites in extinction should not be prematurely dismissed. Moreover, some convincingly documented examples of local extinction or near-extinction caused by disease have been documented (reviewed in de Castro and Bolker, 2005, McCallum and Dobson, 2002), and there is growing evidence that infections by a chytrid fungus is one of the major causes of dramatic amphibian declines observed in the last three decades (Lips et al., 2008).
Even if selection induced by parasites is hard, population survival might not be compromised by the reduction of MHC variation if the variants retained show much of the original functional variation. Indeed, Hedrick et al. (2002) argued that species threatened with extinction tend to retain highly divergent allelic variants. In the red wolf, only four allelic variants are found, but their divergence is high. Simulations based on 27 sequences found in red wolves and in related Mexican wolves and coyotes have shown that this level of divergence was unlikely to have arisen by chance. If this is a general pattern, then the potential of endangered species to respond to pathogen assault may be higher than the small number of retained alleles suggest, i.e. due to the promiscuity of peptide binding, a few divergent alleles may be able to bind antigens from a wide range of parasites.
4. Conclusions
There is ample evidence for long-term balancing selection acting on peptide binding regions of MHC, and associations between MHC types and susceptibility to infection have been documented in numerous cases. Despite balancing selection shaping MHC polymorphism in the long-term, MHC variation is often substantially reduced in bottlenecked and fragmented populations. However, whether such a loss poses a threat to the survival of populations remains unclear. The scarcity of direct evidence for the impact of MHC diversity on the survival prospects of populations, coupled with examples of long-term survival of populations despite reduced MHC polymorphism may suggest that MHC diversity is not as serious a concern in conservation as some authors have suggested. However, as the causes of past extinction events are usually uncertain, the evidence is likely to be unbalanced: it is easier to document survival, than extinction of species with depleted MHC diversity. Furthermore, most convincing examples of long-term survival of species come from northern populations (Babik et al., 2005, Ellegren et al., 1993, Mainguy et al., 2007, Mikko and Andersson, 1995). This may be a consequence parasite pressure decreasing with latitude (Guernier et al., 2004, Mainguy et al., 2007), and so these examples may not be representative of the overall impact of MHC variation on population survival. There is thus an imperative need for data that could indirectly reveal the possible consequences of MHC diversity for population viability. In particular, we need more data on the impact of MHC allelic richness on the abundance of parasites and prevalence of disease in populations. Such data, although indirect, are much easier to obtain than the data relating MHC variation to actual extinction events. As we stressed earlier, efforts should be made to control for genome-wide inbreeding. Complementary research should assess the role of pathogens in shaping population dynamics (see de Castro and Bolker, 2005, Smith et al., 2006). Regarding studies comparing differentiation and levels of variation between MHC and neutral markers, our methodological suggestions are that the researchers should report exact P-values of the F ST outlier tests and correlation coefficients between MHC and neutral allelic richness; this would facilitate future synthesis of the results.
Hughes (1991) argued that captive breeding programs should be designed to protect diversity at MHC. His argument may be summarised as follows: although for a given population size the loss of average heterozygosity may not be very high, variation at some individual loci may be lost. Loss of variation at many loci would not matter, as allelic variants are most often neutral. Therefore, we should concentrate on loci whose adaptive significance is well understood, such as MHC loci. Hedrick (2001), however, pointed out that if captive breeding programs aim to preserve variation at a single locus, families with rare alleles will be over-represented and this may lead to increased genome-wide homozygosity, exposing deleterious mutations and causing inbreeding depression. Thus, he recommended avoidance of inbreeding as the main aim of genetic restoration programs. Given the uncertainty about the role of MHC variation for population viability, this recommendation seems reasonable, especially since inbreeding depression has well documented detrimental effects on fitness, including impairment of the immune response (reviewed above). Thus, it seems rational to recommend inbreeding avoidance as a priority in cases where it would conflict with retention of maximum MHC variation. However, breeding programs designed to avoid inbreeding will tend to retain much of the MHC variation, so that such conflicts will rarely be severe. Furthermore, natural populations harbouring the most MHC variation will also usually have more genome-wide diversity (Table 1), therefore, the protection of both types of genetic diversity can be achieved simultaneously.
Supplementary material
Supplementary information is available at www.environmentalevidence-org/SR65.html.
Acknowledgements
We thank the editor Andrew Pullin and Referees for their advice and comments on earlier versions of the manuscript. This work was supported by the Foundation for Polish Science, professor subsidy 9/2008 to JR.
References
- Acevedo-Whitehouse K., Cunningham A.A. Is MHC enough for understanding wildlife immunogenetics? Trends in Ecology and Evolution. 2006;21:433–438. doi: 10.1016/j.tree.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Acevedo-Whitehouse K., Gulland F., Greig D., Amos W. Disease susceptibility in California sea lions. Nature. 2003;422:35. doi: 10.1038/422035a. [DOI] [PubMed] [Google Scholar]
- Acevedo-Whitehouse K., Vicente J., Gortazar C., Hofle U., Fernandez-De-Mera I.G., Amos W. Genetic resistance to bovine tuberculosis in the Iberian wild boar. Molecular Ecology. 2005;14:3209–3217. doi: 10.1111/j.1365-294X.2005.02656.x. [DOI] [PubMed] [Google Scholar]
- Aguilar A., Garza J.C. A comparison of variability and population structure for major histocompatibility complex and microsatellite loci in California coastal steelhead (Oncorhynchus mykiss Walbaum) Molecular Ecology. 2006;15:923–937. doi: 10.1111/j.1365-294X.2006.02843.x. [DOI] [PubMed] [Google Scholar]
- Aguilar A., Roemer G., Debenham S., Binns M., Garcelon D., Wayne R.K. High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3490–3494. doi: 10.1073/pnas.0306582101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaide M., Edwards S.V., Negro J.J., Serrano D., Tella J.L. Extensive polymorphism and geographical variation at a positively selected MHC class IIB gene of the lesser kestrel (Falco naumanni) Molecular Ecology. 2008;17:2652–2665. doi: 10.1111/j.1365-294X.2008.03791.x. [DOI] [PubMed] [Google Scholar]
- Altizer S., Harvell D., Friedle E. Rapid evolutionary dynamics and disease threats to biodiversity. Trends in Ecology and Evolution. 2003;18:589–596. [Google Scholar]
- Apanius V., Penn D., Slev P.R., Ruff L.R., Potts W.K. The nature of selection on the major histocompatibility complex. Critical Reviews in Immunology. 1997;17:179–224. doi: 10.1615/critrevimmunol.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- Babik W., Durka W., Radwan J. Sequence diversity of the MHC DRB gene in the Eurasian beaver (Castor fiber) Molecular Ecology. 2005;14:4249–4257. doi: 10.1111/j.1365-294X.2005.02751.x. [DOI] [PubMed] [Google Scholar]
- Babik W., Pabijan M., Arntzen J.W., Cogalniceanu D., Durka W., Radwan J. Long-term survival of a urodele amphibian despite depleted major histocompatibility complex variation. Molecular Ecology. 2009;18:769–781. doi: 10.1111/j.1365-294X.2008.04057.x. [DOI] [PubMed] [Google Scholar]
- Babik W., Pabijan M., Radwan J. Contrasting patterns of variation in MHC loci in the Alpine newt. Molecular Ecology. 2008;17:2339–2355. doi: 10.1111/j.1365-294X.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- Beaumont M.A. Adaptation and speciation: what can Fst tell us? Trends in Ecology and Evolution. 2005;20:435–440. doi: 10.1016/j.tree.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Beaumont M.A., Nichols R.A. Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society of London Series B–Biological Sciences. 1996;263:1619–1626. [Google Scholar]
- Bernatchez L., Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? Journal of Evolutionary Biology. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- Biedrzycka A., Radwan J. Population fragmentation and major histocompatibility complex variation in the spotted suslik, Spermophilus suslicus. Molecular Ecology. 2008;17:4801–4811. doi: 10.1111/j.1365-294X.2008.03955.x. [DOI] [PubMed] [Google Scholar]
- Bollmer J.L., Vargas F.H., Parker P.G. Low MHC variation in the endangered Galapagos penguin (Spheniscus mendiculus) Immunogenetics. 2007;59:593–602. doi: 10.1007/s00251-007-0221-y. [DOI] [PubMed] [Google Scholar]
- Bonneaud C., Chastel O., Federici P., Westerdahl H., Sorci G. Complex MHC-based mate choice in a wild passerine. Proceedings of the Royal Society of London B–Biological Sciences. 2006;273:1111–1116. doi: 10.1098/rspb.2005.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneaud C., Perez-Tris J., Federici P., Chastel O., Sorci G. Major histocompatibility alleles associated with local resistance to malaria in a passerine. Evolution. 2006;60:383–389. [PubMed] [Google Scholar]
- Borghans J.A.M., Beltman J.B., De Boer R.J. MHC polymorphism under host-pathogen coevolution. Immunogenetics. 2004;55:732–739. doi: 10.1007/s00251-003-0630-5. [DOI] [PubMed] [Google Scholar]
- Bryja J., Charbonnel N., Berthier K., Galan M., Cosson J.F. Density-related changes in selection pattern for major histocompatibility complex genes in fluctuating populations of voles. Molecular Ecology. 2007;16:5084–5097. doi: 10.1111/j.1365-294X.2007.03584.x. [DOI] [PubMed] [Google Scholar]
- Campos J.L., Posada D., Moran P. Genetic variation at MHC, mitochondrial and microsatellite loci in isolated populations of brown trout (Salmo trutta) Conservation Genetics. 2006;7:515–530. [Google Scholar]
- Coltman D.W., Pilkington J.G., Smith J.A., Pemberton J.M. Parasite-mediated selection against inbred Soay sheep in a free-living, island population. Evolution. 1999;53:1259–1267. doi: 10.1111/j.1558-5646.1999.tb04538.x. [DOI] [PubMed] [Google Scholar]
- Combes C. Parasites, biodiversity and ecosystem stability. Biodiversity and Conservation. 1996;5:953–962. [Google Scholar]
- Combes C. Fitness of parasites: pathology and selection. International Journal for Parasitology. 1997;27:1–10. doi: 10.1016/s0020-7519(96)00168-3. [DOI] [PubMed] [Google Scholar]
- de Bellocq J.G., Charbonnel N., Morand S. Coevolutionary relationship between helminth diversity and MHC class II polymorphism in rodents. Journal of Evolutionary Biology. 2008;21:1144–1150. doi: 10.1111/j.1420-9101.2008.01538.x. [DOI] [PubMed] [Google Scholar]
- de Campos-Lima P.O., Gavioli R., Zhang Q.J., Wallace L.E., Dolcetti R., Rowe M., Rickinson A.B., Masucci M.G. HLA-A11 epitope loss isolates of Epstein–Barr virus from a highly A11+ population. Science. 1993;260:98–100. doi: 10.1126/science.7682013. [DOI] [PubMed] [Google Scholar]
- de Castro F., Bolker B. Mechanisms of disease-induced extinction. Ecology Letters. 2005;8:117–126. [Google Scholar]
- Deter J., Bryja J., Chaval Y., Galan M., Henttonen H., Laakkonen J., Voutilainen L., Vapalahti O., Vaheri A., Salvador A.R., Morand S., Cosson J.F., Charbonnel N. Association between the DQA MHC class II gene and Puumala virus infection in Myodes glareolus, the bank vole. Infection Genetics and Evolution. 2008;8:450–458. doi: 10.1016/j.meegid.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Dionne M., Miller K.M., Dodson J.J., Caron F., Bernatchez L. Clinal variation in MHC diversity with temperature: evidence for the role of host–pathogen interaction on local adaptation in Atlantic salmon. Evolution. 2007;61:2154–2164. doi: 10.1111/j.1558-5646.2007.00178.x. [DOI] [PubMed] [Google Scholar]
- Doherty P.C., Zinkernagel R.M. Enhanced immunological surveillance in mice heterozygous at H-2 gene complex. Nature. 1975;256:50–52. doi: 10.1038/256050a0. [DOI] [PubMed] [Google Scholar]
- Edwards S.V., Potts W.K. Polymorphism of genes in the major histocompatibility complex: implications for conservation genetics of vertebrates. In: Smith T.B., Wayne R.K., editors. Molecular Genetic Approaches in Conservation. Oxford University Press; New York – Oxford: 1996. pp. 214–237. [Google Scholar]
- Ekblom R., Saether S.A., Jacobsson P., Fiske P., Sahlman T., Grahn M., Kalas J.A., Hoglund J. Spatial pattern of MHC class II variation in the great snipe (Gallinago media) Molecular Ecology. 2007;16:1439–1451. doi: 10.1111/j.1365-294X.2007.03281.x. [DOI] [PubMed] [Google Scholar]
- Ellegren H., Hartman G., Johansson M., Andersson L. Major histocompatibility complex monomorphism and low-levels of DNA-fingerprinting variability in a reintroduced and rapidly expanding population of beavers. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:8150–8153. doi: 10.1073/pnas.90.17.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Genetics and extinction. Biological Conservation. 2005;126:131–140. [Google Scholar]
- Garrigan D., Hedrick P.W. Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution. 2003;57:1707–1722. doi: 10.1111/j.0014-3820.2003.tb00580.x. [DOI] [PubMed] [Google Scholar]
- Giese A.R., Hedrick P.W. Genetic variation and resistance to a bacterial infection in the endangered Gila topminnow. Animal Conservation. 2003;6:369–377. [Google Scholar]
- Guernier V., Hochberg M.E., Guegan J.F.O. Ecology drives the worldwide distribution of human diseases. PLoS Biology. 2004;2:740–746. doi: 10.1371/journal.pbio.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley D.J., Rosell F. The beaver’s reconquest of Eurasia: status, population development and management of a conservation success. Mammal Review. 2002;32:153–178. [Google Scholar]
- Hansen M.M., Skaala O., Jensen L.F., Bekkevold D., Mensberg K.L.D. Gene flow, effective population size and selection at major histocompatibility complex genes: brown trout in the Hardanger Fjord, Norway. Molecular Ecology. 2007;16:1413–1425. doi: 10.1111/j.1365-294X.2007.03255.x. [DOI] [PubMed] [Google Scholar]
- Hansson B., Richardson D.S. Genetic variation in two endangered Acrocephalus species compared to a widespread congener: estimates based on functional and random loci. Animal Conservation. 2005;8:83–90. [Google Scholar]
- Hedrick P.W. The major histocompatibility complex (MHC) in declining populations: an example of adaptive variation. In: Holt W.V., Pickard A.R., Rodger J.C., Wildt D.E., editors. Reproduction Science and Integrated Conservation. Cambridge Univ. Press; Cambridge, UK: 2003. pp. 97–113. [Google Scholar]
- Hedrick P. Foxy MHC selection story. Heredity. 2004;93:237–238. doi: 10.1038/sj.hdy.6800539. [DOI] [PubMed] [Google Scholar]
- Hedrick P.W. Conservation genetics: where are we now? Trends in Ecology and Evolution. 2001;16:629–636. [Google Scholar]
- Hedrick P.W., Kim T.J. Genetics of complex polymorphisms: parasites and maintenance of MHC variation. In: Singh R.S., Krimbas C.K., editors. Evolutionary Genetics from Molecules to Morphology. Cambridge University Press; New York: 1999. pp. 204–234. [Google Scholar]
- Hedrick P.W., Lee R.N., Garrigan D. Major histocompatibility complex variation in red wolves: evidence for common ancestry with coyotes and balancing selection. Molecular Ecology. 2002;11:1905–1913. doi: 10.1046/j.1365-294x.2002.01579.x. [DOI] [PubMed] [Google Scholar]
- Hughes A.L. MHC polymorphism and the design of captive breeding programs. Conservation Biology. 1991;5:249–251. [Google Scholar]
- Hughes A.L., Nei M. Maintenance of MHC polymorphism. Nature. 1992;355:402–403. doi: 10.1038/355402b0. [DOI] [PubMed] [Google Scholar]
- Ilmonen P., Penn D.J., Damjanovich K., Clarke J., Lamborn D., Morrison L., Ghotbi L., Potts W.K. Experimental infection magnifies inbreeding depression in house mice. Journal of Evolutionary Biology. 2008;21:834–841. doi: 10.1111/j.1420-9101.2008.01510.x. [DOI] [PubMed] [Google Scholar]
- Janeway C.A., Travers P., Walport D., Shlomchik M.J. Garland Publishing; New York: 2004. Immunobiology: The Immune System in Health and Disease. [Google Scholar]
- Keller L.F., Waller D.M. Inbreeding effects in wild populations. Trends in Ecology and Evolution. 2002;17:230–241. [Google Scholar]
- Klein J. Origin of major histocompatibility complex polymorphism – the transspecies hypothesis. Human Immunology. 1987;19:155–162. doi: 10.1016/0198-8859(87)90066-8. [DOI] [PubMed] [Google Scholar]
- Lachish S., Jones M., Mccallum H. The impact of disease on the survival and population growth rate of the Tasmanian devil. Journal of Animal Ecology. 2007;76:926–936. doi: 10.1111/j.1365-2656.2007.01272.x. [DOI] [PubMed] [Google Scholar]
- Lampert K.P., Bernal X.E., Rand A.S., Mueller U.G., Ryan M.J. No evidence for female mate choice based on genetic similarity in the tungara frog Physalaemus pustulosus. Behavioral Ecology and Sociobiology. 2006;59:796–804. [Google Scholar]
- Landry C., Bernatchez L. Comparative analysis of population structure across environments and geographical scales at major histocompatibility complex and microsatellite loci in Atlantic salmon (Salmo salar) Molecular Ecology. 2001;10:2525–2539. doi: 10.1046/j.1365-294x.2001.01383.x. [DOI] [PubMed] [Google Scholar]
- Lips K.R., Diffendorfer J., Mendelson J.R., Sears M.W. Riding the wave: reconciling the roles of disease and climate change in amphibian declines. PLoS Biology. 2008;6:441–454. doi: 10.1371/journal.pbio.0060072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau C., Richard M., Garnier S., Chastel O., Julliard R., Zoorob R., Sorci G. Diversifying selection on MHC class I in the house sparrow (Passer domesticus) Molecular Ecology. 2009;18:1331–1340. doi: 10.1111/j.1365-294X.2009.04105.x. [DOI] [PubMed] [Google Scholar]
- Mainguy J., Worley K., Cote S.D., Coltman D.W. Low MHC DRB class II diversity in the mountain goat: past bottlenecks and possible role of pathogens and parasites. Conservation Genetics. 2007;8:885–891. [Google Scholar]
- McCallum H., Dobson A. Disease, habitat fragmentation and conservation. Proceedings of the Royal Society of London Series B-Biological Sciences. 2002;269:2041–2049. doi: 10.1098/rspb.2002.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikko S., Andersson L. Low major histocompatibility complex class-II diversity in European and North-American moose. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:4259–4263. doi: 10.1073/pnas.92.10.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milinski M. The major histocompatibility complex, sexual selection, and mate choice. Annual Review of Ecology Evolution and Systematics. 2006;37:159–186. [Google Scholar]
- Miller H.C., Lambert D.M. Genetic drift outweighs balancing selection in shaping post-bottleneck major histocompatibility complex variation in New Zealand robins (Petroicidae) Molecular Ecology. 2004;13:3709–3721. doi: 10.1111/j.1365-294X.2004.02368.x. [DOI] [PubMed] [Google Scholar]
- Miller K.M., Kaukinen K.H., Beacham T.D., Withler R.E. Geographic heterogeneity in natural selection on an MHC locus in sockeye salmon. Genetica. 2001;111:237–257. doi: 10.1023/a:1013716020351. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annual Review of Genetics. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Nikolich-Zugich J., Fremont D.H., Miley M.J., Messaoudi I. The role of MHC polymorphism in anti-microbial resistance. Microbes and Infection. 2004;6:501–512. doi: 10.1016/j.micinf.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Nolet B.A., Rosell F. Comeback of the beaver Castor fiber: an overview of old and new conservation problems. Biological Conservation. 1998;83:165–173. [Google Scholar]
- O’Brien S.J., Evermann F.F. Interactive influence of infectious disease and genetic diversity in natural populations. Trends in Ecology and Evolution. 1988;3:254–259. doi: 10.1016/0169-5347(88)90058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien S.J., Roelke M.E., Marker L., Newman A., Winkler C.A., Meltzer D., Colly L., Evermann J.F., Bush M., Wildt D.E. Genetic basis for species vulnerability in the cheetah. Science. 1985;227:1428–1434. doi: 10.1126/science.2983425. [DOI] [PubMed] [Google Scholar]
- Oliver M.K., Lambin X., Cornulier T., Piertney S.B. Spatio-temporal variation in the strength and mode of selection acting on major histocompatibility complex diversity in water vole (Arvicola terrestris) metapopulations. Molecular Ecology. 2009;18:80–92. doi: 10.1111/j.1365-294X.2008.04015.x. [DOI] [PubMed] [Google Scholar]
- Olsson M., Madsen T., Nordby J., Wapstra E., Ujvari B., Wittsell H. Major histocompatibility complex and mate choice in sand lizards. Proceedings of the Royal Society of London Series B–Biological Sciences. 2003;270:S254–S256. doi: 10.1098/rsbl.2003.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson S., Wilson K., Pemberton J.M. Major histocompatibility complex variation associated with juvenile survival and parasite resistance in a large unmanaged ungulate population (Ovis aries L.) Proceedings of the National Academy of Sciences of the United States of America. 1998;95:3714–3719. doi: 10.1073/pnas.95.7.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearman P.B., Garner T.W.J. Susceptibility of Italian agile frog populations to an emerging strain of Ranavirus parallels population genetic diversity. Ecology Letters. 2005;8:401–408. [Google Scholar]
- Penn D.J. The scent of genetic compatibility: sexual selection and the major histocompatibility complex. Ethology. 2002;108:1–21. [Google Scholar]
- Peters M.B., Turner T.F. Genetic variation of the major histocompatibility complex (MHC class II beta gene) in the threatened Gila trout, Oncorhynchus gilae gilae. Conservation Genetics. 2008;9:257–270. [Google Scholar]
- Piertney S.B., Oliver M.K. The evolutionary ecology of the major histocompatibility complex. Heredity. 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. [DOI] [PubMed] [Google Scholar]
- Radwan J., Kawalko A., Wojcik J.M., Babik W. MHC-DRB3 variation in a free-living population of the European bison, Bison bonasus. Molecular Ecology. 2007;16:531–540. doi: 10.1111/j.1365-294X.2006.03179.x. [DOI] [PubMed] [Google Scholar]
- Radwan J., Tkacz A., Kloch A. MHC and preferences for male odour in the bank vole. Ethology. 2008;114:827–833. [Google Scholar]
- Reid J.M., Arcese P., Keller L.F., Elliott K.H., Sampson L., Hasselquist D. Inbreeding effects on immune response in free-living song sparrows (Melospiza melodia) Proceedings of the Royal Society B–Biological Sciences. 2007;274:697–706. doi: 10.1098/rspb.2006.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman A.D., Herrera L.G., Nash D., Schierup M.H. Relative roles of mutation and recombination in generating allelic polymorphism at an MHC class II locus in Peromyscus maniculatus. Genetical Research. 2003;82:89–99. doi: 10.1017/s0016672303006347. [DOI] [PubMed] [Google Scholar]
- Ross-Gillespie A., O’Riain M.J., Keller L.F. Viral epizootic reveals inbreeding depression in a habitually inbreeding mammal. Evolution. 2007;61:2268–2273. doi: 10.1111/j.1558-5646.2007.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwensow N., Eberle M., Sommer S. Compatibility counts: MHC-associated mate choice in a wild promiscuous primate. Proceedings of the Royal Society B–Biological Sciences. 2008;275:555–564. doi: 10.1098/rspb.2007.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwensow N., Fietz J., Dausmann K.H., Sommer S. Neutral versus adaptive genetic variation in parasite resistance: importance of major histocompatibility complex supertypes in a free-ranging primate. Heredity. 2007;99:265–277. doi: 10.1038/sj.hdy.6800993. [DOI] [PubMed] [Google Scholar]
- Siddle H.V., Kreiss A., Eldridge M.D.B., Noonan E., Clarke C.J., Pyecroft S., Woods G.M., Belov K. Transmission of a fatal clonal tumor by biting occurs due to depleted MHC diversity in a threatened carnivorous marsupial. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16221–16226. doi: 10.1073/pnas.0704580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.F., Acevedo-Whitehouse K., Pedersen A.B. The role of infectious diseases in biological conservation. Animal Conservation. 2009;12:1–12. [Google Scholar]
- Smith K.F., Sax D.F., Lafferty K.D. Evidence for the role of infectious disease in species extinction and endangerment. Conservation Biology. 2006;20:1349–1357. doi: 10.1111/j.1523-1739.2006.00524.x. [DOI] [PubMed] [Google Scholar]
- Snell G.D. The H-2 locus of the mouse: observations and speculations concerning its comparative genetics and its polymorphism. Folia Biologica (Prague) 1968;14:335–358. [PubMed] [Google Scholar]
- Sommer S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Frontiers in Zoology. 2005;2:16. doi: 10.1186/1742-9994-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman D., Brook B.W., Briscoe D.A., Frankham R. Does inbreeding and loss of genetic diversity decrease disease resistance? Conservation Genetics. 2004;5:439–448. [Google Scholar]
- Takahata N., Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Bildt M.W.G., Kuiken T., Visee A.M., Lema S., Fitzjohn T.R., Osterhaus A.D.M.E. Distemper outbreak and its effect on African wild dog conservation. Emerging Infectious Diseases. 2002;8:211–213. doi: 10.3201/eid0802.010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosterhout C., Joyce D.A., Cummings S.M., Blais J., Barson N.J., Ramnarine I.W., Mohammed R.S., Persad N., Cable J. Balancing selection, random genetic drift, and genetic variation at the major histocompatibility complex in two wild populations of guppies (Poecilia reticulata) Evolution. 2006;60:2562–2574. [PubMed] [Google Scholar]
- Weber D.S., Stewart B.S., Schienman J., Lehman N. Major histocompatibility complex variation at three class II loci in the northern elephant seal. Molecular Ecology. 2004;13:711–718. doi: 10.1111/j.1365-294x.2004.02095.x. [DOI] [PubMed] [Google Scholar]
- Wegner K.M., Reusch T.B.H., Kalbe M. Multiple parasites are driving major histocompatibility complex polymorphism in the wild. Journal of Evolutionary Biology. 2003;16:224–232. doi: 10.1046/j.1420-9101.2003.00519.x. [DOI] [PubMed] [Google Scholar]
- Whiteman N.K., Matson K.D., Bollmer J.L., Parker P.G. Disease ecology in the Galapagos Hawk (Buteo galapagoensis): host genetic diversity, parasite load and natural antibodies. Proceedings of the Royal Society of London Series B–Biological Sciences. 2006;273:797–804. doi: 10.1098/rspb.2005.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcove D.S., Rothstein D., Dubow J., Phillips A., Losos E. Quantifying threats to imperiled species in the United States. Bioscience. 1998;48:607–615. [Google Scholar]
- Wolf F.M. Sage Publications; Beverly Hills, CA: 1993. Meta-analysis: Quantitative Methods for Research Synthesis. [Google Scholar]
- Yamazaki K., Boyse E.A., Mike V., Thaler H.T., Mathieson B.J., Abbott J., Boyse J., Zayas Z.A., Thomas L. Control of mating preferences in mice by genes in major histocompatibility complex. Journal of Experimental Medicine. 1976;144:1324–1335. doi: 10.1084/jem.144.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Ruan X.D., Ge Y.F., Wan Q.H., Fang S.G. Low major histocompatibility complex class II DQA diversity in the Giant Panda (Ailuropoda melanoleuca) BMC Genetics. 2007;8:29. doi: 10.1186/1471-2156-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]


