Abstract
Chikungunya virus (CHIKV) is a re-emerging mosquito-transmitted RNA virus causing joint and muscle pain. Although the protein-protein interactions (PPIs) between nonstructural proteins of CHIKV have been extensively established, the complete CHIKV intraviral interactome remains to be elucidated. In this study, we examined all possible CHIKV intraviral PPIs by immunoprecipitation and constructed the intraviral interactome of CHIKV. We reported 19 novel PPIs including the homo-oligomerization of TF. Disulfide bonds promoted the oligomerization of CHIKV TF protein. 2-BP, a palmitoylation inhibitor reduced the palmitoylation of TF and increased TF oligomerization. A quadruple mutant of Cys33, Cys35, Cys41, and Cys43 in TF blocked its palmitoylation and reduced oligomerization. Furthermore, we determined the association of TF with nsP1 and nsP3 in a palmitoylation-dependent manner. Construction of intraviral interactome of CHIKV provides the basis for further studying the function of CHIKV proteins.
Keywords: Chikungunya virus, Protein-protein interaction, TF, Disulfide bond, Palmitoylation
Abbreviations: CHIKV, Chikungunya virus; PPIs, protein-protein interactions; ORFs, open reading frames; TF, transframe; -1 PRF, -1 ribosomal frameshifting; SINV, Sindbis virus; DMEM, Dulbecco's Modified Eagle's Medium; FBS, fetal bovine serum; 2-BP, 2-bromopalmitate; HAM, hydroxylamine; IP, immunoprecipitation; DTT, dithiothreitol; MW, molecular weight
Highlights
-
•
An intraviral interactome of CHIKV proteins is constructed.
-
•
CHIKV TF is homo-oligomerized.
-
•
CHIKV TF is palmitoylated.
-
•
TF interacts with nsP1 and nsP3 in a palmitoylation-dependent manner.
1. Introduction
Chikungunya virus (CHIKV) is transmitted to human by mosquitoes including Aedes albopictus and Aedes aegypti [1]. The common symptoms for CHIKV infection are severe joint and muscle pain, fever, headache, and rash [2]. In recent years, CHIKV causes re-emerging epidemics in countries in the Indian Ocean region, United States and Europe, considering a major threat to public health [3]. There is currently no approved vaccine available to prevent CHIKV infection. Our knowledge about CHIKV infection contains numerous gaps that hamper the development of new therapeutic methods against CHIKV.
CHIKV belongs to the genus Alphavirus in the family Togaviridae, which encodes a 12-kb positive-sense RNA genome including two open reading frames (ORFs) [4]. One ORF encodes four nonstructural proteins (nsP1, nsP2, nsP3 and nsP4) that are involved in genome replication. The other ORF encodes structural proteins including capsid, E3, E2, 6 K, transframe (TF), and E1. Alphavirus TF is generated from 6 K by −1 ribosomal frameshifting (−1 PRF) [5,6]. TF is not essential for virus replication, although disrupting TF production decreases virus particle release. Recently, we demonstrated that CHIKV nsP1 is palmitoylated. Mutations that prevented nsP1 palmitoylation reduced the CHIKV infection [7]. TF from Sindbis virus (SINV) was reported to be palmitoylated [8]. However, whether CHIKV TF is palmitoylated remains unexplored.
A classical approach for investigating viral life cycle is to elucidate intraviral protein-protein interactions (PPIs). Many intraviral interactomes of viruses are available including hepatitis C virus [9], severe acute respiratory syndrome coronavirus [10], hepatitis E virus [11], Kaposi sarcoma-associated herpesvirus [12], varicella-zoster virus [12], herpes simplex virus 1 [13], murine cytomegalovirus [13], Epstein-Barr virus [13], and vaccinia virus [14]. Previously, PPIs between nonstructural proteins of CHIKV have been reported [15]. However, the complete CHIKV intraviral interactome remains to be elucidated. In this study, we constructed the CHIKV intraviral interactome based on immunoprecipitation method. CHIKV TF was identified to be palmitoylated and homo-oligomerized. Our finding could shed important lights on further studying CHIKV biology.
2. Materials and methods
2.1. Cells and reagents
293T cells was cultured in Dulbecco's Modified Eagle's Medium (DMEM, Thermo scientific, Waltham, MA, US) supplemented with 10% fetal bovine serum (FBS), 50 μg/mL gentamicin, and 1% l-glutamine. 2-bromopalmitate (2-BP) and hydroxylamine (HAM) were from Sigma-Aldrich (St. Louis, MO, US).
2.2. Antibodies
Mouse antibodies used in this study are listed: anti-actin (Sigma-Aldrich, Piscataway, NJ, US, catalogue no. A2228), anti-FLAG (Sigma-Aldrich, Piscataway, NJ, US, catalogue no. A2220), anti-GFP (Xuheyuan, Beijing, China, catalogue no. XHY038L). Rabbit antibody used in this study is listed: anti-GFP (Xuheyuan, Beijing, China, catalogue no. XHY026L). Secondary antibodies are horseradish peroxidase (HRP)-conjugated ECL goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA, US, catalogue No. A4416), HRP-conjugated ECL goat anti-rabbit IgG (Sigma-Aldrich, St. Louis, MO, US, catalogue No. A6154).
2.3. Plasmids
CHIKV strain 181/25 plasmid (pSinRep5-181/25ic, plasmid 60078) was provided by Dr. Terence Dermody through Addgene [16]. Constructs encoding for nsP1, nsP2, nsP3, nsP4, E1, E2, E3, 6 K, capsid and TF from pSinRep5-181/25ic were generated using pEGFP-C1 or p3xFlag-CMV-14 expression plasmids. All constructs were sequence verified. Plasmid transfection was performed using FuGENE HD (Promega Promega, Madison, WI, US) according to the manufacturer instructions. Constructs expressing TF-C/A mutants with GFP tag or Flag tag were generated using the Stratagene mutagenesis kit.
2.4. GFP-Trap assays
293T Cells were lysed with lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, protease inhibitor cocktail). Cell lysates were incubated with GFP-Trap_A beads (Chromotech, Planegg-Martinsried, Germany) for 1 h at 4 °C and then the beads were washed three times with wash buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA) and analyzed by Western blotting.
2.5. Immuno-precipitation assays
293T cells were lysed in lysis buffer 1 (1% Triton X-100, 50 mM Tris-HCl pH 7.4, 150 mM NaCl, protease inhibitor cocktail). Cell lysates were precleared with protein A/G beads for 30 min at 4 °C to reduce nonspecific binding affinity. Then, cell lysates were incubated with protein A/G beads pre-bound with 1 μg antibody for 1 h at 4 °C. Samples were washed three times with washing buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% Triton X-100) and analyzed by Western blotting.
2.6. HAM treatment
The cell lysates was treated with HAM buffer (1 M HAM, 1% IGEPAL CA-630, 50 mM Tris-HCl pH 7.2, 150 mM NaCl, 10% Glycerol) or control buffer (1% IGEPAL CA-630, 50 mM Tris-HCl pH 7.2, 150 mM NaCl, 10% Glycerol) for 1 h at room temperature with gentle shaking.
2.7. Western blotting
Protein samples were loaded on 12% or 15% SDS-PAGE gels and transferred to polyvinylidene fluoride membrane. Blots were blocked in 5% nonfat milk in TBS with 0.2% Tween 20 and probed with primary antibodies followed by HRP conjugated secondary antibody. Blots were imaged by ECL detection kit (Millipore Immobilon, Burlington, MA, US) according to manufacturer's instructions.
2.8. Bioinformatics analysis
The potential disulfide bonds in TF from CHIKV strain 181/25 was predicted by DiANNA 1.1 web server (http://clavius.bc.edu/∼clotelab/DiANNA/) [17]. The potential palmitoylation site in TF from CHIKV strain 181/25 was predicted by CSS-Palm (http://csspalm.biocuckoo.org/online.php) [18].
3. Results
3.1. Construction of CHIKV intraviral interactome
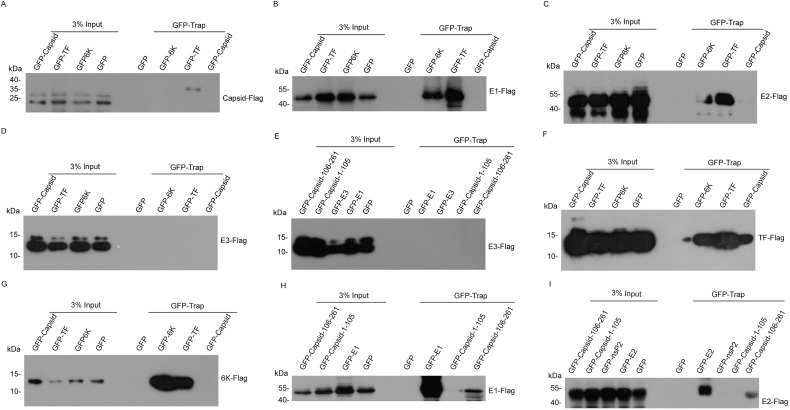
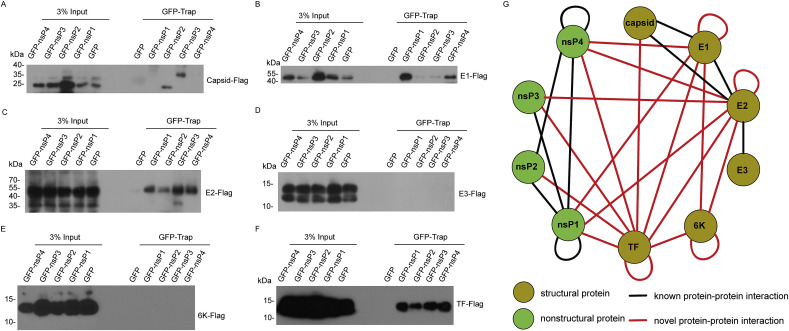
In order to provide the first genome-wide map of intraviral CHIKV PPIs, we performed a co-immuniprecipitation (co-IP) screening of intraviral PPIs for CHIKV proteins. Plasmids expressing each pair of CHIKV proteins with GFP or Flag tag were transfected into 293T cells. Cellular lysates were incubated with GFP-Trap_A beads for immunoprecipitation assay. As shown in Fig. 1, Fig. 2 , Western blotting identified 19 novel PPIs between CHIKV proteins. Combined with published known PPIs, we constructed the intra-CHIKV interactome (Fig. 2G).
Fig. 1.
Identification the protein-protein interactions by immunoprecipitation of GFP tagged structural proteins. (A–I) 293T cells were transfected with plasmids expressing GFP tagged structural proteins and plasmids expressing Flag tagged structural proteins. Cell lysates were immunoprecipitated with GFP-Trap and detected by Western blot.
Fig. 2.
Identification the protein-protein interactions by immunoprecipitation of GFP tagged nonstructural proteins. (A–F) 293T cells were transfected with plasmids expressing GFP tagged nonstructural proteins and plasmids expressing Flag tagged structural proteins. Cell lysates were immunoprecipitated with GFP-Trap and detected by Western blot. (G) Protein-protein interaction network for CHIKV proteins. The connecting line indicated the interactions between viral proteins. The red lines represented the novel PPIs. The black lines represented the known PPIs. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Oligomerization of CHIKV TF
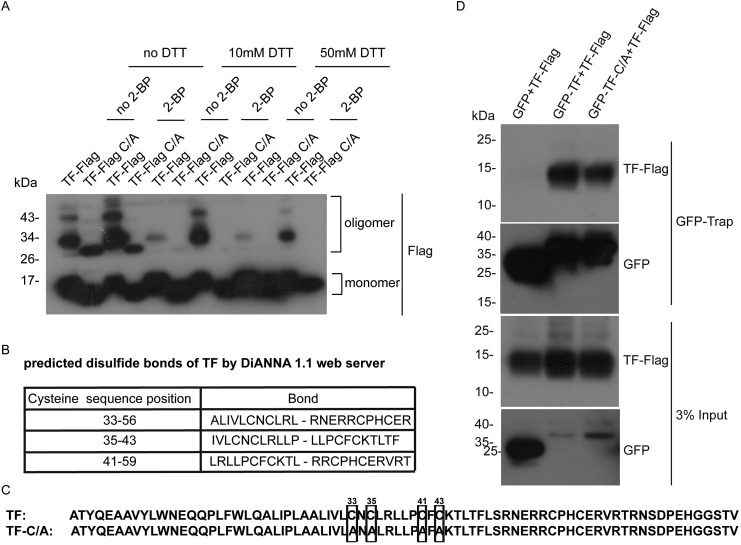
Interestingly, we found the self-interaction of TF, 6 K, E1, and E2. (Fig. 1F, G, 1H, and 1I). CHIKV nsP1 and nsP4 have been reported to be self-interacted [15]. Together, self-interaction was occurred in six CHIKV proteins. TF oligomerization was further confirmed by the appearance of higher molecular weight bands of TF in SDS-PAGE without disulfide reducing agent dithiothreitol (DTT) (Fig. 3 A), indicating that TF oligomerization was dependent on disulfide bonds.
Fig. 3.
CHIKV TF forms homo-oligomer. (A) DTT reduced the oligomerization of CHIKV TF. Constructs expressing TF-Flag or TF-C/A-Flag were transfected to 293T cells. The cells were incubated with 50 μM 2-BP for 6 h before isolation. The cell lysates were loaded with protein loading buffer containing no DTT, 10 mM DTT, or 50 mM DTT for Western blot analysis. (B) Prediction of disulfide bonds in TF. TF from CHIKV strain 181/25 was predicted by DiANNA 1.1 web server (http://clavius.bc.edu/∼clotelab/DiANNA/). (C) Diagram of TF-C/A mutant. (D) TF-C/A mutation reduced the association of TF compared with wild type TF. Constructs expressing GFP, GFP-TF, or GFP-TF-C/A combined with TF-Flag were transfected to 293T cells. GFP-Trap immunoprecipition was performed and accessed by Western blot with indicated antibodies.
In silico analysis (Fig. 3B) identified the cysteines pairs at sites of 33–56, 35–43, and 41–59 in CHIKV TF to be residues potentially involved in disulfide bonds formation. We designed substitution mutations targeting these disulphide-bonding Cysteines at position 33 (C33A), 35 (C35A), 41 (C41A) and 43 (C43A) and tested this TF variant TF-C/A for disulfide bond formation (Fig. 3C). The association between TF-C/A and TF was reduced (Fig. 3D). Thus, we concluded that the cysteines of 33, 35, 41 and 43 in TF are critical for TF oligomerization.
3.3. Palmitoylation of CHIKV TF
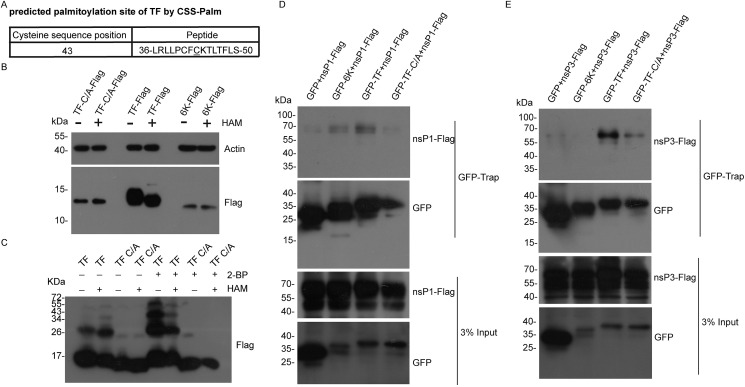
C43 of TF is not only predicted to be involved in disulfide bond formation, but also is the potential site for palmitoylation (Fig. 4 A). To confirm the palmitoylation in TF, we used HAM to remove the thioester-linked palmitate group. In agreement with our prediction, HAM reduced the monomeric TF size on SDS-PAGE but not 6 K, confirming the palmitoylation of TF (Fig. 4B). We next treated TF-expressing 293T cells with a palmitoylation inhibitor 2-BP and analyzed the protein lysates for oligomerization of TF wild type protein or TF-C/A mutant by Western blot. 2-BP increased the higher molecular weight (MW) signal corresponding to the oligomeric TF and reduced the lower MW signal corresponding to the monomeric of TF. However, the oligomerization level of TF-C/A remained the same in the presence of 2-BP (Fig.e 3A and 4C). We reasoned that C43 palmitoylation might occupy the cysteine 43, which in turn blocked formation of disulfide bonds based on cysteine 43.
Fig. 4.
CHIKV TF is palmitoylated. (A) Prediction of palmitoylation site in CHIKV TF. TF from CHIKV strain 181/25 was predicted by CSS-Palm (http://csspalm.biocuckoo.org/online.php). (B) TF was palmitoylated. Constructs expressing TF-Flag, TF-C/A-Flag, or 6 K-Flag were transfected into 293T cells. The cell lysates were treated with HAM buffer (1 M HAM, 1% IGEPAL CA-630, 50 mM Tris-HCl pH 7.2, 150 mM NaCl, 10% Glycerol) or control buffer (1% IGEPAL CA-630, 50 mM Tris-HCl pH 7.2, 150 mM NaCl, 10% Glycerol) for 1 h at room temperature and then analyzed with SDS-PAGE followed by Western blot. (C) 2-BP reduced palmitoylation of CHIKV TF and increased TF oligomerization. Constructs expressing TF-Flag or TF-C/A-Flag were transfected to 293T cells. The cells were incubated with 50 μM 2-BP for 6 h before isolation. The cell lysates were treated with HAM buffer (1 M HAM, 1% IGEPAL CA-630, 50 mM Tris-HCl pH 7.2, 150 mM NaCl, 10% Glycerol) or control buffer (1% IGEPAL CA-630, 50 mM Tris-HCl pH 7.2, 150 mM NaCl, 10% Glycerol) for 1 h at room temperature and then loaded with protein loading buffer containing 10 mM DTT for Western blot analysis. (D) TF associated with nsP1 in a palmitoylation dependent manner. Constructs expressing GFP, GFP-6K, GFP-TF, or GFP-TF-C/A combined with nsP1-Flag were transfected to 293T cells. GFP-Trap immunoprecipition was performed and accessed by Western blot with indicated antibodies. (E) TF associated with nsP3 in a palmitoylation dependent manner. Constructs expressing GFP, GFP-6K, GFP-TF, or GFP-TF-C/A combined with nsP3-Flag were transfected to 293T cells. GFP-Trap immunoprecipition was performed and accessed by Western blot with indicated antibodies.
Although 6 K and TF shared the same N-terminal 49 amino acids, 6 K did not associate with nonstructural proteins, indicating the palmitoylation or C-terminal region of TF is critical for its binding to nonstructural proteins. Compared to wild type TF, TF-C/A associated less efficiently with nsP1 (Fig. 4D). Similarly, interaction between TF-C/A and nsP3 was weaker compared to that of the wild type TF and nsP3 (Fig. 4E). These data suggested that the palmitoylation of TF is important for its association with nonstructural proteins.
4. Discussion
Giving that CHIKV genomes have limited encoding capacity, each viral protein must perform multiple functions in viral life cycles. Better understanding of how CHIKV proteins associate with each other will potentially facilitate the development of antiviral therapy which may block these interactions and consequently inhibit viral replication. Here, we present an extensive study of intraviral PPIs for CHIKV using GFP-Trap immunoprecipitation as the main experimental method. By combining our results with published references, we were able to draw the CHIKV intraviral interactome. Construction of CHIKV intraviral interactome not only advanced our understanding of CHIKV viral proteins, but also provided more insights for antiviral development.
Two major strategies have been applied to build the intraviral or cellular interactome of viral proteins. One strategy is based on yeast two hybrids (Y2H) [11,12,14,19]. However, there was limited validation due to false-positive limitation of Y2H. Another strategy is combined affinity purification with mass spectrometry or Western blot [[20], [21], [22]]. Because CHIKV only encoded 10 viral proteins, we took the second strategy using GFP-Trap method to immunoprecipitate GFP-fusion proteins and their interacting factors.
It is remarkable that structural protein TF associated with most of the CHIKV proteins except E3 (Fig. 2G). Hence, TF may act as a central hub that connects proteins involved in multiple steps of CHIKV life cycle. Why is TF able to interact with so many viral proteins? Protein palmitoylation plays a key role in regulating protein localization and function. For instance, palmitoylation is critical for the plasma membrane localization of nsP1 from CHIKV [7]. Here, we identified that CHIKV TF was palmitoylated to promote its interaction with nonstructural proteins. It is possible that palmitoylated TF located in plasma membrane to associate with other CHIKV proteins for virus assembly.
We observed the self-interaction of TF, 6 K, E1 and E2 by coimmunoprecipitation of GFP- and FLAG-tagged constructs in transiently transfected 293T cells (Fig. 1F, G, 1H, and 1I). Inhibitor and mutagenesis experiments suggested that disulfide bonds formation was required for oligomerization of TF. However, the function of oligomeric state of CHIKV TF remains to be studied. Interestingly, palmitoylation of CHIKV TF reduced the homo-oligomerization of TF, which is consistent with previous reports that less palmitoylation of SNAP-25 and sortilin increased disulfide bonds formation [23,24]. CHIKV TF formed homo-oligomer with an intermolecular disulfide bond at the cysteine 43, which could be palmitoylated. It is plausible that cysteine 43 could be shared by palmitoylation and an intermolecular disulfide bond.
The interactome map in Fig. 2G reflected the interaction between full length viral proteins. We also found that E1 interacted with capsid protein (106–261) (Fig. 1H), which confirms the structure study of CHIKV [25]. In the future, it will be of high interest to use co-immunoprecipitation assay to map protein domains crucial for the intra-CHIKV protein interactions.
In this study, we constructed an intraviral interactome of CHIKV proteins. We showed that CHIKV TF protein oligomerizes and is palmitoylated. TF interacts with nsP1 and nsP3 in a palmitoylation dependent manner. Given that CHIKV proteins are conserved across alphaviruses, many PPIs identified in this study might also exist in other alphaviruses. Our findings will not only set up the basis for studying the function of these proteins in the viral replication cycle, but also provide key targets for the development of potential antiviral drugs.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the National Key Plan for Research and Development of China [2016YFD0500300], the National Natural Science Foundation of China [81871663, 81672035], the Natural Science Foundation of Shandong Province [ZR2017PC017], and the Innovation Project of Shandong Academy of Medical Sciences. We thank Dr. Andres Merits and Dr. Terence Dermody for reagents.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bbrc.2019.04.098.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Weaver S.C., Lecuit M. Chikungunya virus and the global spread of a mosquito-borne disease. N. Engl. J. Med. 2015;372:1231–1239. doi: 10.1056/NEJMra1406035. [DOI] [PubMed] [Google Scholar]
- 2.Couderc T., Lecuit M. Chikungunya virus pathogenesis: from bedside to bench. Antivir. Res. 2015;121:120–131. doi: 10.1016/j.antiviral.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Weaver S.C., Forrester N.L. Chikungunya: evolutionary history and recent epidemic spread. Antivir. Res. 2015;120:32–39. doi: 10.1016/j.antiviral.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 4.Frolov I., Frolova E.I. Molecular virology of Chikungunya virus. Curr. Top. Microbiol. Immunol. 2019 doi: 10.1007/82_2018_146. [DOI] [PubMed] [Google Scholar]
- 5.Firth A.E., Chung B.Y., Fleeton M.N., Atkins J.F. Discovery of frameshifting in Alphavirus 6K resolves a 20-year enigma. Virol. J. 2008;5:108. doi: 10.1186/1743-422X-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramsey J., Mukhopadhyay S. Disentangling the frames, the state of Research on the Alphavirus 6K and TF proteins. Viruses. 2017;9 doi: 10.3390/v9080228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang N., Zhao H., Zhang L. Fatty acid synthase promotes the palmitoylation of Chikungunya virus nsP1. J. Virol. 2019;93 doi: 10.1128/JVI.01747-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramsey J., Renzi E.C., Arnold R.J., Trinidad J.C., Mukhopadhyay S. Palmitoylation of Sindbis virus TF protein regulates its plasma membrane localization and subsequent incorporation into virions. J. Virol. 2017;91 doi: 10.1128/JVI.02000-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagen N., Bayer K., Rosch K., Schindler M. The intraviral protein interaction network of hepatitis C virus. Mol. Cell. Proteomics MCP. 2014;13:1676–1689. doi: 10.1074/mcp.M113.036301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osterman A., Stellberger T., Gebhardt A., Kurz M., Friedel C.C., Uetz P., Nitschko H., Baiker A., Vizoso-Pinto M.G. The Hepatitis E virus intraviral interactome. Sci. Rep. 2015;5:13872. doi: 10.1038/srep13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uetz P., Dong Y.A., Zeretzke C., Atzler C., Baiker A., Berger B., Rajagopala S.V., Roupelieva M., Rose D., Fossum E., Haas J. Herpesviral protein networks and their interaction with the human proteome. Science (New York, N.Y.) 2006;311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 13.Fossum E., Friedel C.C., Rajagopala S.V., Titz B., Baiker A., Schmidt T., Kraus T., Stellberger T., Rutenberg C., Suthram S., Bandyopadhyay S., Rose D., von Brunn A., Uhlmann M., Zeretzke C., Dong Y.A., Boulet H., Koegl M., Bailer S.M., Koszinowski U., Ideker T., Uetz P., Zimmer R., Haas J. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCraith S., Holtzman T., Moss B., Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sreejith R., Rana J., Dudha N., Kumar K., Gabrani R., Sharma S.K., Gupta A., Vrati S., Chaudhary V.K., Gupta S. Mapping interactions of Chikungunya virus nonstructural proteins. Virus Res. 2012;169:231–236. doi: 10.1016/j.virusres.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Mainou B.A., Zamora P.F., Ashbrook A.W., Dorset D.C., Kim K.S., Dermody T.S. Reovirus cell entry requires functional microtubules. mBio. 2013:4. doi: 10.1128/mBio.00405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferre F., Clote P. DiANNA 1.1: an extension of the DiANNA web server for ternary cysteine classification. Nucleic Acids Res. 2006;34:W182–W185. doi: 10.1093/nar/gkl189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction, Protein engineering, design & selection. PEDS. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L., Villa N.Y., Rahman M.M., Smallwood S., Shattuck D., Neff C., Dufford M., Lanchbury J.S., Labaer J., McFadden G. Analysis of vaccinia virus-host protein-protein interactions: validations of yeast two-hybrid screenings. J. Proteome Res. 2009;8:4311–4318. doi: 10.1021/pr900491n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Zhang L., Ke Y. Cellular interactome analysis of vaccinia virus K7 protein identifies three transport machineries as binding partners for K7. Virus Gene. 2017;53:814–822. doi: 10.1007/s11262-017-1504-5. [DOI] [PubMed] [Google Scholar]
- 21.Shah P.S., Link N., Jang G.M., Sharp P.P., Zhu T., Swaney D.L., Johnson J.R., Von Dollen J., Ramage H.R., Satkamp L., Newton B., Huttenhain R., Petit M.J., Baum T., Everitt A., Laufman O., Tassetto M., Shales M., Stevenson E., Iglesias G.N., Shokat L., Tripathi S., Balasubramaniam V., Webb L.G., Aguirre S., Willsey A.J., Garcia-Sastre A., Pollard K.S., Cherry S., Gamarnik A.V., Marazzi I., Taunton J., Fernandez-Sesma A., Bellen H.J., Andino R., Krogan N.J. Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and Zika virus pathogenesis. Cell. 2018;175:1931–1945. doi: 10.1016/j.cell.2018.11.028. e1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batra J., Hultquist J.F., Liu D., Shtanko O., Von Dollen J., Satkamp L., Jang G.M., Luthra P., Schwarz T.M., Small G.I., Arnett E., Anantpadma M., Reyes A., Leung D.W., Kaake R., Haas P., Schmidt C.B., Schlesinger L.S., LaCount D.J., Davey R.A., Amarasinghe G.K., Basler C.F., Krogan N.J. Protein interaction mapping identifies RBBP6 as a negative regulator of ebola virus replication. Cell. 2018;175:1917–1930. doi: 10.1016/j.cell.2018.08.044. e1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foley T.D., Clark A.R., Stredny E.S., Wierbowski B.M. SNAP-25 contains non-acylated thiol pairs that can form intrachain disulfide bonds: possible sites for redox modulation of neurotransmission. Cell. Mol. Neurobiol. 2012;32:201–208. doi: 10.1007/s10571-011-9748-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh S., Mizuno K., Aikawa M., Aikawa E. Dimerization of sortilin regulates its trafficking to extracellular vesicles. J. Biol. Chem. 2018;293:4532–4544. doi: 10.1074/jbc.RA117.000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yap M.L., Klose T., Urakami A., Hasan S.S., Akahata W., Rossmann M.G. Structural studies of Chikungunya virus maturation. Proc. Natl. Acad. Sci. U. S. A. 2017;114:13703–13707. doi: 10.1073/pnas.1713166114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.