Abstract
Introns were found to enhance almost every steps of gene expression except increasing mRNA stability. By analyzing the genome-wide data of mRNA stability published by someone previously, we found that human intron-containing genes have more stable mRNAs than intronless genes, and the Arabidopsis thaliana genes with the most unstable mRNAs have fewer introns than other genes in the genome. After controlling for mRNA length, we found mRNA stability is still positively correlated with intron number in human intron-containing genes. But in yeast Saccharomyces cerevisiae, two different datasets on mRNA half-life gave conflicting results. The components of messenger ribonucleoprotein particles recruited during intron splicing may be retained in cytoplasmic mRNPs and act as signals of mRNA stability or simply insulators to avoid mRNA degradation.
Keywords: mRNA decay rate, mRNA half-life, Intron presence, Intron abundance, Gene ontology
Introns are widespread noncoding sequences in eukaryotic genomes, their cost and benefit to the host are still not established [1], [2], [3]. Recent progress has revealed that splicing out introns from pre-mRNAs can enhance almost every steps of gene expression from transcription to translation [4]. As mRNA accumulation is determined by both synthesis and degradation, mRNA stability is equally important in regulating gene expression as transcription [5], [6]. Do introns also increase the amount of protein produced from a gene by enhancing the mRNA stability?
Ryu and Mertz [7] showed that mutants of virus SV40 late transcript lacking introns are defective in mRNA stability in the nucleus, but not in mRNA stability in the cytoplasm. In maize, inclusion of salT intron can stimulate cat gene expression to 10–18-fold higher than the intronless control gene; but the spliced mRNAs do not have a higher stability than those encoded by the intronless control gene [8]. Nott et al. [9] found that human TPI intron 6 inserted into reporter gene Renilla luciferase can enhance the mRNA accumulation, but they did not observe any significant splicing-dependent alteration in mRNA stability. Chang et al. [10] found that insertion of a 138-bp intron into SARS-CoV spike protein gene can enhance the protein expression in mammalian cells, but the mRNAs exhibited similar decay rates as the intronless control mRNA. Splicing was found to be essential for significant protein expression of human β-globin gene. Absence of introns results in inefficient 3′-end mRNA processing, and the unprocessed β-globin mRNA is substantially less stable than the 3′-end processed mRNA [11]. In addition, the half-life of 3′-end processed β-globin mRNAs encoded by intron-containing gene was 21 ± 7 h while the half-life of the 3′-end processed β-globin mRNAs encoded by intronless gene was 15 ± 3 h [11]. But the authors [11] and another scientist that cited the paper [12] looked it as a minor difference, and so they did not think that introns can alter mRNA stability.
The accumulating data from genome sequencing and large-scale analysis of gene expression make it possible to re-examine or further testing the conclusions of previous experimental studies on specific genes. A successful example is the survey of genes with one or more 3′-untranslated exons to test the rule for termination-codon position in nonsense-mediated mRNA decay [13]. For the present issue, the relationship between mRNA stability and intron presence was not extensively studied in genome scale. Gutierrez et al. [14] compared 100 genes with the most unstable transcripts against genes encoding stable transcripts in Arabidopsis thaliana. They did not find significant differences in intron numbers. Although most of the genes are intronless in yeast Saccharomyces cerevisiae, more than 70% of ribosome protein genes have at least one intron [15]. Five ribosome protein genes were found to encode anomalous unstable mRNAs (half-life < 10 min). Wang et al. [15] noticed that four of the five genes lack introns. The aberrantly decay rates of these genes were attributed to specialized regulatory programs and distinct functions [15].
Here, we collected the genome-wide data on the stability of mRNAs (half-life or decay rate) in Homo sapiens, S. cerevisiae, and A. thaliana from publication supplements and examined their relationships with intron presence/abundance.
Materials and methods
The genome annotation files of H. sapiens (build 35 version 1), yeast S. cerevisiae (updated February 6, 2006), and A. thaliana (updated November 16, 2005) were downloaded from the NCBI genome database (ftp://ftp.ncbi.nih.gov/genomes/). In the case of alternative splicing variants, we retained the longest mRNA for analysis (although similar results were obtained by analyzing the shortest mRNA, data not shown).
The data on mRNA stability were gathered from publication supplements, including the decay rates of human mRNAs in hepatocellular carcinoma cell line HepG2 cells and primary fibroblast cell line Bud8 cells [16], mRNA half-lives in human T lymphocytes stimulated with medium, human T lymphocytes stimulated with an anti-CD3 antibody, and human T lymphocytes stimulated with antibodies anti-CD3 and anti-CD28 [17]. The mRNAs with decay rate ⩽0 h−1 were excluded from our analyses. In assigning faster expressed genes and slower expressed genes and surveying motifs that regulate mRNA decay, we followed the methods/results of Yang et al. [16]. We use GNF GeneAtlas Version 2 [18] to determine human gene expression level. The data on mRNA half-life and gene expression levels of yeast S. cerevisiae strain Y262 [15] and yeast S. cerevisiae strain rpb1-1 [19] were collected from publication supplements deposited by the authors, http://web.wi.mit.edu/young/pub/holstege.html and http://genome-www.stanford.edu/turnover/data.shtml, respectively. Gutierrez et al. studied the mRNA stability of A. thaliana [14]. But they only provided the data of 100 genes with the most unstable transcripts (with half-life ⩽ 60 min). So, these 100 genes were compared with other genes annotated in A. thaliana genome.
The data were normalized by logarithmic transformation using 10 as the base in partial correlation analyses.
Results
In human, the mRNAs of intron-containing genes are more stable
In human HepG2 cells and Bud8 cells, the mRNAs of intron-containing genes have lower decay rates than those of intronless genes (Table 1 ). In human T lymphocytes stimulated with medium and T lymphocytes stimulated with antibodies anti-CD3 and anti-CD28, the mRNAs of intron-containing genes have longer half-lives than those of intronless genes (Table 1). In human T lymphocytes stimulated with anti-CD3 antibody, there is also a significant difference between mRNA half-lives of intron-containing genes and those of intronless genes (P = 0.045). Considered from the average value of mRNA half-lives, the mRNAs of intronless genes are a little more stable than those of the intron-containing genes. But if considered from the median value of mRNA half-lives, the mRNAs of intron-containing genes are more stable than those of the intronless genes. As the nonparametric Mann–Whitney test uses the ranks of the data rather than their raw values to calculate the statistic, the conclusion based on median value is stronger. That is, the mRNAs of intron-containing genes have longer half-lives than those of intronless genes in human T lymphocytes stimulated with anti-CD3 antibody. As a stable mRNA can be measured by lower decay rate or longer half-life, our analyses of different sources of data consistently showed that the mRNAs of intron-containing genes are more stable than those of intronless genes in human cells.
Table 1.
Comparison of mRNA stabilities between intron-containing genes and intronless genes
| Species, cell types/strains | mRNA stability indicator | Gene category | n | Mean ± SEM | Median | P valuea |
|---|---|---|---|---|---|---|
| Homo sapiens | ||||||
| HepG2 | Decay rate (h−1) | Intronless | 43 | 0.421 ± 0.068 | 0.25 | 9.8 × 10−7 |
| Intron-containing | 3414 | 0.168 ± 0.003 | 0.113 | |||
| Bud8 | Decay rate (h−1) | Intronless | 54 | 0.343 ± 0.053 | 0.2 | 1.6 × 10−4 |
| Intron-containing | 3142 | 0.180 ± 0.003 | 0.15 | |||
| T lymphocytes (with medium) | Half-life (min) | Intronless | 78 | 1104 ± 157 | 377.5 | 9.4 × 10−4 |
| Intron-containing | 3933 | 1591 ± 25 | 1045 | |||
| T lymphocytes (with anti-CD3) | Half-life (min) | Intronless | 78 | 987 ± 150 | 156.25 | 0.045 |
| Intron-containing | 3933 | 986 ± 21 | 231 | |||
| T lymphocytes (with anti-CD3 and anti-CD28) | Half-life (min) | Intronless | 78 | 873 ± 139 | 134 | 5.4 × 10−4 |
| Intron-containing | 3933 | 1216 ± 24 | 310 | |||
| Saccharomyces cerevisiae | ||||||
| Strain Y262 | Half-life (min) | Intronless | 4016 | 25.3 ± 0.332 | 20 | 2.1 × 10−5 |
| Intron-containing | 225 | 27.4 ± 1.336 | 24 | |||
| Strain rpb1-1 | Half-life (min) | Intronless | 4745 | 18.8 ± 0.143 | 16 | 6.5 × 10−8 |
| Intron-containing | 223 | 16.1 ± 0.463 | 14 | |||
P values were calculated using the Mann–Whitney test.
We further tested the relationship between intron presence and human mRNA stability by controlling other biological characters to see whether the above relationship is the byproduct of other relationships.
Some evidence suggested that short mRNAs may be more stable [20], [21]. But the intronless genes we analyzed have significantly shorter mRNAs than the intron-containing genes (Supplementary Table S1). So the difference in mRNA stabilities between intron-containing genes and intronless genes could not be attributed to the difference in mRNA lengths.
Human intronless genes are not randomly distributed across molecular function categories [22]. Meanwhile, previous studies on yeast and human showed a strong relationship between physiological function and mRNA turnover rates [15], [16]. Could the difference in mRNA stability between intronless genes and intron-containing genes be attributed to functional differences? According to the observed mRNA decay rates, Yang and coauthors [16] divided the Gene Ontology (GO) categories into three groups: faster, slower, and no significant. We analyzed the distribution of intron-containing genes and intronless genes in the three GO groups by chi-square test. There are no significant differences in the genes analyzed in bud8 cells and those analyzed in T cells (P > 0.10). For the genes analyzed in HepG2 cells, intronless genes appear to be enriched in faster decay group (P = 0.046), but the P value is near the conventional significant borderline of 0.05.
Furthermore, we designed a method to remove the effects of functional difference on mRNA stability. For each human intronless gene, we selected an intron-containing gene with similar function and similar mRNA length to pair with it. The functional similarity was defined as if they share two or more GO terms (http://www.geneontology.org/) and the mRNA length similarity was defined as if the length difference is below 30%. More stringent criteria result in much fewer pairs of genes giving too small a sample to study. In the cases that two or more intron-containing genes are paired with one intronless gene, we used the median value of the mRNA stabilities of the intron-containing genes in the pairwise comparison with the intronless gene. We found that the mRNAs of intronless genes are less stable (i.e. have higher decay rates or shorter half-lives) than the intron-containing genes with similar mRNA lengths and functions (Fig. 1 ). But the differences are not significant in Wilcoxon signed ranks tests of the data from T lymphocytes with anti-CD3 and from T lymphocytes with both anti-CD3 and anti-CD28 (Fig. 1).
Fig. 1.
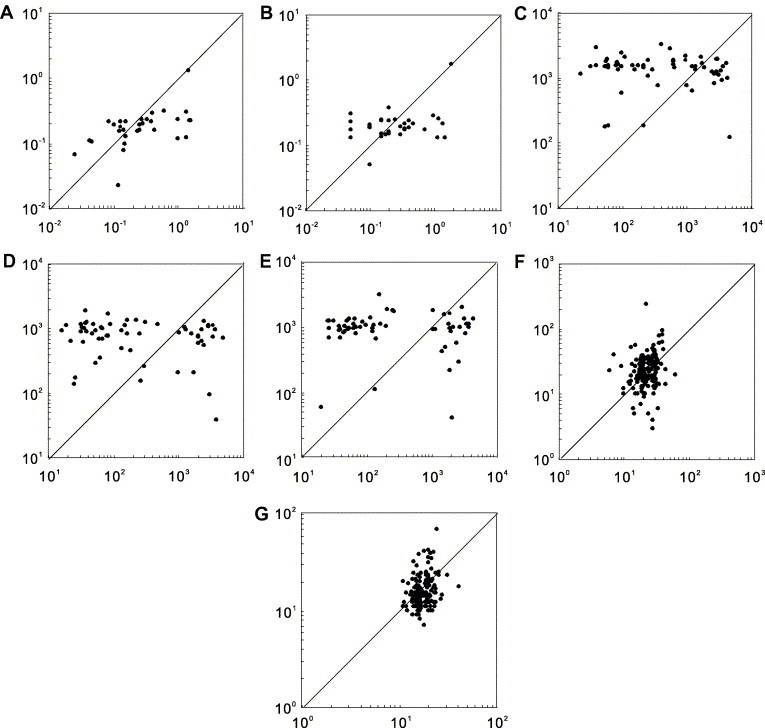
Comparisons of mRNA stabilities between intronless genes and intron-containing genes with similar mRNA lengths and functions. The X axis represents intronless genes, while the Y axis shows their intron-containing counterpart genes. The distribution of dots below and above the right angle bisector intuitively illustrates the comparisons of mRNA stability between intronless genes and intron-containing genes. Meanwhile, we performed Wilcoxon signed ranks test to determine the significance of the difference. (A) mRNA decay rate (h−1) in human HepG2 cells, 31 gene pairs, P = 4.77 × 10−3; (B) mRNA decay rate (h−1) in human Bud8 cells, 34 gene pairs, P = 0.033; (C) mRNA half-life (min) in human T lymphocytes (with medium), 62 gene pairs, P = 0.043; (D) mRNA half-life (min) in human T lymphocytes (with an Anti-CD3), 62 gene pairs, P = 0.666; (E) mRNA half-life (min) in human T lymphocytes (with anti-CD3 and anti-CD28), 62 gene pairs, P = 0.375; (F) mRNA half-life (min) in Saccharomyces cerevisiae strain Y262, 205 gene pairs, P = 3.04 × 10−4; and (G) mRNA half-life (min) in S. cerevisiae strain rpb1-1, 201 gene pairs, P = 3.88 × 10−9.
Some AU-rich elements in the 3′-untranslated region of mRNAs were found to decrease mRNA stability [5], [16]. But we found they are not more abundant in the mRNAs of intronless genes (Supplementary Table S2). On the contrary, they are a little more abundant in the mRNAs of intron-containing genes (Supplementary Table S2).
The presence of introns seems to enhance mRNA stability in human. A further question is whether multiple introns have cumulative effect on mRNA stability in intron-containing genes. After controlling for mRNA length, we found significant negative correlations between intron number and mRNA decay rate in human HepG2 cells and Bud8 cells, and significant positive correlations between intron number and mRNA half-life in human T lymphocytes (Table 2 ). Controlling for mRNA length together with other potential factors like gene expression level and GC content gave similar results (data not shown).
Table 2.
Partial correlations between mRNA stability and intron number controlling for mRNA length
| Species, cell types/strains | mRNA stability indicator | n | r | P |
|---|---|---|---|---|
| Homo sapiens | ||||
| HepG2 | Decay rate | 3414 | −0.184 | 1.8 × 10−27 |
| Bud8 | Decay rate | 3142 | −0.136 | 1.7 × 10−14 |
| T lymphocytes (with medium) | Half-life | 3933 | 0.144 | 1.4 × 10−19 |
| T lymphocytes (with anti-CD3) | Half-life | 3933 | 0.130 | 3.4 × 10−16 |
| T lymphocytes (with anti-CD3 and anti-CD28) | Half-life | 3933 | 0.144 | 1.1 × 10−19 |
| Saccharomyces cerevisiae | ||||
| Strain Y262 | Half-life | 225 | 0.260 | 8.2 × 10−5 |
| Strain rpb1-1 | Half-life | 223 | Not significant | |
Conflicting results on the relationship between introns and mRNA stability in yeast
The mRNAs of intron-containing genes are more stable than those of intronless genes in yeast strain Y262 (Table 1). Strangely, we found that the mRNAs of intronless genes are more stable in yeast strain rpb1-1 (Table 1), although the difference is very small. Different from human genes, the mRNAs of the yeast intronless genes we analyzed are significantly longer than those of intron-containing genes (Supplementary Table S1). We further compared the mRNA stabilities between intronless genes and intron-containing genes with similar mRNA length and similar GO annotations as above (as intron-containing genes are rare in yeast, we selected intronless genes with similar functions and mRNA lengths to pair with each intron-containing gene). The mRNAs of intron-containing genes are still more stable than those of intronless genes in yeast strain Y262, but less stable in yeast strain rpb1-1 (Fig. 1). Controlling for mRNA length, mRNA stability and intron number are positively correlated in strain Y262, but not in strain rpb1-1 (Table 2).
In Arabidopsis, genes with unstable mRNA have fewer introns
We re-examined the relationship between mRNA stability and intron-presence in A. thaliana. In the 100 genes reported to have the most unstable mRNAs [14], 94 genes were found in the annotated genome of A. thaliana. As the authors did not provide a list of genes with stable transcripts, we compared the 94 genes with all other 26448 genes annotated in A. thaliana genome. We found 36.2% of the 94 genes are intronless. Comparably, there are much lower percentage of intronless genes in the other 26448 genes (19.8%, Pearson Chi-square test P = 7.3 × 10−5). Different from previous study [14], we found a significant difference in intron number between the 94 genes with the most unstable transcripts and the other 26448 genes (Table 3 ). Furthermore, the difference in intron number between intron-containing genes with the most unstable transcripts and other intron-containing genes annotated in A. thaliana genome is also significant (Table 3).
Table 3.
Comparison of intron numbers between genes with unstable mRNAs and other genes in Arabidopsis
| n | Mean ± SEM | Median | P valuea | |
|---|---|---|---|---|
| Genes with unstable mRNAs | 94 | 2.41 ± 0.33 | 1 | 9.5 × 10−6 |
| Other genes in the genome | 26448 | 4.37 ± 0.03 | 3 | |
| Intron-containing genes with unstable mRNAs | 60 | 3.78 ± 0.42 | 3 | 9.3 × 10−3 |
| Other intron-containing genes in the genome | 21207 | 5.45 ± 0.04 | 4 |
P values were calculated using the Mann–Whitney test.
Discussion
The data from yeast strain Y262 [15] and from strain rpb1-1 [19] did not give consistent results. We do not exactly know which dataset is more reliable. Considering the date of publishing (the dataset from strain Y262 was published in 2002 while that from strain rpb1-1 was published in 1998), it is likely that the data from strain Y262 [15] may be more accurate because of the advancement of analyzing techniques. For that reason, we are inclined to think that introns and/or their splicing out from pre-mRNAs have enhancing effects on mRNA stability in yeast S. cerevisiae.
The data from human T lymphocytes gave weaker results than those from hepatocellular carcinoma cell line HepG2 cells and primary fibroblast cell line Bud8 cells. Heat shock, hypoxia, and other stresses were reported to cause stabilization of some mRNAs [23], [24]. The stability of mRNAs apparently varies with environmental or physiological changes. Nonetheless we can see a trend (although not very distinct) for the mRNAs of intronless genes to be less stable. The enhancing effect seems to be not very strong and so it may be overwhelmed by environmentally or physiologically induced changes.
Statistical analysis of genome-wide data can reveal significant small differences. As we see, common experimental studies on specific genes are difficult to reveal small differences. Previous experiments [7], [8], [9], [10], [11] did not show the enhancing effect of introns on mRNA stability probably because the effects are not very strong. In addition, there is the possibility that a limited number of genes do not follow the general rules because of some specific reasons.
Then, how can introns enhance the mRNA stability? The components of mRNPs (messenger ribonucleoprotein particles) recruited during intron splicing or deposited onto exon–exon junctions [25], [26] may be retained in cytoplasmic mRNPs and act as signals of mRNA stability, or simply insulators to avoid inter- or intra-RNA base-pairing. Some proteins have been shown to have such insulating effects to prevent RNA:DNA hybrid (i.e. R loop structure) in nucleus, thereby suppressing unwanted DNA recombination [27], [28], [29], [30]. Similarly, a gene having high intron density is expected to have better insulated mRNAs in cytoplasm. The mRNAs of a human gene without introns or with few introns are more likely to form stem-loop which will stall ribosome and trigger endonucleolytic mRNA cleavage [31], [32]. Meanwhile these mRNAs are also more likely to form double-stranded RNA with other RNAs in cytoplasm, being prone to degraded by RNA interference [33]. In addition, there is also evidence that the exon junction complexes of mRNPs promotes mRNA ribosome association [34]. The ribosomes attached on mRNA may have the effect of stabilizing mRNA in a way similar with the protein components of mRNPs.
Acknowledgments
We thank Jie Guo and Da-Yong Zhang for suggestion. This work was supported by the National Natural Science Foundation of China (Grant No. 30270695) and Beijing Normal University.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bbrc.2006.12.184.
Appendix A. Supplementary data
References
- 1.Jeffares D.C., Mourier T., Penny D. The biology of intron gain and loss. Trends Genet. 2006;22:16–22. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Roy S.W., Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 3.Belshaw R., Bensasson D. The rise and falls of introns. Heredity. 2006;96:208–213. doi: 10.1038/sj.hdy.6800791. [DOI] [PubMed] [Google Scholar]
- 4.Le Hir H., Nott A., Moore M.J. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 2003;28:215–220. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 5.Wilusz C.J., Wilusz J. Bringing the role of rnRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Mata J., Marguerat S., Bahler A. Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem. Sci. 2005;30:506–514. doi: 10.1016/j.tibs.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Ryu W.S., Mertz J.E. Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. J. Virol. 1989;63:4386–4394. doi: 10.1128/jvi.63.10.4386-4394.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rethmeier N., Seurinck J., Van Montagu M., Cornelissen M. Intron-mediated enhancement of transgene expression in maize is a nuclear, gene-dependent process. Plant J. 1997;12:895–899. doi: 10.1046/j.1365-313x.1997.12040895.x. [DOI] [PubMed] [Google Scholar]
- 9.Nott A., Muslin S.H., Moore M.J. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–617. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang C.Y., Hong W.W.L., Chong P., Wu S.C. Influence of intron and exon splicing enhancers on mammalian cell expression of a truncated spike protein of SARS-CoV and its implication for subunit vaccine development. Vaccine. 2006;24:1132–1141. doi: 10.1016/j.vaccine.2005.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu S., Cullen B.R. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA. 2003;9:618–630. doi: 10.1261/rna.5260303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rose A.B. The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J. 2004;40:744–751. doi: 10.1111/j.1365-313X.2004.02247.x. [DOI] [PubMed] [Google Scholar]
- 13.Nagy E., Maquat L.E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 14.Gutierrez R.A., Ewing R.M., Cherry J.M., Green P.J. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. USA. 2002;99:11513–11518. doi: 10.1073/pnas.152204099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Liu C.L., Storey J.D., Tibshirani R.J., Herschlag D., Brown P.O. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang E., van Nimwegen E., Zavolan M., Rajewsky N., Schroeder M., Magnasco M., Darnell J.E., Jr. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raghavan A., Ogilvie R.L., Reilly C., Abelson M.L., Raghavan S., Vasdewani J., Krathwohl M., Bohjanen P.R. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M.P., Walker J.R., Hogenesch J.B. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holstege F.C., Jennings E.G., Wyrick J.J., Lee T.I., Hengartner C.J., Green M.R., Golub T.R., Lander E.S., Young R.A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 20.Santiago T.C., Purvis I.J., Bettany A.J., Brown A.J. The relationship between mRNA stability and length in Saccharomyces cerevisiae. Nucleic Acids Res. 1986;14:8347–8360. doi: 10.1093/nar/14.21.8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.L. Feng, D.-K. Niu, Relationship between mRNA stability and length: an old question with a new twist, Biochem. Genet., in press, doi:10.1007/s10528-10006-19059-10525. [DOI] [PubMed]
- 22.Hill A.E., Sorscher E.J. The non-random distribution of intronless human genes across molecular function categories. FEBS Lett. 2006;580:4303–4305. doi: 10.1016/j.febslet.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 23.Knapinska A.M., Irizarry-Barreto P., Adusumalli S., Androulakis L., Brewer G. Molecular mechanisms regulating mRNA stability: physiological and pathological significance. Curr. Genomics. 2005;6:471–486. [Google Scholar]
- 24.Kang J.-H., Kim S.-A., Hong K.-J. Induction of TSP1 gene expression by heat shock is mediated via an increase in mRNA stability. FEBS Lett. 2006;580:510–516. doi: 10.1016/j.febslet.2005.12.065. [DOI] [PubMed] [Google Scholar]
- 25.Kuersten S., Goodwin E.B. Linking nuclear mRNP assembly and cytoplasmic destiny. Biol Cell. 2005;97:469–478. doi: 10.1042/BC20040106. [DOI] [PubMed] [Google Scholar]
- 26.Lejeune F., Ishigaki Y., Li X.J., Maquat L.E. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huertas P., Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 28.Svejstrup J. Keeping RNA and DNA apart during transcription. Mol. Cell. 2003;12:538–539. doi: 10.1016/s1097-2765(03)00354-x. [DOI] [PubMed] [Google Scholar]
- 29.Li X.L., Manley J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Aguilera A. mRNA processing and genomic instability. Nat. Struct. Mol. Biol. 2005;12:737–738. doi: 10.1038/nsmb0905-737. [DOI] [PubMed] [Google Scholar]
- 31.Doma M.K., Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tollervey D. RNA lost in translation. Nature. 2006;440:425–426. doi: 10.1038/440425a. [DOI] [PubMed] [Google Scholar]
- 33.Filipowicz W., Jaskiewicz L., Kolb F.A., Pillai R.S. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 34.Nott A., Le Hir H., Moore M.J. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


