Abstract
The severe acute respiratory syndrome-coronavirus nucleocapsid (N) protein is involved in virus replication and modulation of cell processes. In this latter respect control may in part be achieved through the sub-cellular localisation of the protein. N protein predominately localises in the cytoplasm (the site of virus replication and assembly) but also in the nucleus/nucleolus. Using a combination of live-cell and confocal microscopy coupled to mutagenesis we identified a cryptic nucleolar localisation signal in the central part of the N protein. In addition, based on structural comparison to the avian coronavirus N protein, a nuclear export signal was identified in the C-terminal region of the protein.
Keywords: Coronavirus, Nucleocapsid protein, Nucleolus, Nucleolar localisation signal, Nuclear export signal, Nucleus, Confocal microscopy
The coronavirus and related arterivirus nucleocapsid (N) protein can localise to both the cytoplasm and the nucleolus [1], [2], [3], [4], and may be cell cycle-related [5]. Although the principal site of replication for both viruses is the cytoplasm, interaction with the nucleolus is an emerging paradigm for RNA viruses [6]. The nucleolus is a dynamic sub-nuclear structure involved in ribosome subunit biogenesis, RNA processing, control of cell growth and response to cell stress [7], [8], [9]. Trafficking between the two compartments may be determined by appropriate signals [10]. N protein is a phosphoprotein [11], [12] with roles in replication and interacting with host cell processes. Three conserved regions (I, II and III) can be identified in the coronavirus N protein, and may play a role in different functions such as RNA binding [13]. The coronavirus infectious bronchitis virus (IBV) N protein contains an eight-amino acid nucleolar localisation signal (NoLS) in region 1 [14], and the arterivirus porcine reproductive and respiratory syndrome virus N protein contains an NLS which is required for efficient replication [15]. If the N proteins traffic to the nucleolus then they must contain appropriate signals in order to return to the cytoplasm, to play a principal role in virus biology. Indeed IBV N protein contains a non-CRM1 dependent nuclear export signal (NES) in region 3 [14], [16].
The severe acute respiratory syndrome coronavirus (SARS-CoV) N protein has also been reported to localise to the nucleolus/nucleus [17], [18], [19]. However, nucleolar localisation was not observed in virus infected cells [20], [21]. Over expression of SARS-CoV N protein indicated that nucleolar localisation was less than compared to IBV N protein [20]. However, SARS-CoV N protein could be translocated to the cytoplasm from the nucleus via binding to the 14-3-3 protein [22]. Taken together the data suggested that the SARS-CoV N protein may contain a strong NES. The signals which mediate the potential trafficking of SARS-CoV N protein are unknown [20]. Deletion mutagenesis revealed that regions 1 and 2 could direct a fluorescent fusion protein to the nucleus and nucleus/nucleolus, respectively. Region 3 could direct a fusion protein to the cytoplasm [20]. The predicted NES in region 2 was also found to be non-functional [20]. This study investigated the potential signals involved in trafficking of the SARS-CoV N protein to the nucleolus and its export from the nucleus.
Materials and methods
Plasmids. The expression cDNAs used in this study were generated by PCR and cloning into pEGFP-C2 and pECFP-C1 (Clontech) (which express the fusion protein C-terminal of enhanced green and cyan fluorescence proteins, respectively), utilizing non-templated restriction sites in sequence specific primers [14], [23]. The plasmids generated were; pECFP-SARSCoV-NIIAB, pECFP-SARSCoV-NIIBC, pEGFP-SARSCoV-NIIA, pEGFP-SARSCoV-NIIB and pEGFP-SARSCoV-NII. The potential NES in SARS-CoV N protein was deleted by overlapping PCR using primers NewNESfor and NewNESrev (details of primers available on request). All constructs were verified by sequencing in both directions and Western blot (data not shown).
Transfection and imaging. Cos7 cells were grown and transfected using Lipofectamine (Invitrogen) as described previously [20], [23]. Live-cell imaging was performed on a Nikon Eclipse TS 100 microscope utilising the appropriate filter for EGFP and ECFP. Fluorescence and bright-field images were captured. Cells were processed for confocal microscopy as described [14], [23].
Results and discussion
SARS-CoV N protein region II contains motifs capable of directing nucleolar localisation
To investigate whether SARS-CoV N protein contained motifs capable of directing nucleolar localisation, deletion mutagenesis was performed on region II to delineate any potential amino acids involved in this process. Previously, we have shown that region II (amino acids 157–299) can direct ECFP to the nucleolus [20]. Following the approach which was used to identify the NoLS in IBV N protein [14], region II of SARS-CoV N protein was subdivided into five fragments AB, BC and A, B and C which spanned amino acids 157–246, 210–299, 157–209, 210–247 and 247–299, respectively. These fragments were cloned C-terminal of either the ECFP or the EGFP reporter protein, creating plasmids pECFP-SARSCoV-NIIAB, pECFP-SARSCoV-NIIBC, pEGFP-SARSCoV-NIIA, pEGFP-SARSCoV-NIIB and pEGFP-SARSCoV-NIIC, which when transfected into mammalian cells led to the expression of the appropriate fusion protein. The localisation of these proteins was compared to region II which had been previously cloned downstream of ECFP (referred to in this study as pECFP-SARSCoV-NII [20]), at 24-h post-transfection using both live-cell and confocal microscopy (Note we have shown previously that EGFP, ECFP and DsRed localise to the cytoplasm and nucleus with no preferential accumulation in either compartment [14], [16], [20]). The data indicated that ECFP-SARSCoV-NII protein localised predominately to the nucleolus with some nuclear localisation (as described previously [20]) when analysed using live-cell and confocal microscopy (Fig. 1 ). Relative fluorescent intensity analysis of ECFP-SARSCoV-NII and the nucleolar marker protein B23.1 (plasmid pDsRedB23.1 [20]) indicated that ECFP-SARSCoV-NII was predominately nucleolar (Fig. 1).
Fig. 1.
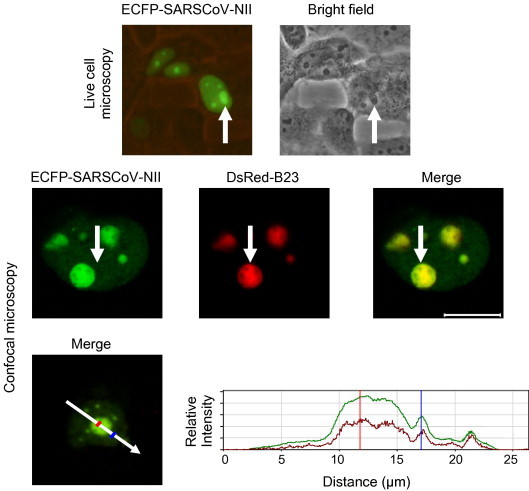
Live-cell imaging of the distribution of ECFP-SARSCoV-NII protein in Cos7 cells; fluorescent and bright view of the same image is presented, the nucleolus is indicated. Below is shown a confocal image of the nucleus of a Cos7 cell expressing ECFP-SARSCoV-NII protein (green) and DsRedB23.1 (red). A merged image is also presented and any co-localisation is shown in yellow. A nucleolus is indicated. In the merged image the line is 10 μm. Relative fluorescent intensity analysis of the distribution of ECFP-SARSCoV-NII protein (green) and DsRedB23.1 (red) in the nucleus. The white line on the merged image to the left is the region analysed. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this paper.)
To investigate whether region II contained a potential NoLS, cells were co-transfected with pDsRedB23.1 and pECFP-SARSCoV-NIIAB, pECFP-SARSCoV-NIIBC, pEGFP-SARSCoV-NIIA, pEGFP-SARSCoV-NIIB and pEGFP-SARSCoV-NIIC and analysed 24-h post-transfection using confocal microscopy and quantitative florescence (Fig. 2 ). The data indicated that ECFP-SARSCoV-NIIAB localised predominately to the cytoplasm and the nucleolus, ECFP-SARSCoV-NIIBC localised predominately to the nucleus with some cytoplasmic localisation. EGFP-SARSCoV-NIIA and EGFP-SARSCoV-NIIB similar to EGFP-SARSCoV-NIIAB localised predominately to the cytoplasm and nucleolus. EGFP-SARSCoV-NIIA also co-localised with DsRedB32.1, pointing to nucleolar localisation. EGFP-SARSCoV-NIIC localised to the cytoplasm and nucleus. If no trafficking signals were present, then the fusion protein would be distributed evenly between the cytoplasm and nucleus. This data indicated that there were no defined short amino acid sequences that directed nucleolar localisation—as is the case with the eight-amino acid motif in IBV N protein [14]. None of these fragments reproduced the predominately nucleolar localisation when ECFP was fused to region II (Fig. 1), suggesting that a cryptic NoLS(s) may encompass SARS-CoV N protein region II.
Fig. 2.
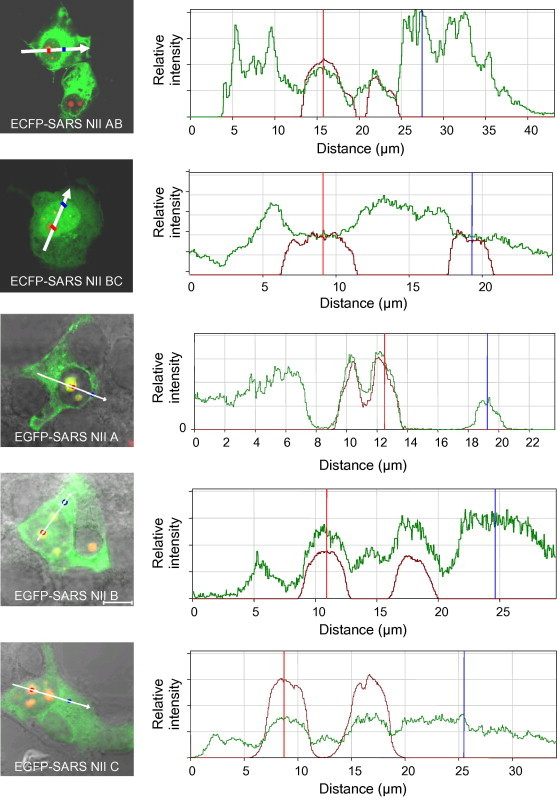
Relative fluorescent intensity analysis of the distribution of fragments of region II of SARS-CoV N protein fused C-terminal of ECFP/EGFP (green) (described in the text and indicated in the appropriate image) and DsRedB23.1 (red). The white line on the merged confocal image to the left is the region analysed. For aid, the red and blue dots represent the vertical red and blue lines for orientation. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this paper.)
Nucleolar localisation of SARS-CoV N protein may therefore be a case of molecular mimicry, in which amino acids are ordered such that they resemble a NoLS [24], and account for the low frequency of observation [20]. Conversely many NoLS are larger than eight amino acids, for example the survivin-ΔEx3 protein is 17 amino acids [25] and the herpes saimiri virus ORF57 protein contains two distinct nuclear localisation signals which together act as an NoLS [26]. Analysis of the human nucleolar proteome suggested certain amino acids and motifs are over and under represented [9]. For example, nucleolar proteins contain more lysine and arginine residues than generic cellular proteins (12%). For example, by weight KR makes up 17.5%, 20.21% and 16.12% of regions 1, 2 and 3, respectively. Overall KR contributes 17.98% of SARS-CoV N protein by weight. Nucleolar proteins also appear to have less cysteine and histidine residues than cellular or nuclear proteins [9]. SARS-CoV N protein contains no cysteine residues and 1.45% histidine by weight, generic cellular proteins are approximately 2% and 3%, respectively. GRG and RGG motifs are also frequent in nucleolar proteins and one third of the human nucleolar proteome contains an RGG motif at least once [9]. Amino acid sequence analysis indicates that SARS-CoV N protein contains no GRG motif but does contain two RGG motifs located at amino acids 96–98 in region 1 and 178–180 in region 2 (and thus in fragment A). However, SARS-CoV N protein region 1 directs a fusion protein to the nucleus but not nucleolus [20]. Taken together the presence of a RGG motif and a higher percentage by weight of KR residues in region II may result in any nucleolar localisation of the protein.
SARS-CoV N protein region III contains a novel NES
Recently, we have shown that the avian coronavirus N protein contains a leucine rich motif in the C-terminal region of the protein which acts as an NES [14], [16]. Therefore the hypothesis was tested that region III of SARS-CoV N protein contained a motif which could act as an NES. To identify such a motif the structure of the C-terminal end of SARS-CoV N protein (amino acids 270–370) [27] was compared that of the C-terminal part of IBV N protein (amino acids 220–333) [28] using the Deep View structural alignment algorithm. The data indicated that the two structures could be superimposed and that the NES identified in the IBV N protein mapped to amino acids 324-EVTPSGTWLT-334 on the SARS-CoV N protein (Fig. 3 ). Amino acid sequence alignment using AlignX (Vector NTI) in the context of the wild-type full-length proteins indicated several conserved amino acids at this region (Fig. 3). To investigate whether this motif was involved in nuclear export of SARS-CoV N protein deletion mutagenesis was used to remove the motif in the context of SARS-CoV N protein fused to ECFP, creating plasmid pECFP-SARSCoV-NΔNES. Cells were co-transfected with pDsRedB23.1 and either pECFP-SARSCoV-N [23] (which expressed wild-type SARS-CoV N protein C-terminally fused to ECFP) and pECFP-SARSCoV-NΔNES and examined using live cell and confocal microscopy 24-h post-transfection (Fig. 4 ). The data indicated that in contrast to ECFP-SARSCoV-N protein which localised to the cytoplasm, with little signal in the nucleus (Fig. 4), ECFP-SARSCoV-NΔNES localised to the cytoplasm and nucleus (Fig. 4), which would be predicted if nuclear export had been disrupted.
Fig. 3.
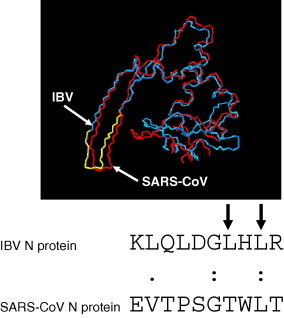
Comparison of the structure of the C-terminal region of IBV and SARS-CoV N protein (indicated). The IBV N protein NES motif is indicated in yellow. Below is shown the sequence alignment of the relevant SARS-CoV N protein sequence with the IBV N protein NES motif. Arrows indicate key residues involved in the activity of the NES. (.) indicates a conserved amino acid and (:) indicates an identical amino acid. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this paper.)
Fig. 4.
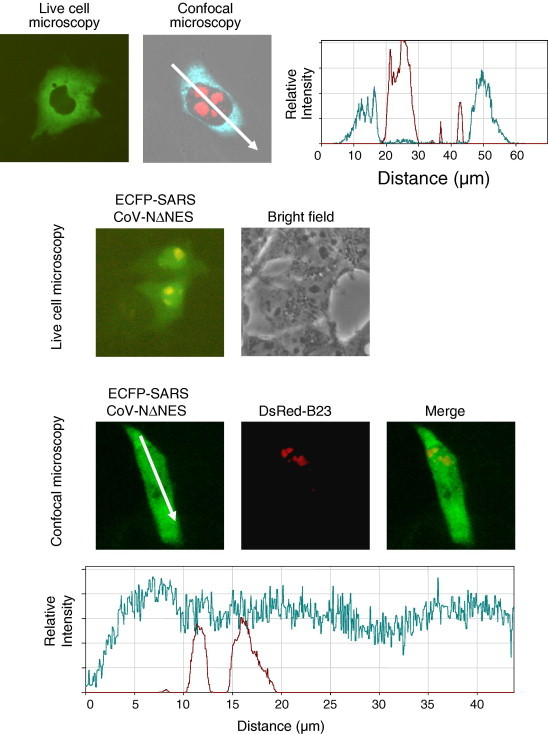
Live-cell and confocal microscopy of the distribution of ECFP-SARS-CoV N protein in cells. Also shown in the confocal image is the expression of DSRedB23.1, a relative fluorescent intensity profile of this cell is also shown. Below are shown live-cell and confocal microscopy of the distribution of ECFP-SARSCoV-NdNES and relative fluorescent intensity are shown. Cells were also co-expressing DsRedB23.1.
Clearly, SARS-CoV N protein contains amino acid motifs which would allow its trafficking to the nucleus. Given that this protein is observed in the cytoplasm of infected cells and over-expression studies [21], [23], the possibility arises that either N protein is inefficiently imported into the nucleus or efficiently exported. The former possibility may occur through structural alteration of the protein exposing the potential NoLS in region II, perhaps through differential phosphorylation or multi-merisation. Deletion and mutagenesis analysis in this and previous studies [20] indicates that the NES motif identified between amino acids 324-EVTPSGTWLT-334 in SARS-CoV N protein is the dominant signal in determining N protein localisation. This motif does not conform to a classical leucine-rich NES and thus represents a novel motif involved in nuclear export.
References
- 1.Hiscox J.A., Wurm T., Wilson L., Cavanagh D., Britton P., Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2001;75:506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wurm T., Chen H., Britton P., Brooks G., Hiscox J.A. Localisation to the nucleolus is a common feature of coronavirus nucleoproteins and the protein may disrupt host cell division. J. Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowland R.R., Kerwin R., Kuckleburg C., Sperlich A., Benfield D.A. The localisation of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 1999;64:1–12. doi: 10.1016/s0168-1702(99)00048-9. [DOI] [PubMed] [Google Scholar]
- 4.Tijms M.A., van der Meer Y., Snijder E.J. Nuclear localization of non-structural protein 1 and nucleocapsid protein of equine arteritis virus. J. Gen. Virol. 2002;83:795–800. doi: 10.1099/0022-1317-83-4-795. [DOI] [PubMed] [Google Scholar]
- 5.Cawood R., Harrison S.M., Dove B.K., Reed M.L., Hiscox J.A. Cell cycle dependent localisation of the coronavirus nucleocapsid protein. Cell Cycle. 2007;6:863–867. doi: 10.4161/cc.6.7.4032. [DOI] [PubMed] [Google Scholar]
- 6.Hiscox J.A. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 2007;5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez-Verdun D. Nucleolus: from structure to dynamics. Histochem. Cell. Biol. 2006;125:127–137. doi: 10.1007/s00418-005-0046-4. [DOI] [PubMed] [Google Scholar]
- 8.Matthews D.A., Olson M.O. What is new in the nucleolus?: workshop on the nucleolus: new perspectives. EMBO Rep. 2006;7:870–873. doi: 10.1038/sj.embor.7400786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung A.K., Andersen J.S., Mann M., Lamond A.I. Bioinformatic analysis of the nucleolus. Biochem. J. 2003;376:553–569. doi: 10.1042/BJ20031169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmo-Fonseca M., Mendes-Soares L., Campos I. To be or not to be in the nucleolus. Nat. Cell Biol. 2000;2:E107–E112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- 11.Chen H., Gill A., Dove B.K., Emmett S.R., Kemp F.C., Ritchie M.A., Dee M., Hiscox J.A. Mass spectroscopic characterisation of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding using surface plasmon resonance. J. Virol. 2005;79:1164–1179. doi: 10.1128/JVI.79.2.1164-1179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calvo E., Escors D., Lopez J.A., Gonzalez J.M., Alvarez A., Arza E., Enjuanes L. Phosphorylation and subcellular localization of transmissible gastroenteritis virus nucleocapsid protein in infected cells. J. Gen. Virol. 2005;86:2255–2267. doi: 10.1099/vir.0.80975-0. [DOI] [PubMed] [Google Scholar]
- 13.Spencer K.A., Hiscox J.A. Characterisation of the RNA binding properties of the coronavirus infectious bronchitis virus nucleocapsid protein amino-terminal region. FEBS Lett. 2006;580:5993–5998. doi: 10.1016/j.febslet.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed M.L., Dove B.K., Jackson R.M., Collins R., Brooks G., Hiscox J.A. Delineation and modelling of a nucleolar retention signal in the coronavirus nucleocapsid protein. Traffic. 2006;7:833–848. doi: 10.1111/j.1600-0854.2006.00424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee C., Hodgins D., Calvert J.G., Welch S.K., Jolie R., Yoo D. Mutations within the nuclear localization signal of the porcine reproductive and respiratory syndrome virus nucleocapsid protein attenuate virus replication. Virology. 2006;346:238–250. doi: 10.1016/j.virol.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reed M.L., Howell G., Harrison S.M., Spencer K.A., Hiscox J.A. Characterisation of the nuclear export signal in the coronavirus infectious bronchitis virus nucleocapsid protein. J. Virol. 2007;81:4298–4304. doi: 10.1128/JVI.02239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F.Q., Xiao H., Tam J.P., Liu D.X. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005;579:2387–2396. doi: 10.1016/j.febslet.2005.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Timani K.A., Liao Q., Ye L., Zeng Y., Liu J., Zheng Y., Yang X., Lingbao K., Gao J., Zhu Y. Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res. 2005;114:23–34. doi: 10.1016/j.virusres.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qinfen Z., Jinming C., Xiaojun H., Huanying Z., Jicheng H., Ling F., Kunpeng L., Jingqiang Z. The life cycle of SARS coronavirus in Vero E6 cells. J. Med. Virol. 2004;73:332–337. doi: 10.1002/jmv.20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.You J.-H., Dove B.K., Enjuanes L., DeDiego M.L., Alvarez E., Howell G., Heinen P., Zambon M., Hiscox J.A. Sub-cellular localisation of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Gen. Virol. 2005;86:3303–3310. doi: 10.1099/vir.0.81076-0. [DOI] [PubMed] [Google Scholar]
- 21.Rowland R.R., Chauhan V., Fang Y., Pekosz A., Kerrigan M., Burton M.D. Intracellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein: absence of nucleolar accumulation during infection and after expression as a recombinant protein in vero cells. J. Virol. 2005;79:11507–11512. doi: 10.1128/JVI.79.17.11507-11512.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Surjit M., Kumar R., Mishra R.N., Reddy M.K., Chow V.T., Lal S.K. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J. Virol. 2005;79:11476–11486. doi: 10.1128/JVI.79.17.11476-11486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.You J.H., Reed M.L., Dove B.K., Hiscox J.A. Three-dimensional reconstruction of the nucleolus using meta-confocal microscopy in cells expressing the coronavirus nucleoprotein. Adv. Exp. Med. Biol. 2006;581:313–318. doi: 10.1007/978-0-387-33012-9_55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rowland R.R.R., Yoo D. Nucleolar-cytoplasmic shuttling of PRRSV nucleocapsid protein: a simple case of molecular mimicry or the complex regulation by nuclear import, nucleolar localization and nuclear export signal sequences. Virus Res. 2003;95:23–33. doi: 10.1016/S0168-1702(03)00161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song Z., Wu M. Identification of a novel nucleolar localization signal and a degradation signal in survivin-deltaEx3: a potential link between nucleolus and protein degradation. Oncogene. 2005;24:2723–2734. doi: 10.1038/sj.onc.1208097. [DOI] [PubMed] [Google Scholar]
- 26.Boyne J.R., Whitehouse A. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. USA. 2006;103:15190–15195. doi: 10.1073/pnas.0604890103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu I.M., Oldham M.L., Zhang J., Chen J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J. Biol. Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jayaram H., Fan H., Bowman B.R., Ooi A., Jayaram J., Collisson E.W., Lescar J., Prasad B.V. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


