Abstract
Recent outbreak of Severe Acute Respiratory Syndrome (SARS) that caused almost 800 victims requires a development of efficient inhibitor against SARS coronavirus (SCV). In this study, RNA aptamers against SCV NTPase/Helicase (nsP10) were isolated from RNA library containing random sequences of 40 nts using in vitro selection technique. Nucleotide sequences of enriched RNA aptamer pool (ES15 RNA) contain AG-rich conserved sequence of 10–11 nucleotides [AAAGGR(G)GAAG; R, purine base] and/or additional sequence of 5 nucleotides [GAAAG], which mainly reside at the loop region in all the predicted secondary structures. Isolated RNAs were observed to efficiently inhibit double-stranded DNA unwinding activity of the helicase by up to ∼85% with an IC50 value of 1.2 nM but show a slight effect on ATPase activity of the protein in the presence of cofactor, poly (rU). These results suggest that the pool of selected aptamers might be potentially useful as anti-SCV agents.
Keywords: SARS coronavirus, nsP10 NTPase/Helicase, RNA aptamer, SELEX
Severe acute respiratory syndrome (SARS) that was caused by a novel coronavirus, SARS coronavirus (SCV) claimed almost 800 deaths between 2002 and 2003 [1]. SCV is an enveloped, single-stranded RNA positive-strand virus with a genome of ∼30 kb [2], [3]. There have been several reports of anti-SCV drug development against viral main protease (3CL protease) [4], [5] and SCV NTPase/Helicase (nsP10) [6], [7]. Tertiary structure of the SCV NTPase/Helicase has not been experimentally verified, but the structure prediction of the protein was recently reported [8]. SCV nsP10 has been shown to contain helicase activity of unwinding double-stranded DNA with a 5′ to 3′ polarity [9]. Furthermore, it has been observed that RNA homopolymer significantly stimulated ATPase activity of the helicase [9]. The viral helicase has been identified as a potential target for therapy in other viruses due to its indispensability in viral genome replication [10], [11], [12]. Thus, SCV NTPase/Helicase (nsP10) that was recently purified and characterized is an attractive target for development of anti-SCV agent [9].
RNA ligands (aptamers) that were identified using systematic evolution of ligands by exponential enrichment (SELEX) as an in vitro selection strategy can adopt complex structures to bind target proteins with high affinities [13], [14]. SELEX is an efficient method to isolate high affinity DNA and RNA ligands for target molecules including proteins, organic dyes, and other small molecules [15]. RNA aptamers have been isolated using the SELEX against several viral proteins, such as HIV Tat [16] and reverse transcriptase [17], and HCV NS3 protease [18]/helicase [19], and NS5B RNA-dependent RNA polymerase [20], [21], all of which could inhibit the enzyme activity in vitro. However, no studies have been undertaken for the isolation of RNA aptamers against SCV nsP10 to inhibit NTPase/Helicase activity of the protein. In the present study, we utilized the SELEX to isolate RNA aptamers against SCV NTPase/Helicase from RNA library containing random sequences of 40 nts. After 15 successive rounds of SELEX, we have isolated enriched RNA aptamer pool that can efficiently inhibit nucleic acid unwinding activity of the helicase with a slight effect on ATPase activity of the protein.
Materials and methods
Expression and purification of the His-tagged SCV NTPase/Helicase. The protein expression plasmid harboring the SCV NTPase/Helicase domain (pHelA12, kindly provided by Dr. Huang, J.-D. University of Hong Kong, China), which was constructed in the previous study [9], was transformed into E. coli RosettaTM competent cells (Novagen). The recombinant His-tagged SCV NTPase/Helicase was purified as described previously [9] with a following modification; after purification with a nickel-charged HiTrap chelating column (GE Healthcare) chromatography, fractions containing the protein, which was determined by 10% SDS–PAGE, were ultrafiltrated with Amicon stirred cell (YM-30). During the ultrafiltration, desalting and buffer exchange was accompanied for the next column containing 180 ml of Sephadex G-100 resin (Sigma), which was washed with Buffer A (25 mM Tris–HCl; pH 7.5, and 0.3 mM NaCl). After the protein sample (2 ml) was applied to column, it was eluted with the same buffer at a flow rate 0.3 ml/min. Pure fractions determined by SDS–PAGE were combined, and the pooled fractions were ultrafiltrated with Amicon stirred cell (YM-30) for volume down. The purified protein in 30% (v/v) glycerol was frozen at −80 oC for long term storage. Protein concentration was determined using a Bio-Rad protein assay system (Bio-Rad) with bovine serum albumin as the standard. Typical yields were ∼5 mg of purified protein/6 liter of bacterial culture.
Preparation of random RNA library. The RNA library used for in vitro selection was generated by in vitro transcription, using T7 RNA polymerase and 109 bp DNA template containing 40 random nucleotides (Fig. 1 A). The DNA template for in vitro transcription was generated by 15 cycles of amplification of single stranded DNA with forward primer containing T7 promotor (underlined sequence) (5′-GATAATACGACTCACTATAGGGTTCACTGCAGACTTGACGAA-3′) and reverse primer (5′-GAATTCGTAGATGTGGATCCATT-3′). In PCR-amplified DNA template the random regions were flanked by defined sequences comprising the T7 promotor and restriction sites for in vitro transcription and cloning purposes (Fig. 1A). The amplified DNA was purified by phenol and chloroform, and transcribed with T7 RNA polymerase. The produced RNA was gel-purified on a 12% Urea-denaturing gel. The sequence of the generated RNA is 5′- GGGUUCACUGCAGACUUGACGAAGCUUN40-AAUGGAUCCACAUCUACGAAUUC-3′, where N40 represents 40 nt with equimolar incorporation of A, G, C, and U at each position.
Fig. 1.
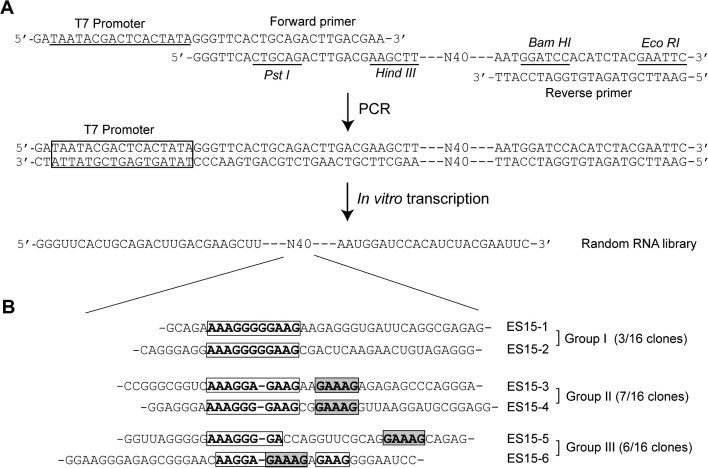
The sequence of RNA pool for in vitro selection and selected RNA aptamers against SCV nsP10. (A) The RNA library was produced by in vitro transcription of the DNA template containing 40 random nucleotides. (B) The 6 different RNA sequences identified in the ES15 RNA pool are shown. These RNAs of three groups were identified to contain a AG-rich conserved sequence of 10–11 nucleotides (in white boxes) in the middle of core region. Group II and III are observed to include an additional conserved sequence (in gray boxes).
In vitro selection of RNA aptamers. In vitro selection was carried out with the purified His-tagged SCV NTPase/Helicase and the generated RNA library. First, 5 μg of the RNA library was preincubated with 100 μl of Ni–NTA sepharose beads in 100 μl of binding buffer (30 mM Tris–HCl; pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 2 mM dithiothreitol (DTT) and 1% (w/v) BSA) for 20 min at room temperature with occasional shaking. The RNA-bead complexes were precipitated and discarded for removing RNAs with nonspecific binding activity to sepharose bead. In each cycle, precleared supernatant was incubated with 2 μg of His-tagged SCV protein in 100 μl binding buffer for 30 min at room temperature. One-hundred microliters of Ni–NTA sepharose was added and incubation was continued for another 20 min. The reaction mixture was centrifuged to remove RNA molecules that do not bind the protein, and pellets were washed five times with 500 μl of the binding buffer. The helicase complexed with RNA was dissociated from the Ni–NTA beads by eluting with the elution buffer (binding buffer components plus 250 mM imidazole). RNAs bound to the protein were recovered from the supernatant by phenol–chloroform extraction and ethanol precipitation. Recovered RNAs were reverse transcribed with ImProm-II™ reverse transcriptase (Promega), amplified by PCR with Taq DNA polymerase, and used for next round of selection. Eight consecutive rounds of selection were performed in the same manner. However, starting from the round nine, a more stringent condition was employed by reducing the protein concentration at every one or second round: 1 μg (round 9), 0.5 μg (rounds 10–11), 0.25 μg (rounds 12–13), and 0.125 μg (rounds 14–15). After the 15th round, cDNA was amplified by PCR and cloned into a linearized pGEM T vector (Promega). After subcloning and transformation into E. coli, plasmid DNA was isolated from individual clones and the DNA sequences of clones were analyzed. The secondary structure of selected RNA aptamers was predicted by the MFold program based on the Zuker algorithm [22].
ATP hydrolysis by SCV NTPase/Helicase. ATPase activity of SCV NTPase/Helicase was investigated in the presence of either original RNA library or enriched RNA pool from the SELEX round #15 (ES15 RNA) for monitoring the inhibition of ATPase activity by RNA aptamers. ATPase activity of the SCV helicase was assayed by measuring the amount of phosphate released from adenosine triphosphate. Inorganic phosphate was quantified by using color-developing reagent that is specific for Pi [23]. ATP hydrolysis reaction was carried out by mixing 0.1 μM SCV helicase with 0.5 mM ATP, 50 mM NaCl, 5 mM MgCl2, and various concentrations of RNA aptamer pool (or random RNA library) with or without 100 nM Poly (rU) (∼290 nts, purchased from GE healthcare) in 50 mM Tris–HCl buffer (pH 6.8) in a final volume of 30 μl. The reactions were incubated for 3 min at 37 oC and were stopped by adding 70 μl of the color-developing reagent (1 part of 10% (w/v) ascorbic acid and 6 part of 0.42% (w/v) Ammonium molybdate in 1 N H2SO4). The quenched reaction then was incubated for 1 h at 37 oC and developed cyan-blue color was read at 820 nm. ATPase activity was assayed by the amount of phosphate released and quantified by comparison with a standard phosphate calibration curve.
FRET-based helicase activity assay. Fluorescence resonance energy transfer (FRET) was used to measure the 5′ to 3′ dsDNA unwinding activity of the SCV helicase. Two oligomers that are end-labeled with fluorescent dye, which are designed to contain 25 base pairs of complementary part and 20 nts of 5′-overhang, were synthesized and purified by PAGE (Bioneer, Korea): T20D25Tam (5′-TTTTTTTTTTTTTTTTTTTTGAGCGGATTACTATACTACATTAGA(TAMRA)-3′), and T0D25Flu (5′-(Fluorescein)TCTAATGTAGTATAGTAATCCGCTC-3′). To prepare dsDNA substrate, the two oligomers were annealed by mixing 1.2:1 mixing ratio of T20D25Tam:T0D25Flu at a concentration of 5.0 μM (of T0D25Flu), heating to 95 oC, and then cooling slowly to 37 oC over an hour. The dsDNA unwinding reaction was carried out by initially mixing 100 nM dsDNA substrate with 2 mM ATP, 0.1 M NaCl, 2.5 mM MgCl2, 20 nM SCV helicase, and 20 mM HEPES (pH 7.4) in a 50 μl volume at 37 oC. After 5 min incubation of the reaction mixture, the trap DNA (500 nM) without or with various amounts of the aptamer RNA (or random RNA) was added to the reaction mixture. The reaction mixture was further incubated for 5 min at 37 oC, and quenched with 50 μl of termination buffer (0.2 M EDTA, 0.2 M NaCl, and 20 mM HEPES (pH 7.4)). The fluorescence was monitored by using the spectrofluorometer (Ex = 485 nm/ Em = 535 nm).
MALDI-TOF MS analysis of RNA aptamer pool. The mass analysis of the RNA aptamer pool was performed using Autoflex III MALDI-TOF mass spectrometer (Brucker Daltonics Inc, Billerica, MA, USA). The spectrum was acquired in a linear mode with a Smartbeam™ laser as a desorption/ionization source. Negative ions were subjected to 20 kV accelerating voltage with 0 V set to the sample plate. The spectrum was accumulated from 5 points with 200 per point. One microliter of a matrix solution, which was prepared by mixing equal volume of hydroxypicolinic acid (20 mg/ml in acetonitrile/water = 1:1) and ammonium citrate (50 mg/ml in water), was mixed with 0.1 μg of the RNA aptamer pool.
Results and discussion
In vitro selection of RNA aptamers for SCV NTPase/Helicase
In order to use the SELEX procedure for isolation of high affinity RNA ligands against the SCV NTPase/Helicase, an RNA library pool (∼1014 molecules) containing 40 nts of random core sequences flanked with defined regions was generated (Fig. 1A). By decreasing the protein concentration successively after the round #8, the RNA pool was more enriched in RNA ligands with high binding affinity and specificity for the SCV helicase. After 15 cycles of selection, the bound RNAs (ES15 RNA pool) were amplified by RT-PCR, and the resulting cDNAs were cloned. The nucleotide sequences of 16 subclones were determined and 4 different sequences were identified with a AG-rich conserved sequence of 10–11 nucleotides [AAAGGR(G)GAAG; R, purine base] in the middle of random core region (Fig. 1B). These RNAs were categorized as groups I and II, depending on the presence of the additional sequence of 5 nucleotides [GAAAG] that follows the AG-rich conserved sequence. Besides these groups, 6 clones among 16 subclones were identified to contain the modified AG-rich conserved sequence with the [GAAAG] sequence (Group III).
The 6 different RNA sequences identified in the ES15 RNA pool were analyzed for their secondary structures by the MFold program [22]. It was found that the predicted secondary structure of every RNA aptamer contains several stem-loop structures, in which the AG-rich conserved sequence and the [GAAAG] sequence mainly reside at the loop region in all the structures (Fig. 2 ). This result implies that the exposed [AAAGGR(G)GAAG] and [GAAAG] sequences in RNA aptamers are important for interaction with the SCV NTPase/Helicase. It has been previously reported that RNA aptamers for hepatitis C virus NS3 protease contains a conserved sequence of 9 nucleotides in the loop structure [18]. Thus, the loop structure containing a conserved sequence in RNA could constitute a binding motif structure to SCV RNA helicase whereas the stem structure of any sequence could have a stabilizing function.
Fig. 2.
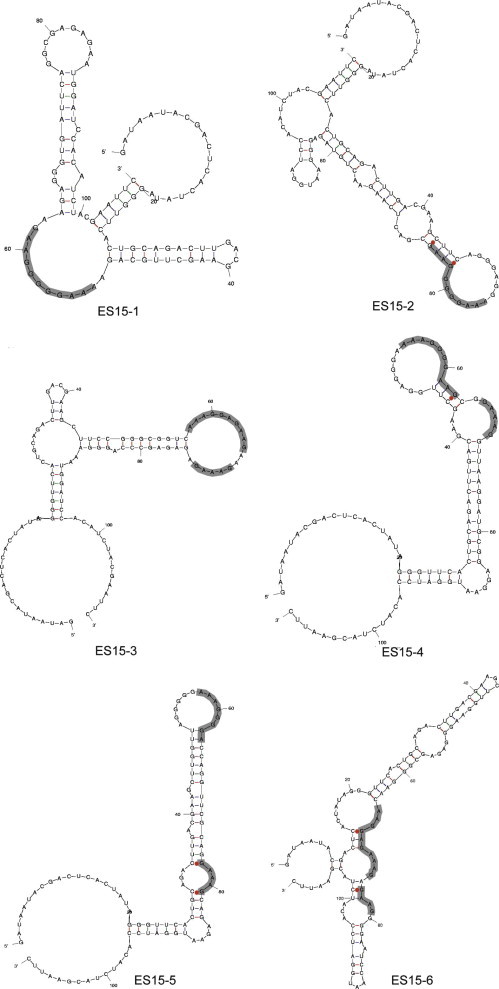
Secondary structures of RNA aptamers derived from the ES15 RNA pool. The secondary structures of 6 different RNA sequences identified in the ES15 RNA pool were predicted using the MFold program. The conserved motifs (gray-colored sequences) mostly reside at the loop region.
Effect of RNA aptamer pool on ATPase activity of the SCV NTPase/Helicase
The enriched and selected RNA aptamer pool at SELEX round #15 (ES15 RNA) was tested for its effect on the ATPase activity of the SCV helicase. ATPase activity of the SCV helicase has been previously reported to be highly stimulated by single-stranded RNA [9]. We have confirmed the RNA-stimulated ATPase activity of the purified SCV helicase using RNA homopolymer (poly (rU)), and RNA concentration dependence on the ATPase activity was investigated (data not shown). Since the RNA aptamer pool constitutes a mixture of RNAs of 90 nts length, it may stimulate the ATPase activity of the protein if the protein recognizes the RNA pool as a genuine cofactor for ATP hydrolysis activity. As shown in Fig. 3 A, ES15 RNA stimulated the ATPase activity in a dose-dependent manner with a K 1/2 value of 2.34 ng/μl. This dose-dependent stimulation of ATPase activity was also observed with the random RNA library, suggesting that the SCV helicase hydrolyzes ATP by utilizing ES15 RNA as a RNA cofactor for ATPase activity regardless of RNA sequences and binding affinity. We next investigated whether the RNA aptamer pool could inhibit ATPase activity of the helicase in the presence of 100 nM poly (rU) RNA. As shown in Fig. 3B, ES15 RNA failed to significantly inhibit the poly (rU)-stimulated ATPase activity even at the highest amount tested. At a high dosage of the RNA aptamer pool slight decrease of the ATPase activity was observed. This is probably because the RNA aptamers might take over the binding of poly (rU) to the helicase and maximal stimulation of the ATP hydrolysis by ES15 RNA is less than that by the poly (rU) (data not shown). Averaged molecular weight of the aptamer pool (ES15 RNA) was precisely determined to be 30.2 kDa by MALDI-TOF mass spectrometry (Fig. 3C). Using the averaged molecular weight and K 1/2 value of ES15 RNA for ATPase stimulation shown in Fig. 3A, apparent ATPaseIC50 for ES15 RNA was calculated to be 77 nM. This value indirectly reflects a binding affinity of the ES15 RNA to putative site for ATP hydrolysis stimulation in the SCV helicase. In the previous report of the RNA aptamers against HCV NS3 helicase domain, RNA aptamers specific to the helicase with high-affinity interfered with the substrate RNA binding without stimulation of the ATPase activity [19]. On the contrary, in our study both original random RNA library and RNA aptamer pool stimulated ATPase activity of the protein, indicating that the RNA aptamer pool against the SCV helicase can substitute RNA substrate in a competitive manner and play a role as a cofactor for ATP hydrolysis.
Fig. 3.
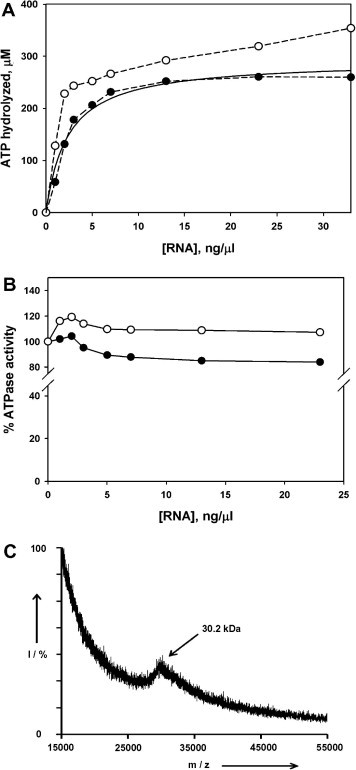
Effects of ES15 RNA pool on the ATPase activity of SCV NTPase/Helicase. (A) ATP hydrolysis reaction by the protein was performed in the presence of either initial random RNA pool (○) or ES15 RNA pool (●). ATP hydrolysis reaction was carried out as described in “Materials and methods”. Hyperbolic fit (solid line) of the ES15 RNA-stimulation profile provided K1/2 value of 2.34 ng/μl for RNA-stimulated ATP hydrolysis by the SCV NTPase/Helicase. (B) Poly (rU) (100 nM)-stimulated ATP hydrolysis reaction by the protein was performed in the presence of either initial random RNA pool (○) or ES15 RNA pool (●). Values shown are averages of measurements performed in triplicate (graphs in panel A and B). (C) Averaged molecular weight of ES15 RNA pool was determined to be 30.2 kDa by MALDI TOF-MS.
Inhibitory effect of RNA aptamer pool on helicase activity of the SCV NTPase/Helicase
We then tested if ES15 RNA could inhibit DNA unwinding activity of the SCV helicase. dsDNA unwinding activity of the protein was measured by a fluorometric assay based on the FRET from the Fluorescein to the TAMRA (Fig. 4 A). A similar approach for the helicase activity assay has been reported recently in the HCV helicase and the SCV helicase [7], [24]. Previously, it has been shown that the SCV helicase requires 5′-overhang of the dsDNA substrate for its 5′ to 3′ directionality in unwinding [9]. Thus, we have designed dsDNA substrate with a 20 nts of 5′-oligo(dT) overhang as described in Materials and methods. When the two oligomers are in very close proximity due to annealing, the FRET occurs from the Fluorescein to TAMRA, emitting a very weak fluorescence from Fluorescein. Whereas, an increase of fluorescence in Fluorescein dye may be observed due to the absence of FRET after the duplex has been unwound either by the heat denaturation or by the helicase (Fig. 4A). To ensure that the unwound oligoDNAs do not reanneal, an excess amount of trap DNA that is identical to the shorter strand (T0D25Flu) but does not contain the Fluorescein was included in the reaction. Thus, reannealing process has very little effect on the fluorescence of Fluorescein. Reaction condition was chosen to ensure that the helicase undergoes its dsDNA unwinding catalysis in a steady-state. As 5 min incubation of the dsDNA substrate and the helicase in the presence of ATP and Mg2+ is sufficient enough for the steady-state of DNA unwinding, effect of the presence of various concentrations of ES15 RNA or random RNA library was tested in the middle of steady-state phase of helicase reaction and compared to the control reaction where no RNA was added. Trap oligoDNA was added along with the testing RNAs to augment the fluorescence signal from 5′-end Fluorescein of the displaced DNA strand.
Fig. 4.
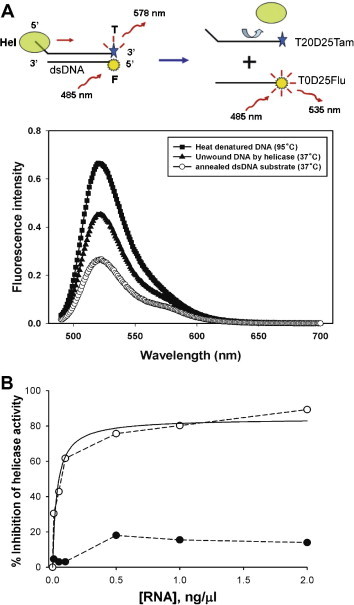
Principles of the FRET-based helicase assay and inhibition of the SCV helicase activity by the ES15 RNA. (A) Schematic drawing of the FRET-based dsDNA unwinding assay of helicase activity. Each T20D25 and T0D25 single stranded DNA labeled with TAMRA and Fluorescein on 3′-end and 5′-end, respectively, were annealed and then incubated at 37 °C or 95 °C (heat denaturation) in the absence or presence of SCV helicase for 10 min. The unwound DNA by heat treatment (■) emitted higher fluorescence than other cases. (B) Inhibition of the helicase activity of the SCV helicase in the presence of various concentrations of the ES15 RNA. Data of the ES15 RNA (○) were fitted to the hyperbolic equation (solid line), which provide an IC50 of 0.035 ng/μl (equivalent to 1.2 nM) and the extent of maximal inhibition (85%). For a comparison, random RNA pool (●) was also tested for the same dsDNA unwinding assay. Points shown in graph are the average of triplicate experiments performed separately.
As shown in Fig. 4B, more efficient inhibition of SCV helicase activity was observed with ES15 RNA by up to ∼85%, whereas the random RNA library exhibited little inhibition. ES15 RNA inhibited the helicase activity in a dose-dependent manner, yielding helicaseIC50 value of 0.035 ng/μl (=1.2 nM) from the hyperbolic fit (solid line in graph, Fig. 4B). Remarkably, the helicaseIC50 value appeared to be smaller than the ATPaseIC50 value, indicating that the RNA aptamers more strongly bind to the helicase when the protein is complexed with dsDNA substrate rather than the helicase without dsDNA substrate. The random RNA library did not show a significant inhibition of the helicase activity, suggesting that strong binding affinity with defined sequences (i.e., conserved sequences in SE15 RNAs) to the helicase is necessary to inhibit dsDNA unwinding activity. Although the binding site of SE15 RNA on the SCV helicase is not defined in this study, the isolated ES15 RNA against the SCV NTPase/Helicase is a fully effective inhibitor of dsDNA unwinding activity of the helicase with IC50 of 1.2 nM, but not ATPase. Thus, we could speculate that the aptamers interact with the helicase either at a substrate binding domain for ATP hydrolysis or at an allosteric site to impede the helicase activity.
Acknowledgments
This research was supported by the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korea government (MOST) (R01-2006-000-10617-0) and the Korea Research Foundation Grant funded by the Korean government (MOEHRD, Basic Research Promotion Fund) (KRF-2006-312-C00594). Y.-J.J. was supported by Seoul R&BD Program (10580) and the research program 2007 of Kookmin University in Korea.
References
- 1.World Health Organization, Summary of probable SARS cases with onset of illness from 1st November 2002 to 31st July 2003, http://www.who.int/csr/sars/country/ (2003).
- 2.Marra M.A. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 3.Rota P.A. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Bacha U., Barrila J., Velazquez-Campoy A., Leavitt S.A., Freire E. Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry. 2004;43:4906–4912. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]
- 5.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 6.Kao R.Y. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem. Biol. 2004;11:1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanner J.A. The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chem. Biol. 2005;12:303–311. doi: 10.1016/j.chembiol.2005.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernini A. Tertiary structure prediction of SARS coronavirus helicase. Biochem. Biophys. Res. Commun. 2006;343:1101–1104. doi: 10.1016/j.bbrc.2006.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanner J.A. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borowski P., Schalinski S., Schmitz H. Nucleotide triphosphatase/helicase of hepatitis C virus as a target for antiviral therapy. Antiviral Res. 2002;55:397–412. doi: 10.1016/s0166-3542(02)00096-7. [DOI] [PubMed] [Google Scholar]
- 11.Crute J.J. Herpes simplex virus helicase-primase inhibitors are active in animal models of human disease. Nat. Med. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- 12.Kleymann G. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat. Med. 2002;8:392–398. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- 13.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 14.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 15.Jayasena S.D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 16.Yamamoto R. A novel RNA motif that binds efficiently and specifically to the Tat protein of HIV and inhibits the trans-activation by Tat of transcription in vitro and in vivo. Genes Cells. 2000;5:371–388. doi: 10.1046/j.1365-2443.2000.00330.x. [DOI] [PubMed] [Google Scholar]
- 17.Tuerk C., MacDougal S., Gold L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda K. Isolation and characterization of RNA aptamers specific for the hepatitis C virus nonstructural protein 3 protease. Eur. J. Biochem. 2000;267:3685–3694. doi: 10.1046/j.1432-1327.2000.01400.x. [DOI] [PubMed] [Google Scholar]
- 19.Hwang B. Isolation of specific and high-affinity RNA aptamers against NS3 helicase domain of hepatitis C virus. RNA. 2004;10:1277–1290. doi: 10.1261/rna.7100904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biroccio A., Hamm J., Incitti I., De Francesco R., Tomei L. Selection of RNA aptamers that are specific and high-affinity ligands of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 2002;76:3688–3696. doi: 10.1128/JVI.76.8.3688-3696.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vo N.V., Oh J.W., Lai M.M. Identification of RNA ligands that bind hepatitis C virus polymerase selectively and inhibit its RNA synthesis from the natural viral RNA templates. Virology. 2003;307:301–316. doi: 10.1016/s0042-6822(02)00095-8. [DOI] [PubMed] [Google Scholar]
- 22.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- 23.Piper J.M., Lovell S.J. One-step molybdate method for rapid determination of inorganic phosphate in the presence of protein. Anal. Biochem. 1981;117:70–75. doi: 10.1016/0003-2697(81)90693-x. [DOI] [PubMed] [Google Scholar]
- 24.Boguszewska-Chachulska A.M. Direct fluorometric measurement of hepatitis C virus helicase activity. FEBS Lett. 2004;567:253–258. doi: 10.1016/j.febslet.2004.04.072. [DOI] [PubMed] [Google Scholar]


